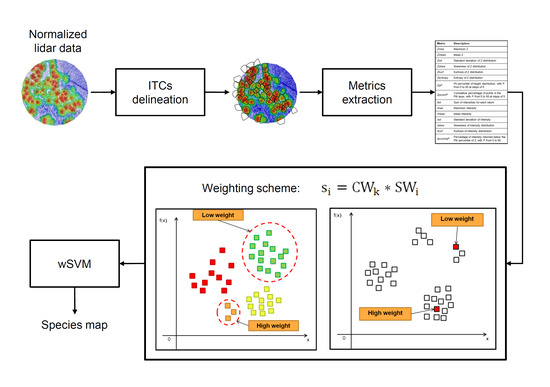
A Weighted SVM-Based Approach to Tree Species Classification at Individual Tree Crown Level Using LiDAR Data
Abstract
1. Introduction
- Class imbalance (or biased sampling) problem: In a forest, not all the tree species are present in the same amount. There are always majority species that represent usually the dominant species and cover the majority of the canopy, and minority species for which, in the extreme cases, only few trees per hectare are present. This results in a class imbalanced training set that, for minority classes, leads to (i) poor estimations of the true underlying distributions of the samples and (ii) reduced information given to the classification algorithm by the considered training samples.
- Field data positions accuracy: Localization of the exact position of a tree in a forest is a particularly difficult task, and it is usually done by using a global positioning system (GPS) device. In some cases, the accuracy could be particularly low, especially in a dense forest or in mountain areas, where the GPS accuracy is usually low. When mapping tree species at tree level, an error of more than one meter could lead to inaccurate classification results.
- Errors in ITCs delineation: Automatic ITCs delineation methods are not perfect, and they are usually associated with a delineation error that could be quite high in the case of broadleaves trees. Moreover, the quality of the delineated ITCs depends on the considered remote-sensing data, i.e., low spatial resolution images or low point density LiDAR data could provide inaccurate delineations.
- Matching errors between field data and remote sensing data: Trees measured in the field should be associated to an ITC delineated on the remote-sensing data. This procedure is subject to possible errors due to misalignments between the field positions and the remote-sensing data, and also because multiple adjacent trees measured in the field could be identified as just one crown by the automatic ITC delineation algorithm.
2. Materials and Methods
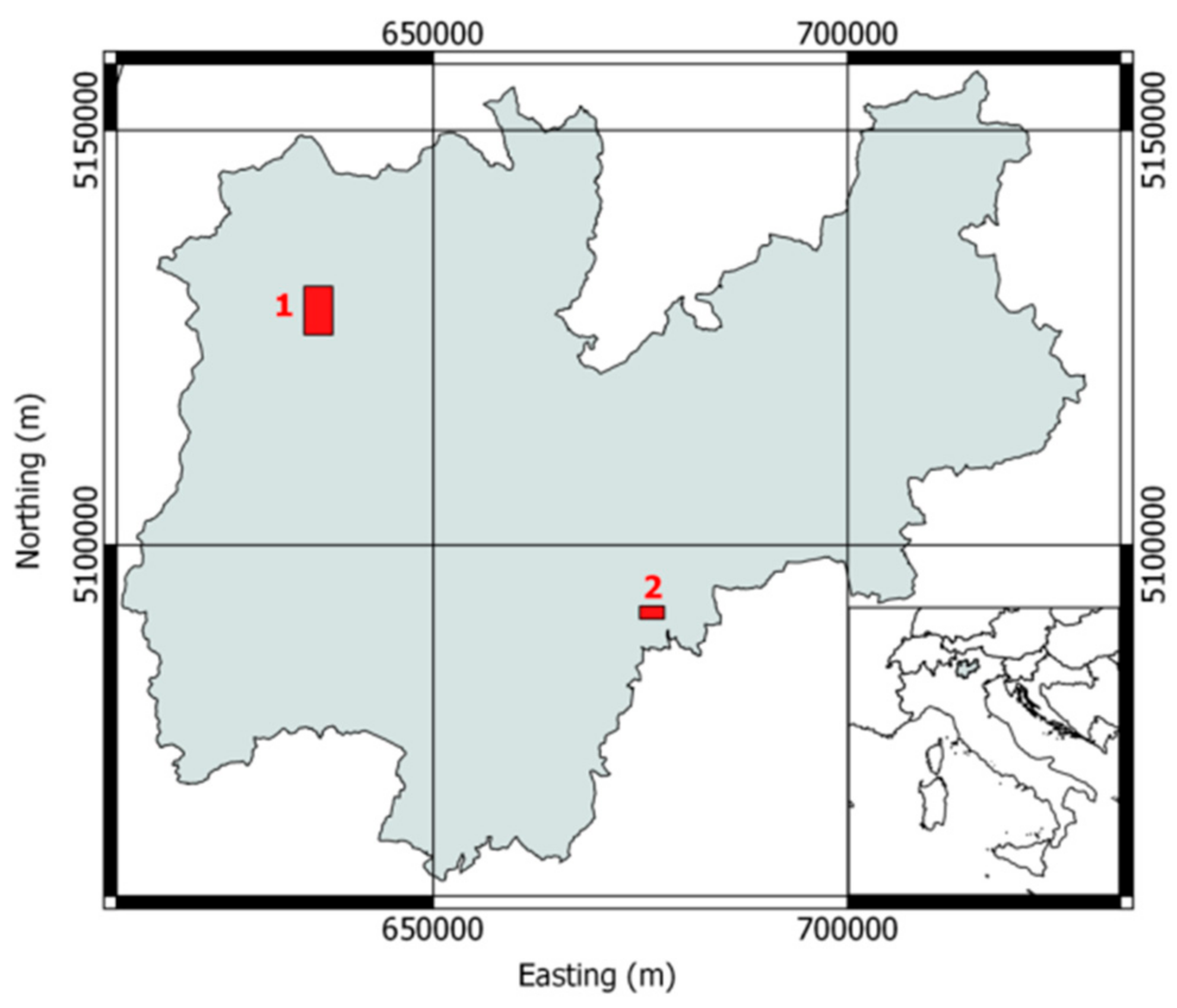
2.1. Datasets Description
2.1.1. Dataset 1: Pellizzano
2.1.2. Dataset 2: Lavarone
2.2. LiDAR Data Preprocessing
2.3. ITCs Delineation
2.4. Feature Extraction
3. Proposed Weighted SVM-Based Approach for Tree Species Classification
3.1. Interclass Weight
3.2. Intraclass Weight
3.2.1. Intraclass Weight Based on k-Means Clustering
- The k-means clustering is applied to the samples of each class in the training set independently from the samples of other classes to identify a set of G-clusters. The number of clusters is defined as the square root of the half of the total number of samples in that class (as suggested in [30]);
- For each cluster , a weight is determined as the number of samples located in that cluster. In this way, labeled samples that fall into the high-density clusters are more important for the classification problem and vice versa;
- Weights are normalized in the interval .
3.2.2. Intraclass Weight Based on Unlabeled Data
- The Euclidean distance between each training sample and all the available unlabeled samples is computed;
- The mean value of the distance between each training sample and the nearest unlabeled samples in the feature space is computed. The value of P varies depending on the class and is estimated as the square root of the total number of samples of each class in the training set;
- Weights are assigned according to the mean distance calculated during step 2: a small distance is related to the fact that the density around that sample is high, and thus a high value of the weight is assigned to that sample;
- Weights are normalized in the range .
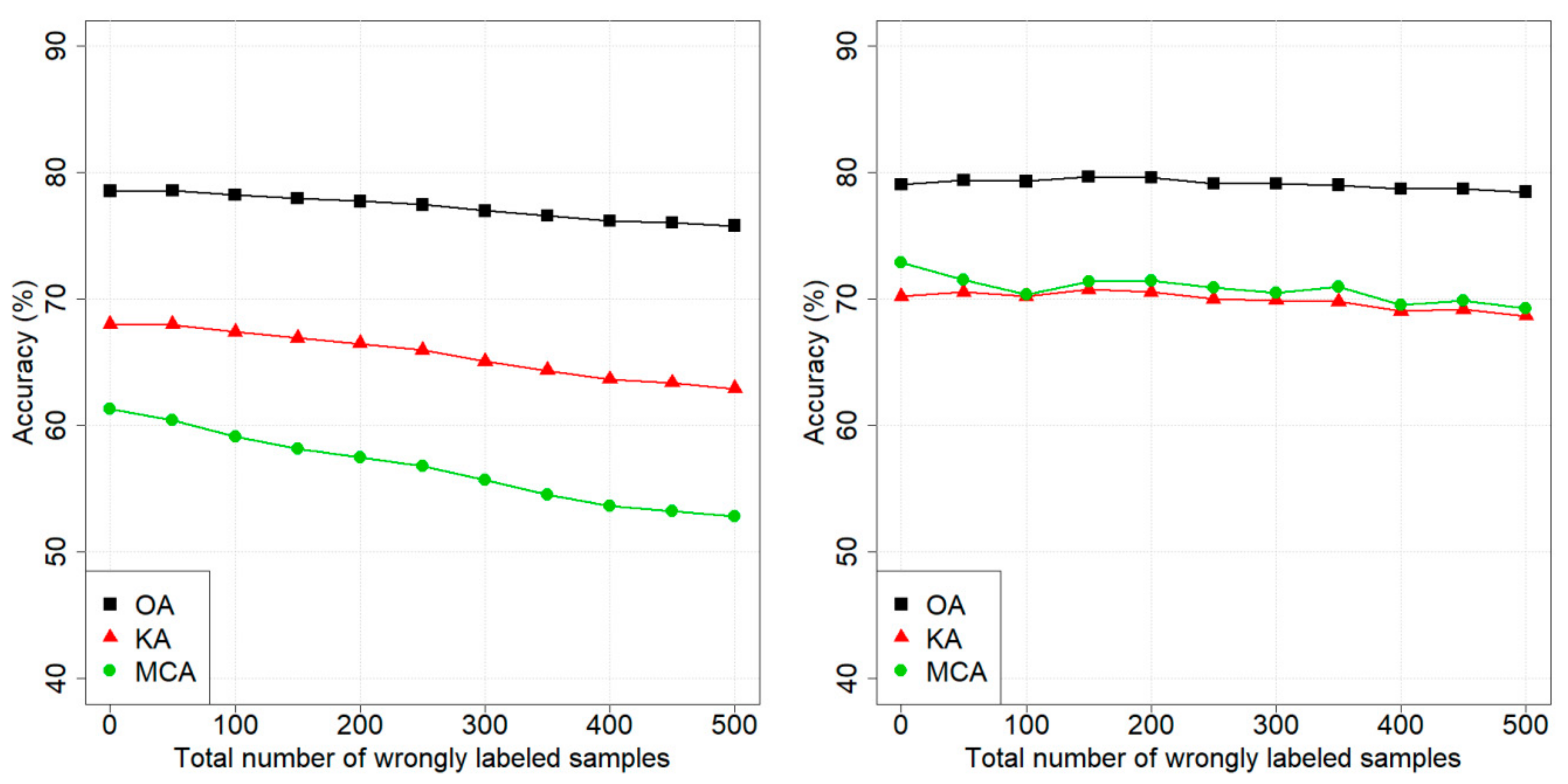
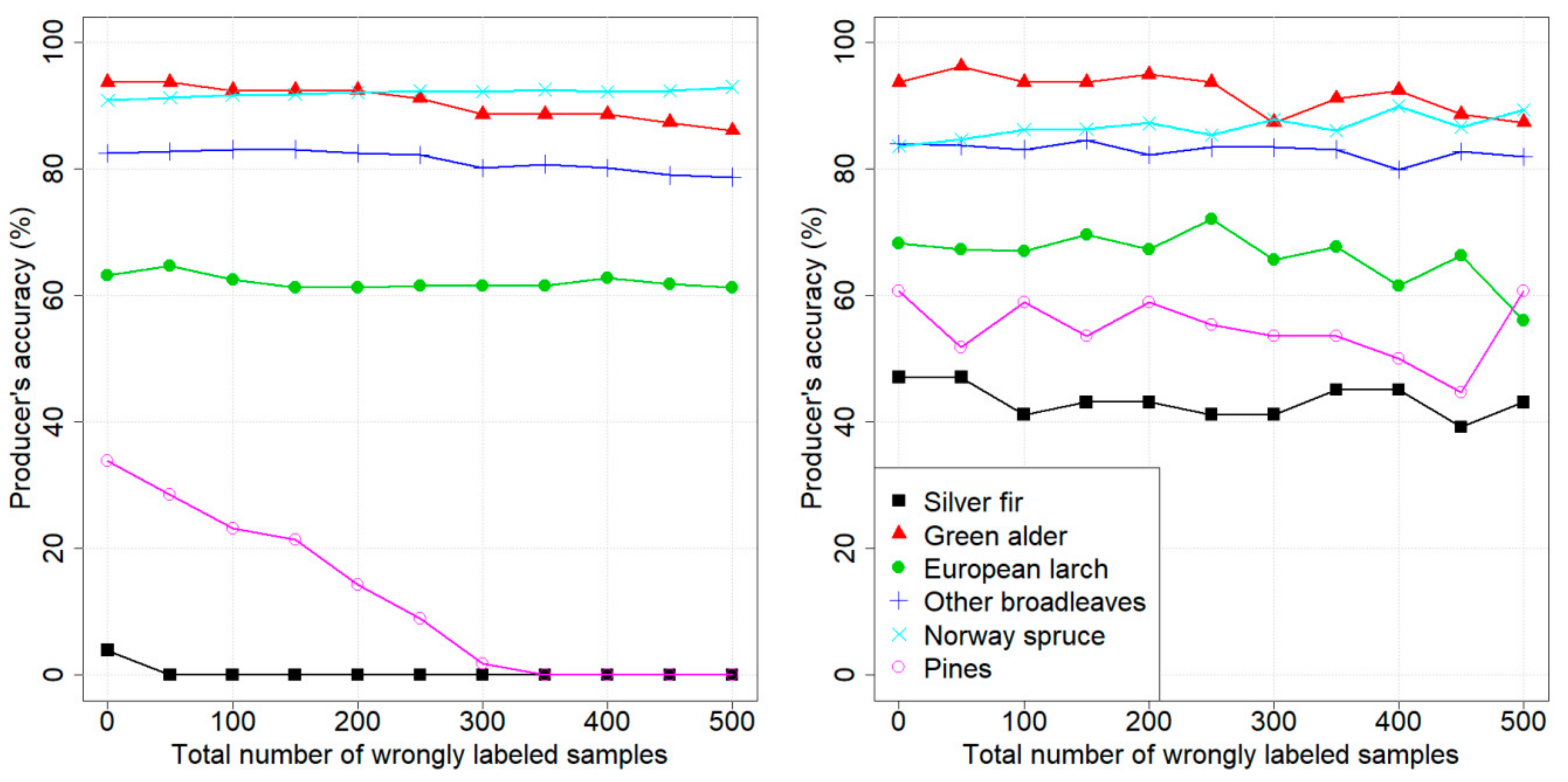
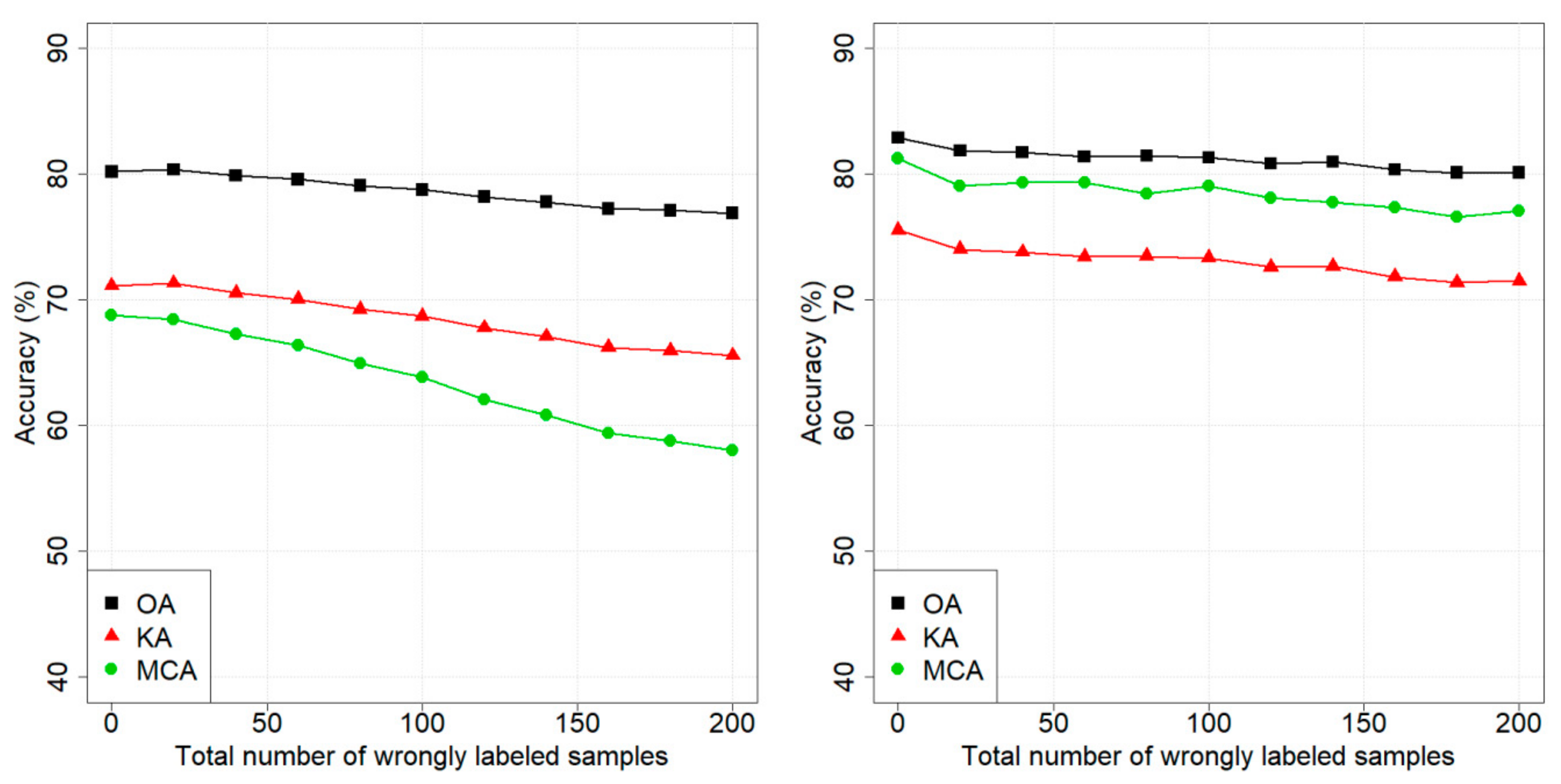
4. Results
4.1. Design of Experiments
4.2. Dataset 1: Pellizzano
4.3. Dataset 2: Lavarone
5. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tabacchi, G.; Di Cosmo, L.; Gasparini, P. Aboveground tree volume and phytomass prediction equations for forest species in Italy. Eur. J. For. Res. 2011, 130, 911–934. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Heinzel, J.; Koch, B. Investigating multiple data sources for tree species classification in temperate forest and use for single tree delineation. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 101–110. [Google Scholar] [CrossRef]
- Shang, X.; Chazette, P. Interest of a full-waveform flown UV lidar to derive forest vertical structures and aboveground carbon. Forests 2014, 5, 1454–1480. [Google Scholar] [CrossRef]
- Graves, S.J.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Colgan, M.S.; Kalantari, L.; Bohlman, S.A. Tree Species Abundance Predictions in a Tropical Agricultural Landscape with a Supervised Classification Model and Imbalanced Data. Remote Sens. 2016, 8, 161. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Gobakken, T.; Gianelle, D.; Næsset, E. Tree species classification in boreal forests with hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2632–2645. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Neumann, C.; Forster, M.; Buddenbaum, H.; Ghosh, A.; Clasen, A.; Joshi, P.K.; Koch, B. Comparison of Feature Reduction Algorithms for Classifying Tree Species with Hyperspectral Data on Three Central European Test Sites. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2547–2561. [Google Scholar] [CrossRef]
- Kim, S.; McGaughey, R.J.; Andersen, H.-E.; Schreuder, G. Tree species differentiation using intensity data derived from leaf-on and leaf-off airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1575–1586. [Google Scholar] [CrossRef]
- Wasser, L.; Day, R.; Chasmer, L.; Taylor, A. Influence of Vegetation Structure on Lidar-derived Canopy Height and Fractional Cover in Forested Riparian Buffers During Leaf-Off and Leaf-On Conditions. PLoS ONE 2013, 8, e54776. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.O.; Hyyppä, J.; Heikkinen, V.; Tokola, T. Range and AGC normalization in airborne discrete-return LiDAR intensity data for forest canopies. ISPRS J. Photogramm. Remote Sens. 2010, 65, 369–379. [Google Scholar] [CrossRef]
- Hovi, A.; Korhonen, L.; Vauhkonen, J.; Korpela, I. LiDAR waveform features for tree species classification and their sensitivity to tree- and acquisition related parameters. Remote Sens. Environ. 2016, 173, 224–237. [Google Scholar] [CrossRef]
- Korpela, I.; Ole Ørka, H.; Maltamo, M.; Tokola, T.; Hyyppä, J. Tree species classification using airborne LiDAR—Effects of stand and tree parameters, downsizing of training set, intensity normalization, and sensor type. Silva Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef]
- Lin, C.; Thomson, G.; Popescu, S. An IPCC-Compliant Technique for Forest Carbon Stock Assessment Using Airborne LiDAR-Derived Tree Metrics and Competition Index. Remote Sens. 2016, 8, 528. [Google Scholar] [CrossRef]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Classifying species of individual trees by intensity and structure features derived from airborne laser scanner data. Remote Sens. Environ. 2009, 113, 1163–1174. [Google Scholar] [CrossRef]
- Li, J.; Hu, B.; Noland, T.L. Classification of tree species based on structural features derived from high density LiDAR data. Agric. For. Meteorol. 2013, 171–172, 104–114. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å. Identifying species of individual trees using airborne laser scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Ko, C.; Sohn, G.; Remmel, T.K. Tree genera classification with geometric features from high-density airborne LiDAR. Can. J. Remote Sens. 2013, 39, S73–S85. [Google Scholar] [CrossRef]
- Vaughn, N.R.; Moskal, L.M.; Turnblom, E.C. Tree Species Detection Accuracies Using Discrete Point Lidar and Airborne Waveform Lidar. Remote Sens. 2012, 4, 377–403. [Google Scholar] [CrossRef]
- Harikumar, A.; Bovolo, F.; Bruzzone, L. An Internal Crown Geometric Model for Conifer Species Classification with High-Density LiDAR Data. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2924–2940. [Google Scholar] [CrossRef]
- Kim, S.; Hinckley, T.; Briggs, D. Classifying individual tree genera using stepwise cluster analysis based on height and intensity metrics derived from airborne laser scanner data. Remote Sens. Environ. 2011, 115, 3329–3342. [Google Scholar] [CrossRef]
- Brandtberg, T. Classifying individual tree species under leaf-off and leaf-on conditions using airborne lidar. ISPRS J. Photogramm. Remote Sens. 2007, 61, 325–340. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.; Zhang, L. Trends in Automatic Individual Tree Crown Detection and Delineation—Evolution of LiDAR Data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Lindberg, E.; Holmgren, J. Individual Tree Crown Methods for 3D Data from Remote Sensing. Curr. For. Rep. 2017, 3, 19–31. [Google Scholar] [CrossRef]
- Dalponte, M.; Ene, L.T.; Marconcini, M.; Gobakken, T.; Næsset, E. Semi-supervised SVM for individual tree crown species classification. ISPRS J. Photogramm. Remote Sens. 2015, 110, 77–87. [Google Scholar] [CrossRef]
- Dalponte, M.; Ene, L.T.; Ørka, H.O.; Gobakken, T.; Næsset, E. Unsupervised Selection of Training Samples for Tree Species Classification Using Hyperspectral Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3560–3569. [Google Scholar] [CrossRef]
- Dalponte, M.; Coomes, D.A. Tree-centric mapping of forest carbon density from airborne laser scanning and hyperspectral data. Methods Ecol. Evol. 2016, 7, 1236–1245. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Litkey, P.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Single-Sensor Solution to Tree Species Classification Using Multispectral Airborne Laser Scanning. Remote Sens. 2017, 9, 108. [Google Scholar] [CrossRef]
- Zhao, K.; Suarez, J.C.; Garcia, M.; Hu, T.; Wang, C.; Londo, A. Utility of multitemporal lidar for forest and carbon monitoring: Tree growth, biomass dynamics, and carbon flux. Remote Sens. Environ. 2018, 204, 883–897. [Google Scholar] [CrossRef]
- Demir, B.; Bruzzone, L. A multiple criteria active learning method for support vector regression. Pattern Recognit. 2014, 47, 2558–2567. [Google Scholar] [CrossRef]
- Schiffner, J.; Hillebrand, S. schiffner/locClass: Collection of Local Classification Methods. 2017. Available online: https://rdrr.io/github/schiffner/locClass/ (accessed on 8 December 2019).
- Dalponte, M.; Ene, L.; Gobakken, T.; Næsset, E.; Gianelle, D. Predicting Selected Forest Stand Characteristics with Multispectral ALS Data. Remote Sens. 2018, 10, 586. [Google Scholar] [CrossRef]
- Lin, Y.; Hyyppä, J. A comprehensive but efficient framework of proposing and validating feature parameters from airborne LiDAR data for tree species classification. Int. J. Appl. Earth Obs. Geoinf. 2016, 46, 45–55. [Google Scholar] [CrossRef]
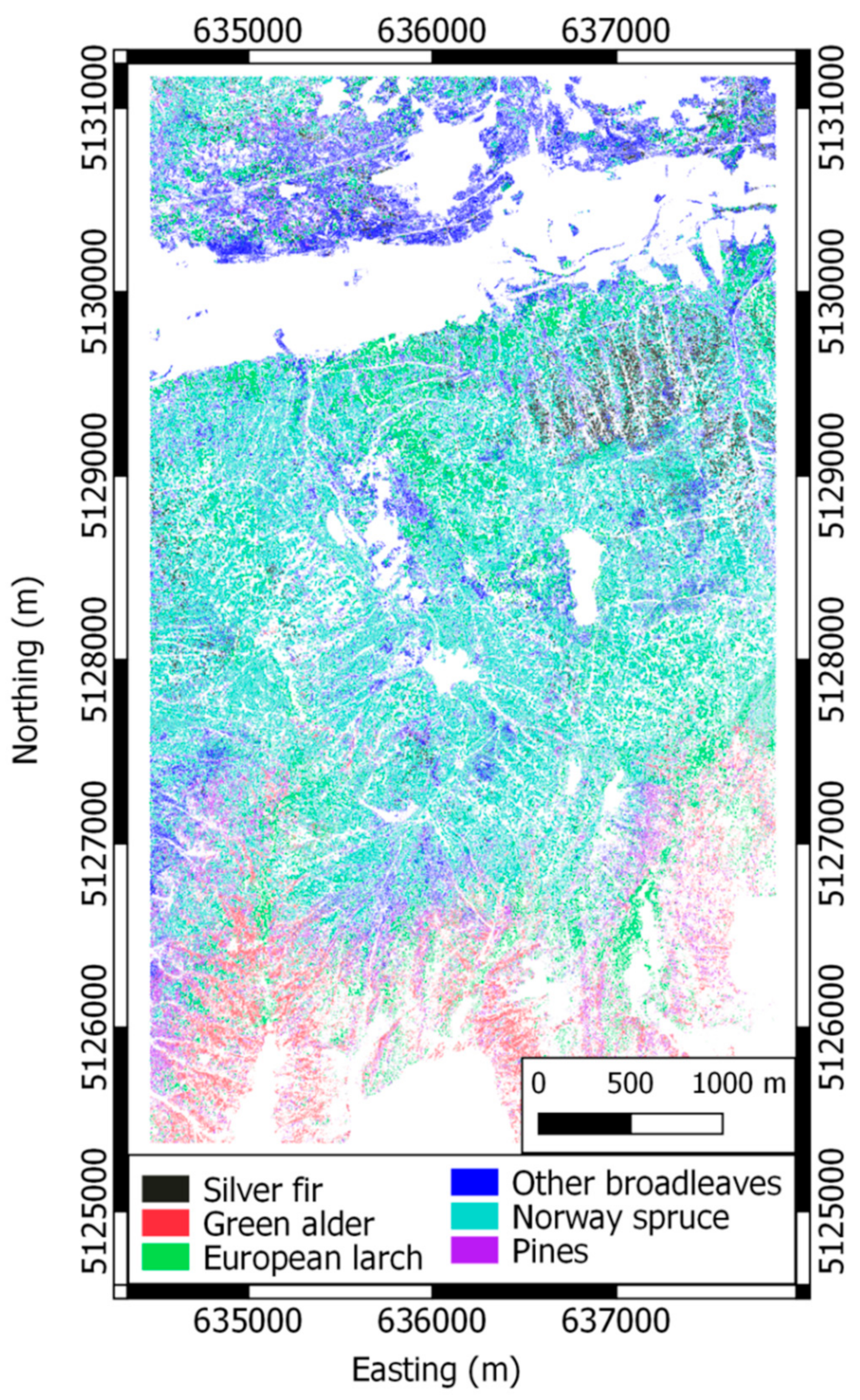

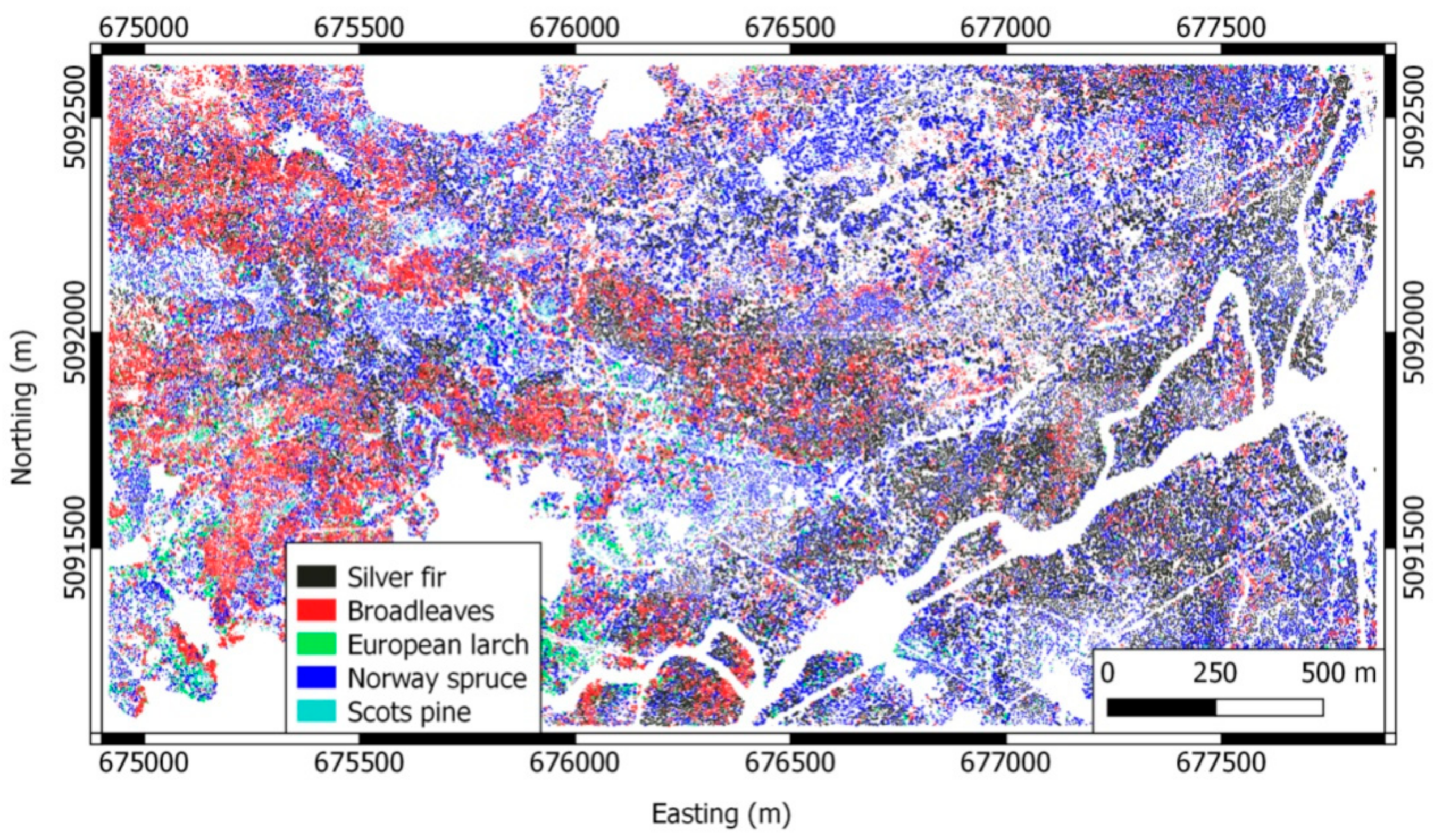
| Dataset 1: Pellizzano | Dataset 2: Lavarone | ||||
|---|---|---|---|---|---|
| Class Name | Training Set | Test Set | Class Name | Training Set | Test Set |
| Sliver fir | 52 | 51 | Sliver fir | 232 | 231 |
| Green alder | 80 | 79 | European larch | 35 | 34 |
| European larch | 313 | 312 | Broadleaves | 108 | 108 |
| Other broadleaves | 343 | 343 | Norway spruce | 253 | 252 |
| Norway spruce | 697 | 696 | Scots pine | 37 | 36 |
| Pines | 56 | 56 | |||
| Metric | Description |
|---|---|
| Zmax | Maximum Z |
| Zmean | Mean Z |
| Zsd | Standard deviation of Z distribution |
| Zskew | Skewness of Z distribution |
| Zkurt | Kurtosis of Z distribution |
| Zentropy | Entropy of Z distribution |
| ZqP | Ph percentile of height distribution, with P from 5 to 95 at steps of 5 |
| ZpcumP | Cumulative percentage of points in the Pth layer, with P from 5 to 95 at steps of 5 |
| Itot | Sum of intensities for each return |
| Imax | Maximum intensity |
| Imean | Mean intensity |
| Isd | Standard deviation of intensity |
| Iskew | Skewness of intensity distribution |
| Ikurt | Kurtosis of intensity distribution |
| IpcumzqP | Percentage of intensity returned below the Pth percentile of Z, with P from 5 to 95 |
| pRth | Percentage of Rth return, with R from 1 to 4 |
| Slope | Properties of the tangent plane to the surface |
| DTM | Digital terrain model value (m) |
| aspect | Compass direction in which the slope faces to |
| Classifier | OA | MCA | KA | Processing Time |
|---|---|---|---|---|
| 79.2 | 62.2 | 69.2 | 57.96 | |
| 80.7 | 71.4 | 72.1 | 60.70 | |
| 79.1 | 72.9 | 70.2 | 58.25 | |
| 80.8 | 71.5 | 72.4 | 898.87 |
| Classifier | Sliver Fir | Green Alder | European Larch | Other Broadleaves | Norway Spruce | Pines |
|---|---|---|---|---|---|---|
| 5.9 | 91.1 | 67.9 | 82.8 | 89.9 | 35.7 | |
| 39.2 | 94.9 | 68.6 | 83.7 | 88.2 | 53.6 | |
| 47.1 | 93.7 | 68.3 | 84.0 | 83.6 | 60.7 | |
| 39.2 | 93.7 | 71.2 | 84.5 | 87.1 | 53.6 |
| SF | GA | EL | OB | NS | P | SF | GA | EL | OB | NS | P | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SF | 3 | 0 | 0 | 1 | 0 | 2 | SF | 20 | 0 | 6 | 7 | 19 | 3 |
| GA | 0 | 72 | 0 | 4 | 0 | 0 | GA | 0 | 75 | 2 | 4 | 1 | 1 |
| EL | 5 | 0 | 212 | 15 | 33 | 23 | EL | 3 | 0 | 214 | 12 | 28 | 11 |
| OB | 5 | 6 | 16 | 284 | 36 | 8 | OB | 3 | 3 | 16 | 287 | 30 | 9 |
| NS | 38 | 1 | 81 | 37 | 626 | 3 | NS | 24 | 1 | 66 | 25 | 614 | 2 |
| P | 0 | 0 | 3 | 2 | 1 | 20 | P | 1 | 0 | 8 | 8 | 4 | 30 |
| SF | GA | EL | OB | NS | P | SF | GA | EL | OB | NS | P | ||
| SF | 24 | 0 | 7 | 9 | 24 | 3 | SF | 20 | 0 | 3 | 5 | 14 | 3 |
| GA | 0 | 74 | 2 | 5 | 0 | 1 | GA | 0 | 74 | 1 | 4 | 0 | 0 |
| EL | 6 | 0 | 213 | 11 | 45 | 10 | EL | 5 | 0 | 222 | 15 | 36 | 12 |
| OB | 3 | 3 | 20 | 288 | 37 | 8 | OB | 3 | 4 | 17 | 290 | 36 | 9 |
| NS | 17 | 1 | 61 | 22 | 582 | 0 | NS | 22 | 1 | 61 | 22 | 606 | 2 |
| P | 1 | 1 | 9 | 8 | 8 | 34 | P | 1 | 0 | 8 | 7 | 4 | 30 |
| Classifiers | OA | MCA | Kappa | Processing Time (s) |
|---|---|---|---|---|
| 80.2 | 68.8 | 71.1 | 20.86 | |
| 81.4 | 77.8 | 73.5 | 19.72 | |
| 82.9 | 81.2 | 75.5 | 21.49 | |
| 82.6 | 76.9 | 74.9 | 530.75 |
| Classifiers | Sliver Fir | Broadleaves | European Larch | Norway Spruce | Scots Pine |
|---|---|---|---|---|---|
| 86.1 | 84.3 | 38.2 | 82.5 | 52.8 | |
| 87.0 | 78.7 | 73.5 | 80.2 | 69.4 | |
| 84.4 | 80.6 | 82.4 | 83.7 | 75.0 | |
| 87.9 | 78.7 | 67.6 | 83.7 | 66.7 |
| SF | B | EL | NS | SP | SF | B | EL | NS | SP | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SF | 199 | 9 | 0 | 28 | 0 | SF | 201 | 12 | 1 | 23 | 0 |
| B | 3 | 91 | 2 | 8 | 2 | B | 4 | 85 | 0 | 8 | 2 |
| EL | 0 | 1 | 13 | 2 | 0 | EL | 2 | 2 | 25 | 11 | 1 |
| NS | 28 | 7 | 18 | 208 | 15 | NS | 23 | 7 | 7 | 202 | 8 |
| SP | 1 | 0 | 1 | 6 | 19 | SP | 1 | 2 | 1 | 8 | 25 |
| SF | B | EL | NS | SP | SF | B | EL | NS | SP | ||
| SF | 195 | 7 | 0 | 19 | 0 | SF | 203 | 13 | 0 | 23 | 0 |
| B | 4 | 87 | 0 | 8 | 1 | B | 4 | 85 | 0 | 8 | 1 |
| EL | 1 | 3 | 28 | 7 | 0 | EL | 0 | 2 | 23 | 5 | 0 |
| NS | 31 | 11 | 5 | 211 | 8 | NS | 24 | 7 | 10 | 211 | 11 |
| SP | 0 | 0 | 1 | 7 | 27 | SP | 0 | 1 | 1 | 5 | 24 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, H.M.; Demir, B.; Dalponte, M. A Weighted SVM-Based Approach to Tree Species Classification at Individual Tree Crown Level Using LiDAR Data. Remote Sens. 2019, 11, 2948. https://doi.org/10.3390/rs11242948
Nguyen HM, Demir B, Dalponte M. A Weighted SVM-Based Approach to Tree Species Classification at Individual Tree Crown Level Using LiDAR Data. Remote Sensing. 2019; 11(24):2948. https://doi.org/10.3390/rs11242948
Chicago/Turabian StyleNguyen, Hoang Minh, Begüm Demir, and Michele Dalponte. 2019. "A Weighted SVM-Based Approach to Tree Species Classification at Individual Tree Crown Level Using LiDAR Data" Remote Sensing 11, no. 24: 2948. https://doi.org/10.3390/rs11242948
APA StyleNguyen, H. M., Demir, B., & Dalponte, M. (2019). A Weighted SVM-Based Approach to Tree Species Classification at Individual Tree Crown Level Using LiDAR Data. Remote Sensing, 11(24), 2948. https://doi.org/10.3390/rs11242948