Sampling Strategies for Soil Property Mapping Using Multispectral Sentinel-2 and Hyperspectral EnMAP Satellite Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Remote Sensing Data
2.3. Preliminary Investigation
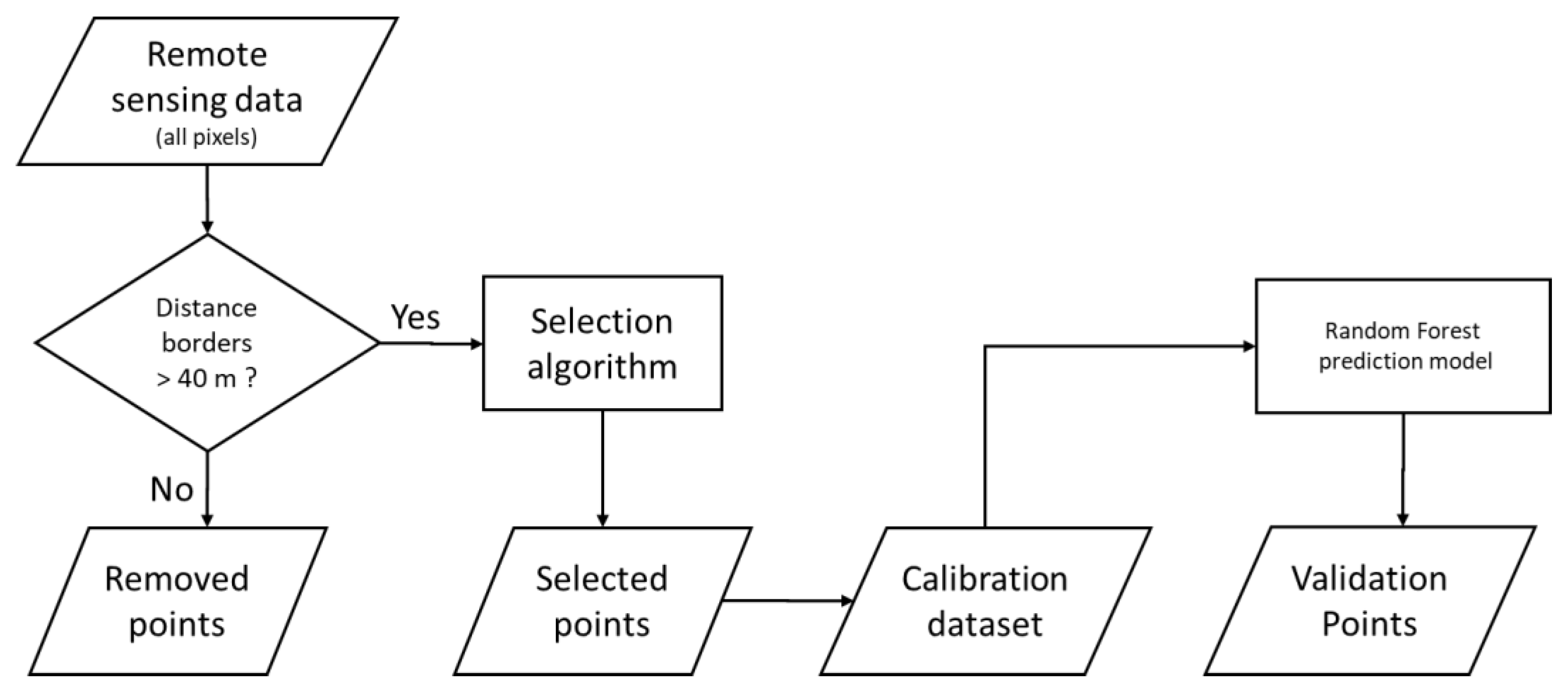
2.4. Soil Sampling Strategies
2.4.1. Selection Algorithms with a Predefined Sample size
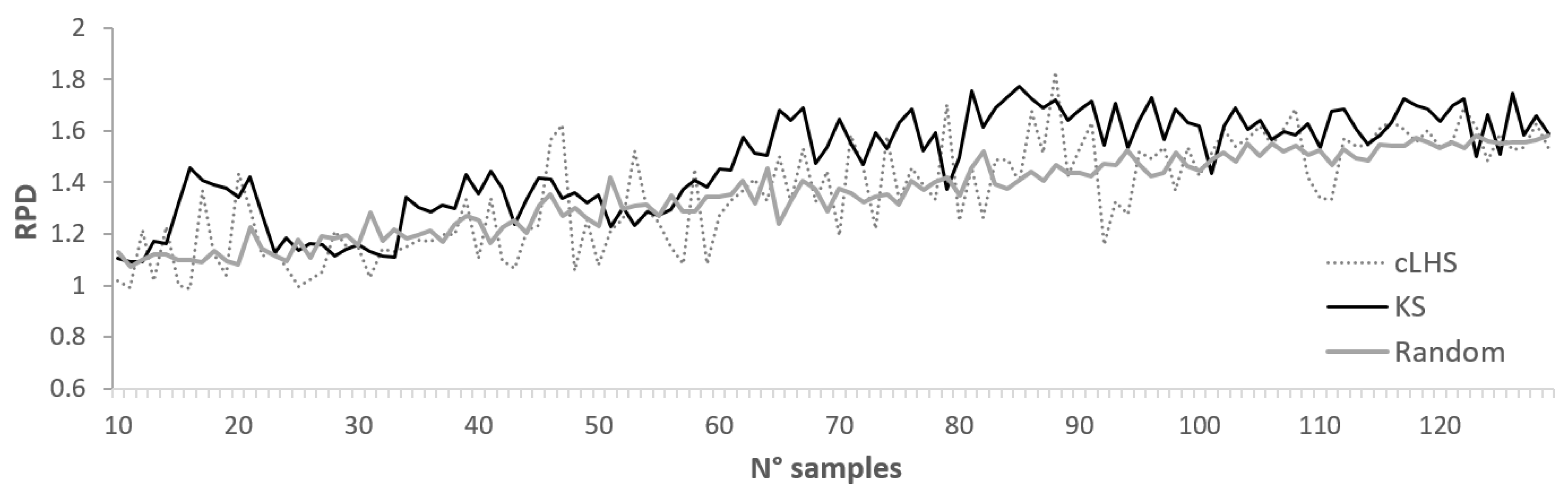
2.4.2. Selection Algorithms Providing the Ideal Number of Samples
2.5. Calibration and Validation of the Prediction Model
2.5.1. Field Tests
2.5.2. Regional Tests
3. Results
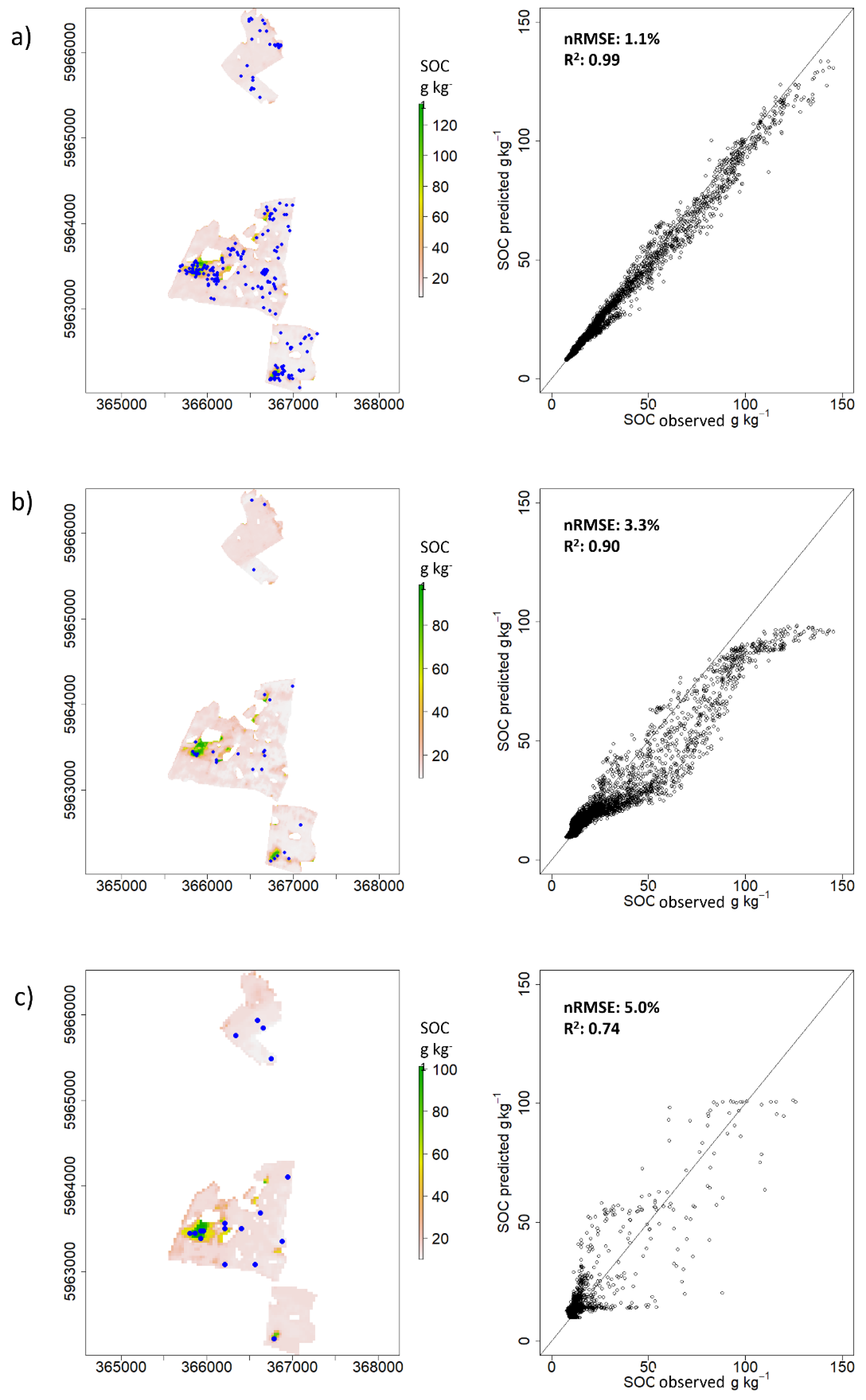
3.1. Validation at Field Scale
3.2. Validation at Regional Scale Based on Sentinel-2 Spectra
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lame, F.P.J.; Defize, P.R. Sampling of contaminated soil: Sampling error in relation to sample size and segregation. Environ. Sci. Technol. 1993, 27, 2035–2044. [Google Scholar] [CrossRef]
- Brus, D.J.; de Gruijter, J.J. Random sampling or geostatistical modelling? Choosing between design-based and model-based sampling strategies for soil (with discussion). Geoderma 1997, 80, 1–44. [Google Scholar] [CrossRef]
- Burgess, T.M.; Webster, R. Optimal interpolation and isarithmic mapping of soil properties. J. Soil Sci. 1980, 31, 315–331. [Google Scholar] [CrossRef]
- Goovaerts, P. Geostatistics for Natural Resources Evaluation; Oxford University Press: Oxford, UK, 1997; ISBN 0195115384. [Google Scholar]
- Hengl, T.; Rossiter, D.; Stein, A. Soil sampling strategies for spatial prediction by correlation with auxiliary maps. Aust. J. Soil Res. 2003, 41, 1408–1422. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A.B. A conditioned Latin hypercube method for sampling in the presence of ancillary information. Comput. Geosci. 2006, 32, 1378–1388. [Google Scholar] [CrossRef]
- Brus, D.J.; Heuvelink, G.B.M. Optimization of sample patterns for universal kriging of environmental variables. Geoderma 2007, 138, 86–95. [Google Scholar] [CrossRef]
- Brus, D.J.; Kempen, B.; Heuvelink, G.B.M. Sampling for validation of digital soil maps. Eur. J. Soil Sci. 2011, 62, 394–407. [Google Scholar] [CrossRef]
- Biswas, A.; Zhang, Y. Sampling Designs for Validating Digital Soil Maps: A Review. Pedosphere 2018, 28, 1–15. [Google Scholar] [CrossRef]
- Hedger, R.D.; Atkinson, P.M.; Malthus, T.J. Optimizing sampling strategies for estimating mean water quality in lakes using geostatistical techniques with remote sensing. Lakes Reserv. Res. Manag. 2001, 6, 279–288. [Google Scholar] [CrossRef]
- Clark, R.N. Spectroscopy of rocks and minerals, and principles of spectroscopy. In Manual of Remote Sensing; Rencz, A.N., Ed.; JohnWiley and Sons: New York, NY, USA, 1999; pp. 3–58. [Google Scholar]
- Castaldi, F.; Chabrillat, S.; Jones, A.; Vreys, K.; Bomans, B.; van Wesemael, B. Soil organic carbon estimation in croplands by hyperspectral remote APEX data using the LUCAS topsoil database. Remote Sens. 2018, 10, 153. [Google Scholar] [CrossRef]
- Castaldi, F.; Palombo, A.; Pascucci, S.; Pignatti, S.; Santini, F.; Casa, R. Reducing the Influence of Soil Moisture on the Estimation of Clay from Hyperspectral Data: A Case Study Using Simulated PRISMA Data. Remote Sens. 2015, 7, 15561–15582. [Google Scholar] [CrossRef]
- Ben-Dor, E.; Inbar, Y.; Chen, Y. The reflectance spectra of organic matter in the visible near-infrared and short wave infrared region (400–2500 nm) during a controlled decomposition process. Remote Sens. Environ. 1997, 61, 1–15. [Google Scholar] [CrossRef]
- Castaldi, F.; Chabrillat, S.; Chartin, C.; Genot, V.; Jones, A.R.; van Wesemael, B. Estimation of soil organic carbon in arable soil in Belgium and Luxembourg with the LUCAS topsoil database. Eur. J. Soil Sci. 2018, 69, 592–603. [Google Scholar] [CrossRef]
- Ramirez-Lopez, L.; Schmidt, K.; Behrens, T.; van Wesemael, B.; Demattê, J.A.M.; Scholten, T. Sampling optimal calibration sets in soil infrared spectroscopy. Geoderma 2014, 226–227, 140–150. [Google Scholar] [CrossRef]
- Nawar, S.; Mouazen, A.M. Optimal sample selection for measurement of soil organic carbon using on-line vis-NIR spectroscopy. Comput. Electron. Agric. 2018, 151, 469–477. [Google Scholar] [CrossRef]
- Guanter, L.; Kaufmann, H.; Segl, K.; Foerster, S.; Rogass, C.; Chabrillat, S.; Kuester, T.; Hollstein, A.; Rossner, G.; Chlebek, C.; et al. The EnMAP Spaceborne Imaging Spectroscopy Mission for Earth Observation. Remote Sens. 2015, 7, 8830–8857. [Google Scholar] [CrossRef]
- Pignatti, S.; Acito, N.; Amato, U.; Casa, R.; Castaldi, F.; Coluzzi, R.; De Bonis, R.; Diani, M.; Imbrenda, V.; Laneve, G.; et al. Environmental products overview of the Italian hyperspectral prisma mission: The SAP4PRISMA project. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 3997–4000. [Google Scholar]
- Zacharias, S.; Bogena, H.; Samaniego, L.; Mauder, M.; Fuß, R.; Pütz, T.; Frenzel, M.; Schwank, M.; Baessler, C.; Butterbach-Bahl, K.; et al. A Network of Terrestrial Environmental Observatories in Germany. Vadose Zone J. 2011, 10, 955. [Google Scholar] [CrossRef]
- Gerighausen, H.; Menz, G.; Kaufmann, H. Spatially Explicit Estimation of Clay and Organic Carbon Content in Agricultural Soils Using Multi-Annual Imaging Spectroscopy Data. Appl. Environ. Soil Sci. 2012, 2012, 1–23. [Google Scholar] [CrossRef]
- NutzungsdifferenzierteBodenübersichtskarte der Bundesrepublik Deutschland 1:1.000.000 (BÜK 1000 N2. 3)—Auszugskarten Acker. Available online: https://www.bgr.bund.de/DE/Themen/Boden/Informationsgrundlagen/Bodenkundliche_Karten_Datenbanken/BUEK1000/buek1000_node.html (accessed on 25 November 2018).
- Blasch, G.; Spengler, D.; Itzerott, S.; Wessolek, G. Organic Matter Modeling at the Landscape Scale Based on Multitemporal Soil Pattern Analysis Using RapidEye Data. Remote Sens. 2015, 7, 11125–11150. [Google Scholar] [CrossRef]
- Blasch, G.; Spengler, D.; Hohmann, C.; Neumann, C.; Itzerott, S.; Kaufmann, H. Multitemporal soil pattern analysis with multispectral remote sensing data at the field-scale. Comput. Electron. Agric. 2015, 113, 1–13. [Google Scholar] [CrossRef]
- Mueller-Wilm, U.; Devignot, O.; Pessiot, L. Sen2Cor Configuration and User Manual. S2-PDGS-MPC-L2A-SUM-V2.5.5. 2018. Available online: http://step.esa.int/thirdparties/sen2cor/2.5.5/docs/S2-PDGS-MPC-L2A-SUM-V2.5.5_V2. pdf (accessed on 1 July 2018).
- Brell, M.; Rogass, C.; Segl, K.; Bookhagen, B.; Guanter, L. Improving Sensor Fusion: A Parametric Method for the Geometric Coalignment of Airborne Hyperspectral and Lidar Data. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3460–3474. [Google Scholar] [CrossRef]
- Richter, R.; Schläpfer, D. Atmospheric/Topographic Correction for Airborne Imagery; Technical Report DLR-IB565-02; ReSe Applications LLC: Wil, Switzerland, 2016. [Google Scholar]
- Segl, K.; Guanter, L.; Rogass, C.; Kuester, T.; Roessner, S.; Kaufmann, H.; Sang, B.; Mogulsky, V.; Hofer, S. EeteS—The EnMAP End-to-End Simulation Tool. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 522–530. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Puchwein, G. Selection of calibration samples for near-infrared spectrometry by factor analysis of spectra. Anal. Chem. 1988, 60, 569–573. [Google Scholar] [CrossRef]
- Shenk, J.S.; Westerhaus, M.O. Population Definition, Sample Selection, and Calibration Procedures for Near Infrared Reflectance Spectroscopy. Crop Sci. 1991, 31, 469. [Google Scholar] [CrossRef]
- Miscellaneous Functions for Processing and Sample Selection of vis-NIR Diffuse Reflectance Data-12-11. 2015. Available online: https://rdrr.io/cran/prospectr/man/prospectr.html (accessed on 25 November 2018).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: http://www.R-project.org/ (accessed on 1 November 2018).
- Castaldi, F.; Hueni, A.; Chabrillat, S.; Ward, K.; Buttafuoco, G.; Bomans, B.; Vreys, K.; Brell, M.; van Wesemael, B. Evaluating the capability of the Sentinel 2 data for soil organic carbon prediction in croplands. ISPRS J. Photogramm. Remote Sens. 2019, 147, 267–282. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pascucci, S.; Casa, R.; Belviso, C.; Palombo, A.; Pignatti, S.; Castaldi, F. Estimation of soil organic carbon from airborne hyperspectral thermal infrared data: A case study. Eur. J. Soil Sci. 2014, 65. [Google Scholar] [CrossRef]
- Steinberg, A.; Chabrillat, S.; Stevens, A.; Segl, K.; Foerster, S. Prediction of Common Surface Soil Properties Based on Vis-NIR Airborne and Simulated EnMAP Imaging Spectroscopy Data: Prediction Accuracy and Influence of Spatial Resolution. Remote Sens. 2016, 8, 613. [Google Scholar] [CrossRef]
- Castaldi, F.; Castrignanò, A.; Casa, R. A data fusion and spatial data analysis approach for the estimation of wheat grain nitrogen uptake from satellite data. Int. J. Remote Sens. 2016, 37, 4317–4336. [Google Scholar] [CrossRef]
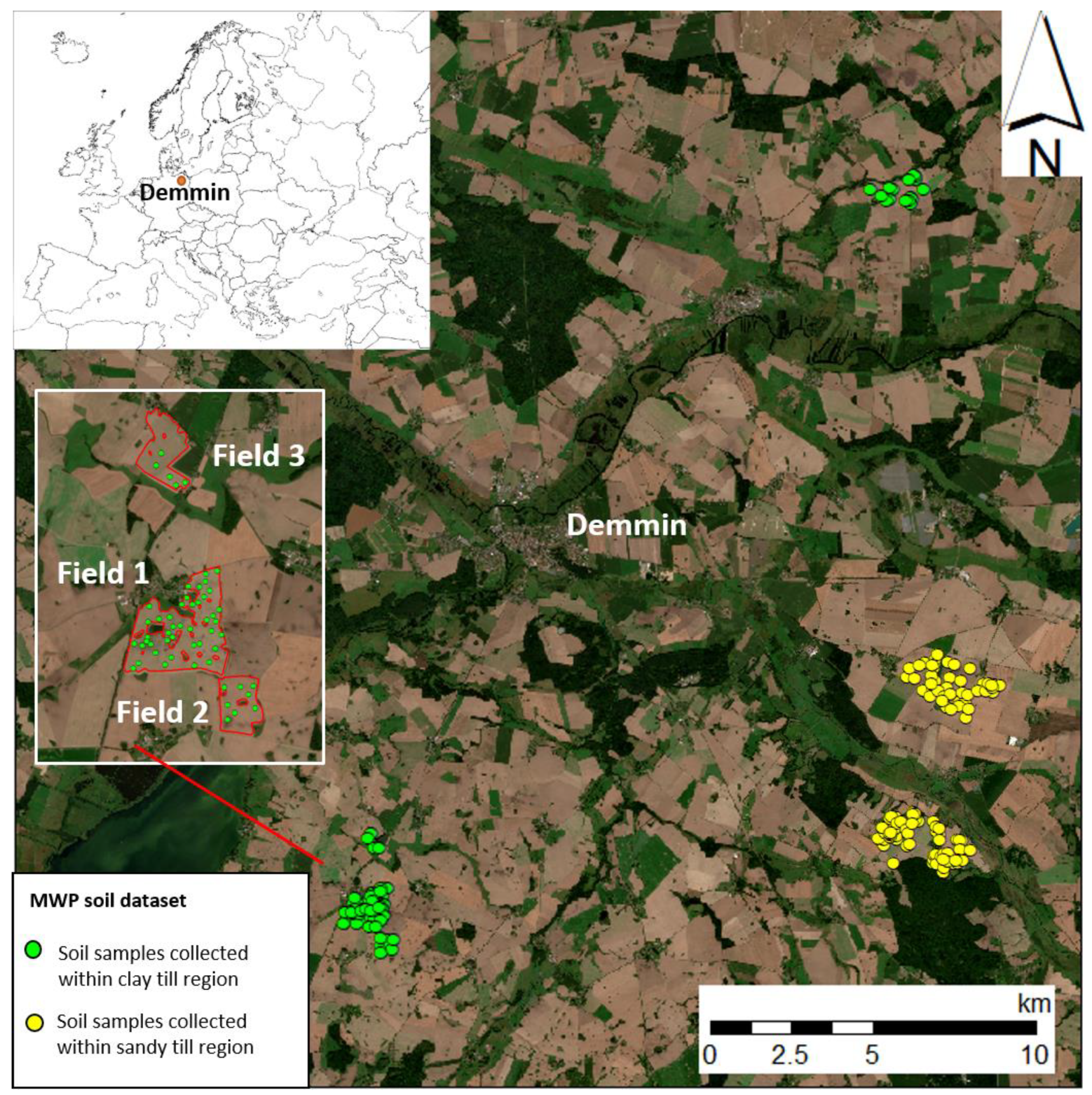
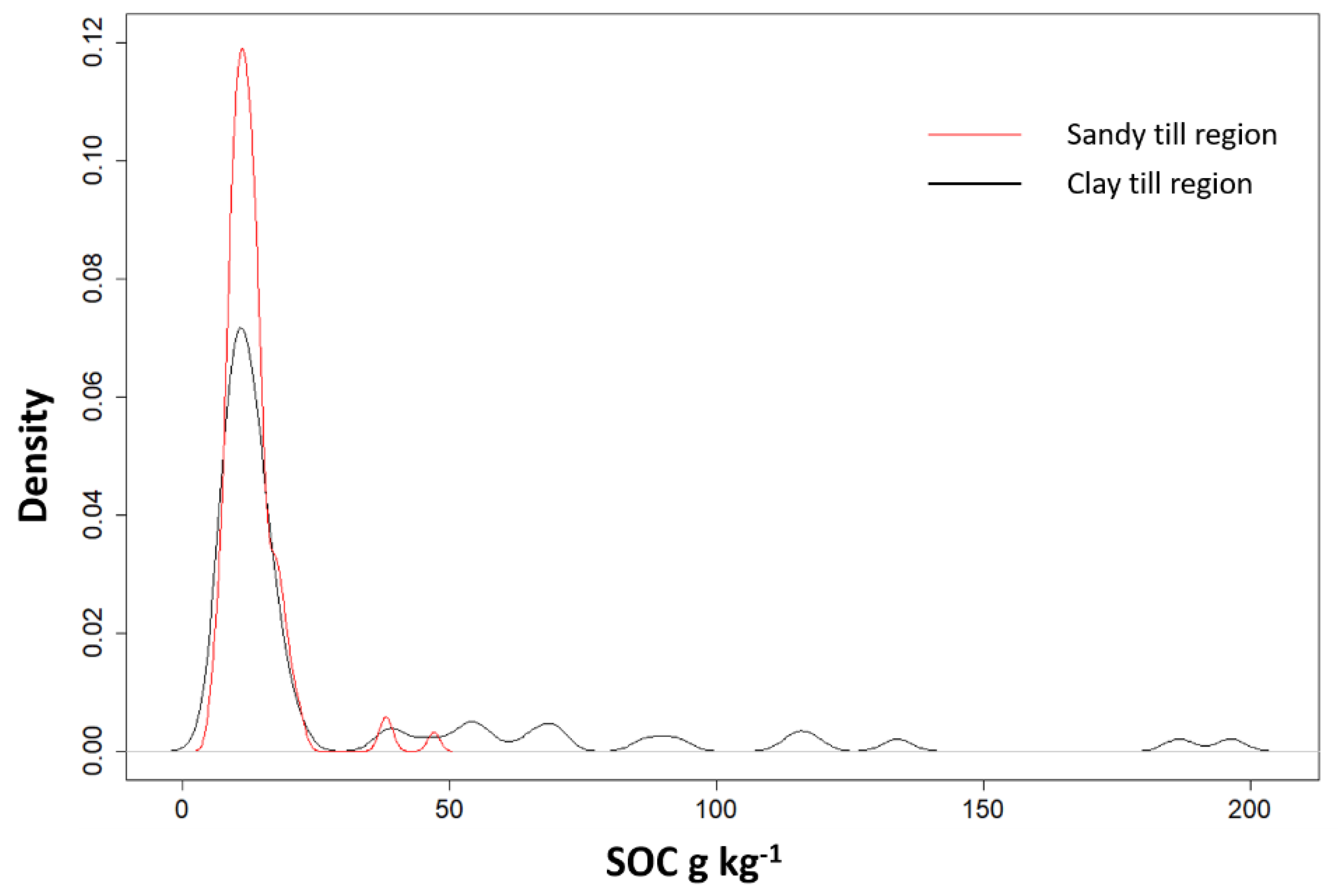
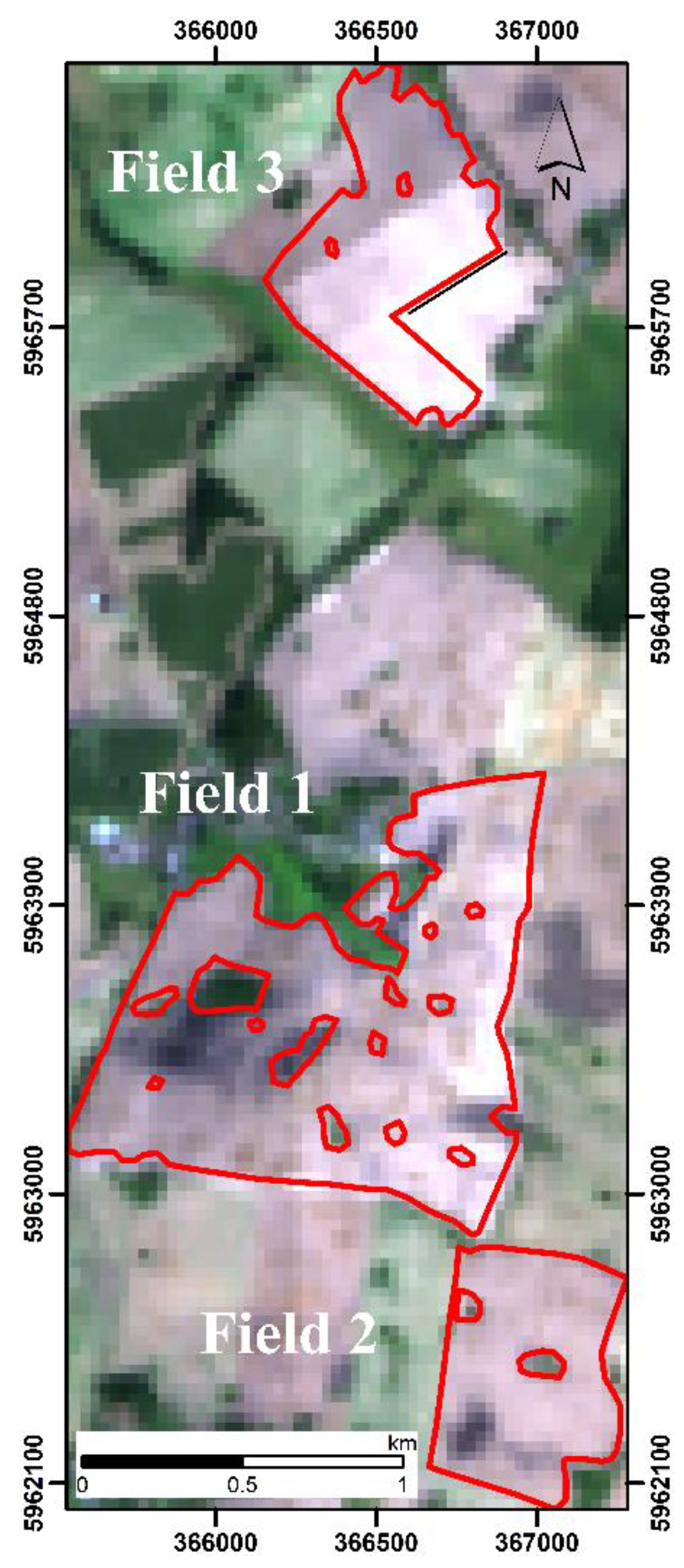
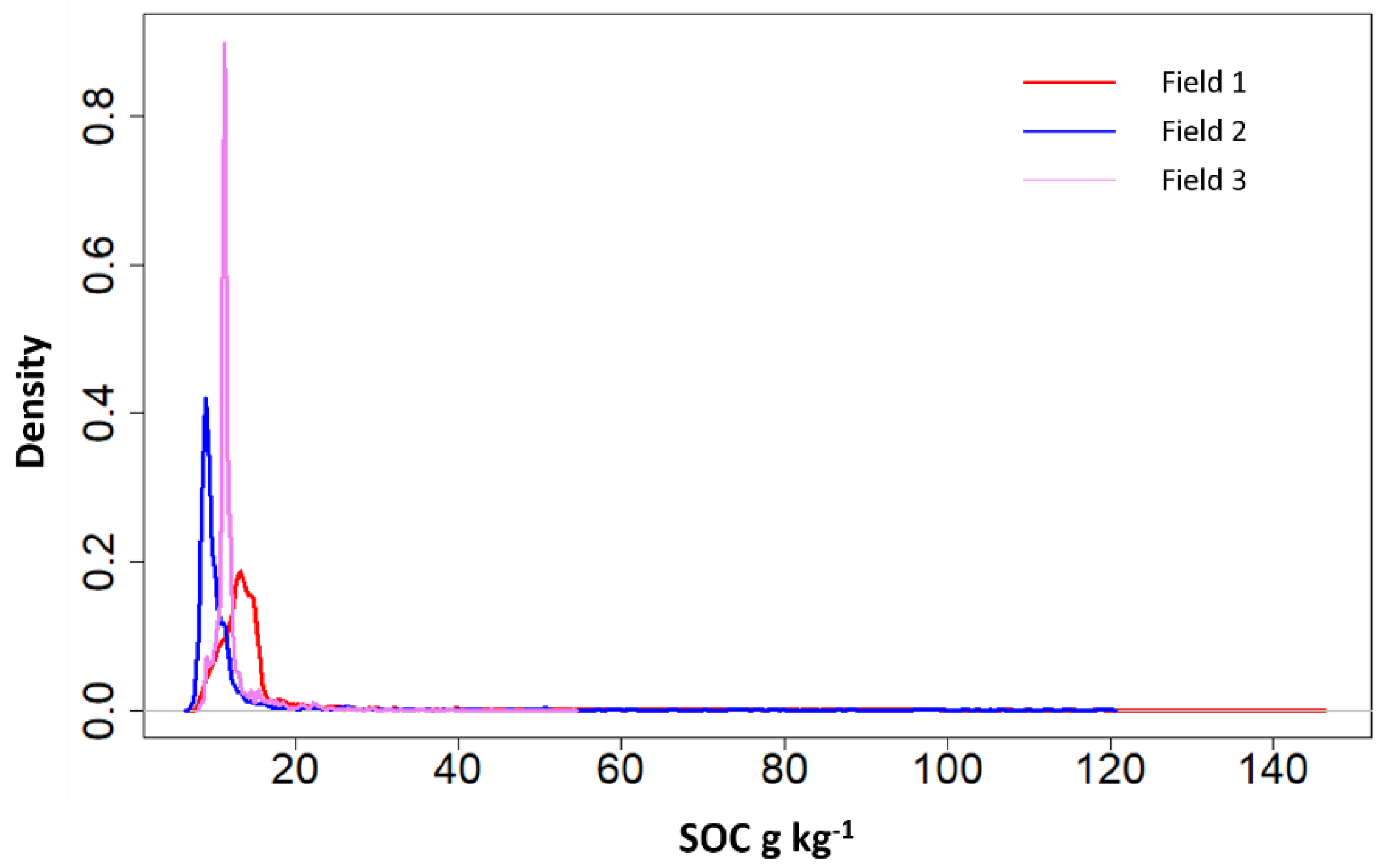
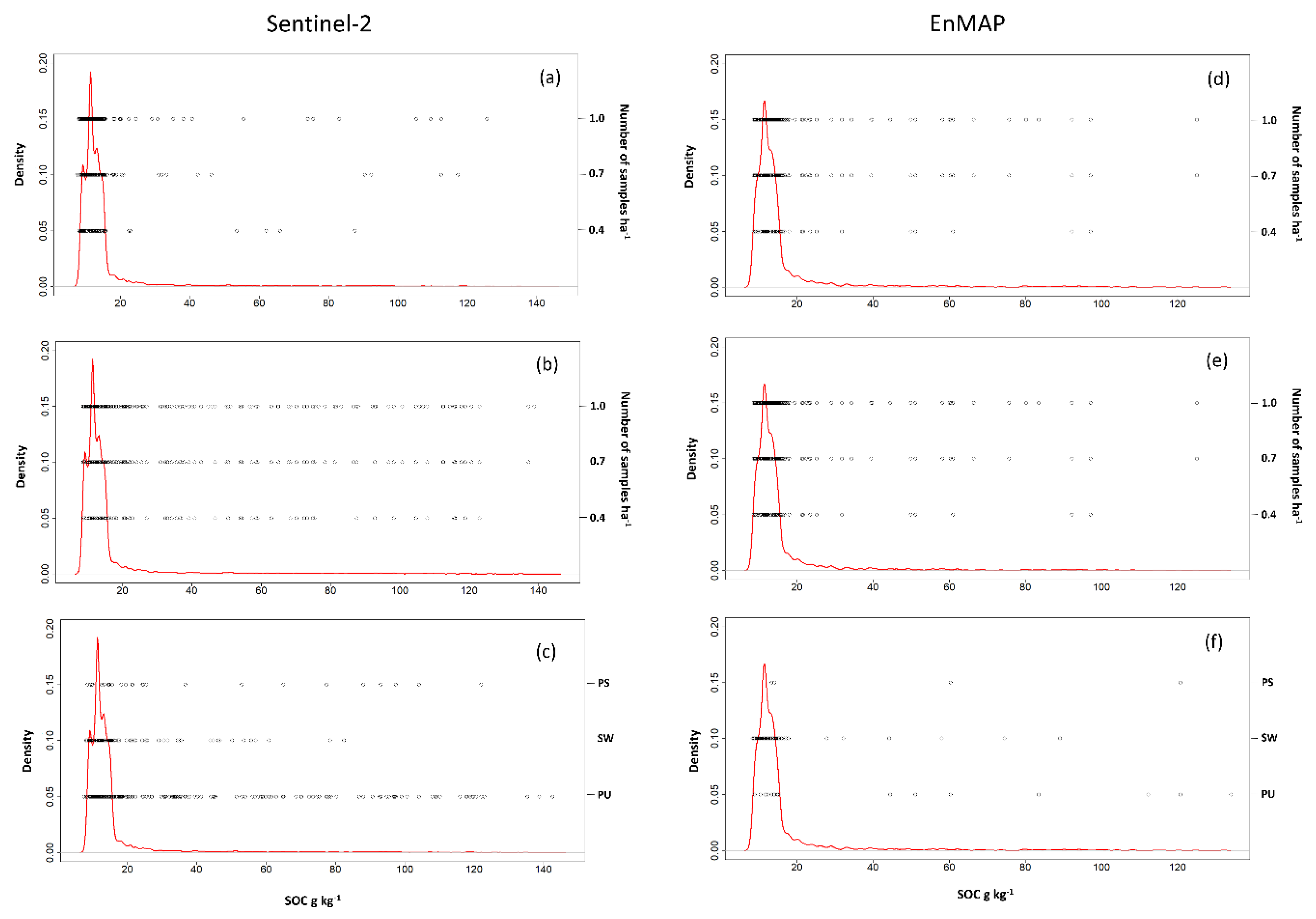

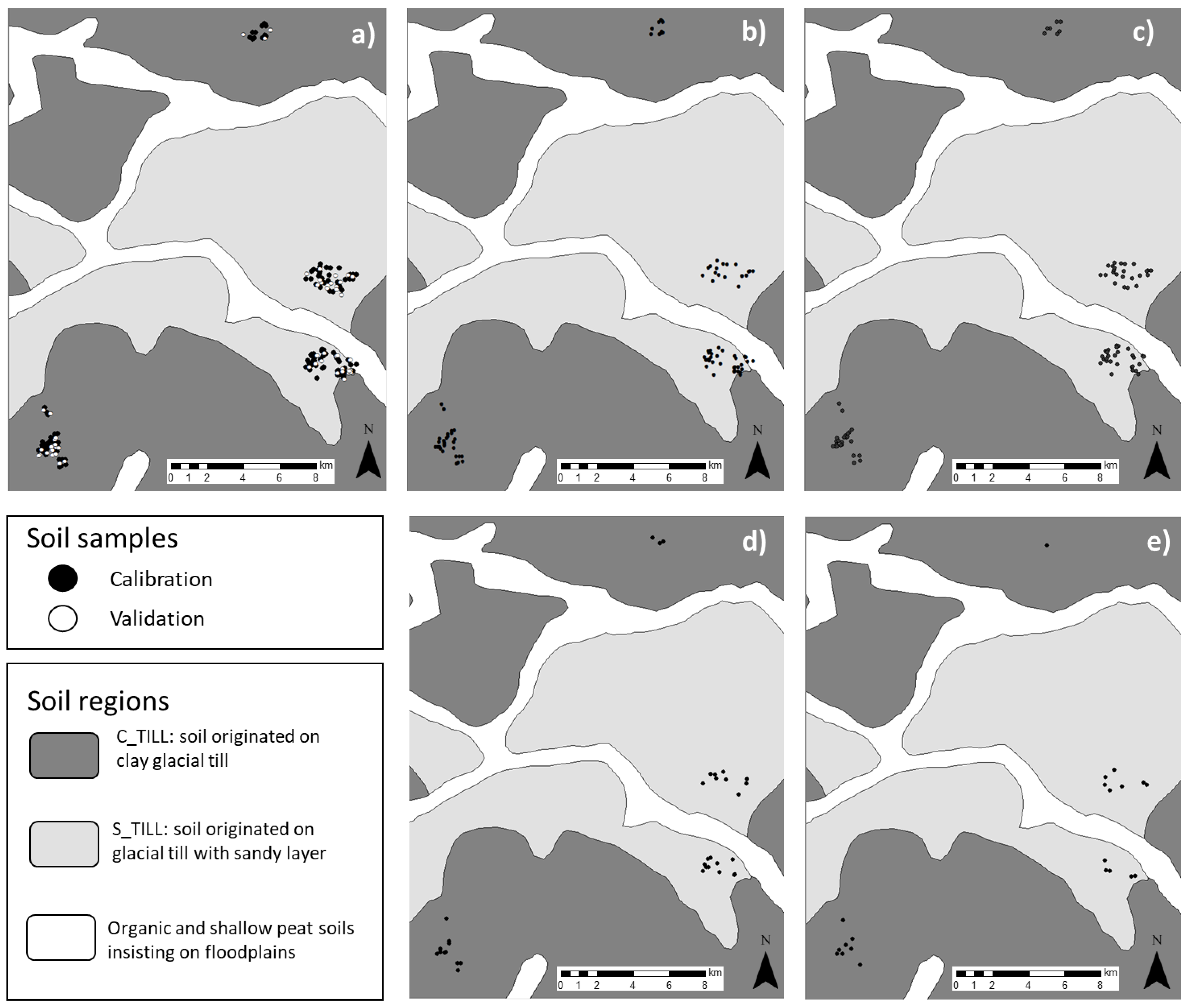
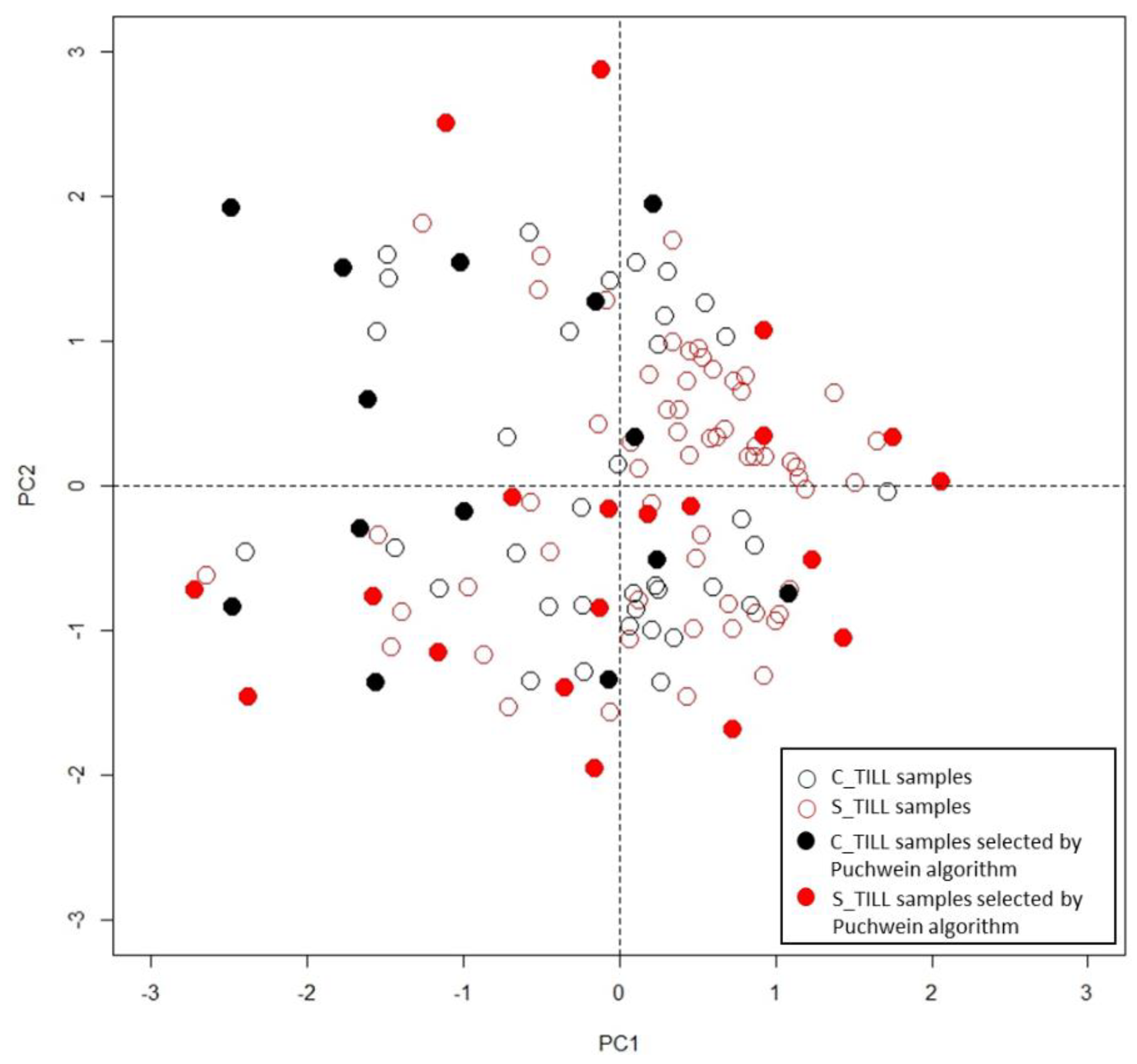
| Map | Field Size ha | Min g kg−1 | Max g kg−1 | Mean g kg−1 | Median g kg−1 | SD g kg−1 |
|---|---|---|---|---|---|---|
| Field 1 | 113.9 | 7.9 | 145.4 | 17.8 | 13.5 | 16.4 |
| Field 2 | 36.1 | 7.4 | 119.8 | 13.0 | 9.5 | 13.7 |
| Field 3 | 55.7 | 8.3 | 54.2 | 12.1 | 11.4 | 3.0 |
| All fields | 205.7 | 7.4 | 145.5 | 15.7 | 12.2 | 14.3 |
| Sensor | Method | Calibration | Validation | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n° | n°ha−1 | std g kg−1 | Min g kg−1 | Max g kg−1 | nRMSE % | RPD | Ef | ||
| Sentinel-2 | R | 81 | 0.4 | 14.5 | 8.1 | 102.1 | 17.8 ± 1.9 | 1.3 ± 0.2 | |
| R | 144 | 0.7 | 14.8 | 8.0 | 111.7 | 15.2 ± 1.9 | 1.5 ± 0.2 | 2.1 | |
| R | 205 | 1.0 | 14.7 | 7.9 | 115.9 | 13.8 ± 1.6 | 1.6 ± 0.2 | 1.6 | |
| cLHS | 81 | 0.4 | 12.6 | 8.1 | 87.6 | 18.3 | 1.2 | ||
| cLHS | 144 | 0.7 | 15.9 | 7.4 | 117.3 | 16.1 | 1.4 | 2.0 | |
| cLHS | 205 | 1.0 | 16.7 | 8.8 | 77.6 | 16.2 | 1.4 | 1.4 | |
| KS | 81 | 0.4 | 31.9 | 8.6 | 122.9 | 11.1 | 2.0 | 5.1 | |
| KS | 144 | 0.7 | 33.5 | 8.6 | 137.1 | 9.9 | 2.2 | 3.1 | |
| KS | 205 | 1.0 | 33.2 | 8.6 | 138.7 | 9.2 | 2.4 | 2.4 | |
| PU | 231 | 1.1 | 34.0 | 7.4 | 142.6 | 8.7 | 2.5 | 2.2 | |
| SW | 114 | 0.6 | 14.1 | 8.0 | 82.4 | 14.4 | 1.5 | 2.7 | |
| PS | 26 | 0.1 | 35.5 | 8.4 | 122.0 | 12.7 | 1.7 | 13.4 | |
| Simulated EnMap | R | 81 | 0.4 | 13.6 | 8.1 | 99.5 | 18.2 ± 2.1 | 1.2 ± 0.2 | |
| R | 144 | 0.7 | 13.1 | 7.9 | 105.9 | 16.4 ± 2.1 | 1.4 ± 0.2 | 2.0 | |
| R | 205 | 1.0 | 14.0 | 7.8 | 114.1 | 14.1 ± 1.8 | 1.6 ± 0.2 | 1.6 | |
| cLHS | 81 | 0.4 | 11.6 | 8.6 | 80.9 | 18.8 | 1.2 | ||
| cLHS | 144 | 0.7 | 11.0 | 7.9 | 79.7 | 15.5 | 1.4 | 2.0 | |
| cLHS | 205 | 1.0 | 14.4 | 7.6 | 125.0 | 14.0 | 1.6 | 1.6 | |
| KS | 81 | 0.4 | 15.1 | 8.6 | 97.1 | 18.5 | 1.2 | ||
| KS | 144 | 0.7 | 17.0 | 8.6 | 125.0 | 14.8 | 1.5 | 2.1 | |
| KS | 205 | 1.0 | 16.3 | 8.6 | 125.0 | 14.8 | 1.5 | 1.5 | |
| PU | 18 | 0.1 | 42.8 | 8.9 | 134.0 | 11.0 | 2.0 | 22.9 | |
| SW | 76 | 0.4 | 13.2 | 8.3 | 89.1 | 17.8 | 1.2 | ||
| PS | <10 | ||||||||
| Method | Calibration | Validation | |||||||
|---|---|---|---|---|---|---|---|---|---|
| n° | n° C_TILL | n° S_TILL | Min g kg−1 | Max g kg−1 | Mean g kg−1 | std g kg−1 | nRMSE % | RPD | |
| CAL | 130 | 51 | 79 | 6 | 196.4 | 21.3 | 29.4 | 8.7 | 1.6 |
| VAL | 51 | 20 | 31 | 6.6 | 117.8 | 14.8 | 15.3 | ||
| R | 123 | 28.9 | 8.7 | 1.6 | |||||
| cLHS | 88 | 37 | 51 | 6 | 186.8 | 21.4 | 28.7 | 7.5 | 1.8 |
| KS | 85 | 31 | 54 | 6 | 196.4 | 24.8 | 35.2 | 7.8 | 1.8 |
| PU | 34 | 14 | 20 | 7.8 | 196.4 | 42.5 | 41.9 | 8.7 | 1.6 |
| SW | 21 | 9 | 12 | 8.2 | 196.4 | 27 | 42.6 | 11.8 | 1.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castaldi, F.; Chabrillat, S.; van Wesemael, B. Sampling Strategies for Soil Property Mapping Using Multispectral Sentinel-2 and Hyperspectral EnMAP Satellite Data. Remote Sens. 2019, 11, 309. https://doi.org/10.3390/rs11030309
Castaldi F, Chabrillat S, van Wesemael B. Sampling Strategies for Soil Property Mapping Using Multispectral Sentinel-2 and Hyperspectral EnMAP Satellite Data. Remote Sensing. 2019; 11(3):309. https://doi.org/10.3390/rs11030309
Chicago/Turabian StyleCastaldi, Fabio, Sabine Chabrillat, and Bas van Wesemael. 2019. "Sampling Strategies for Soil Property Mapping Using Multispectral Sentinel-2 and Hyperspectral EnMAP Satellite Data" Remote Sensing 11, no. 3: 309. https://doi.org/10.3390/rs11030309
APA StyleCastaldi, F., Chabrillat, S., & van Wesemael, B. (2019). Sampling Strategies for Soil Property Mapping Using Multispectral Sentinel-2 and Hyperspectral EnMAP Satellite Data. Remote Sensing, 11(3), 309. https://doi.org/10.3390/rs11030309






