Assessment of Phytoecological Variability by Red-Edge Spectral Indices and Soil-Landscape Relationships
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Phytoecological Relationships
2.2. Taxonomic Classification of Phytophysiognomies
2.3. Remote Sensing Data and Processing
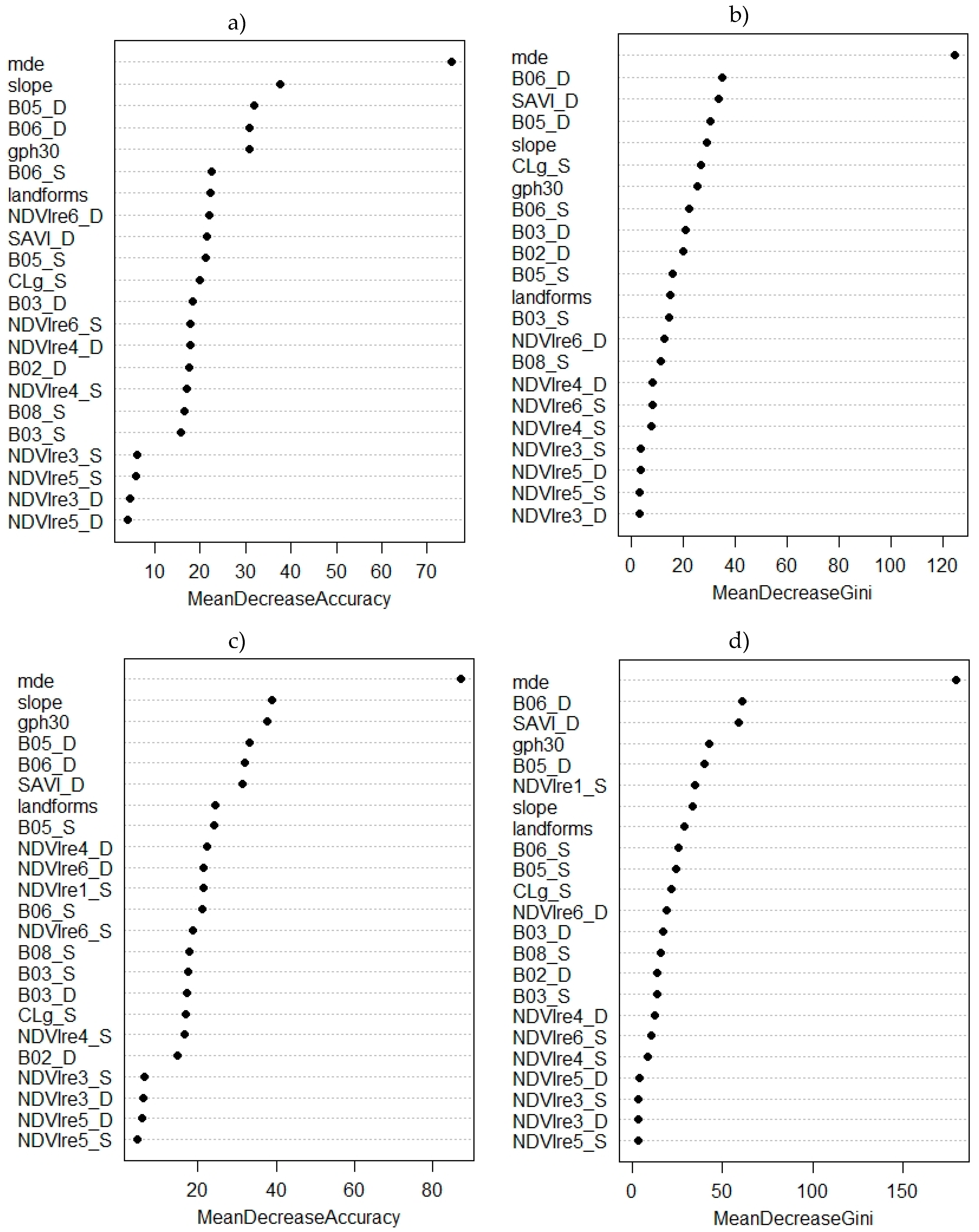
2.4. Supervised Phytophysiognomies Classification
3. Results and Discussion
3.1. Phytophysiognomies and Landscape Relationships
3.2. Mapping Phytophysiognomies
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oliveira-Filho, A.T. Classificação das fitofisionomias da América do Sul Cisandina Tropical e Subtropical: Proposta de um novo sistema-prático e flexível-ou uma injeção a mais de caos? Rodriguésia 2009, 60, 237–258. [Google Scholar] [CrossRef]
- Oliveira-Filho, A.T.; Fontes, M.A.L.; Viana, P.L.; Valente, A.S.M.; Salimena, F.R.G.; Ferreira, F.M. O mosaico de fitofisionomias do Parque Estadual do Ibitipoca. In Flora do Parque Estadual do Ibitipoca e seu Entorno, 1st ed.; Forzza, R.C., Menini Neto, L., Salimena, F.R.G., Zappi, D., Eds.; Editora UFJF: Juiz de Fora, Brazil, 2013; pp. 53–93. ISBN 978857672187-2. [Google Scholar]
- Moreira, B.; Carvalho, F.A.; Neto, L.M.; Salimena, F.R.G. Phanerogamic flora and phytogeography of the Cloud Dwarf Forests of Ibitipoca State Park, Minas Gerais, Brazil. Biota Neotrop. 2018, 18, 1–20. [Google Scholar] [CrossRef]
- Bruijnzeel, L.A.; Scatena, F.N.; Hamilton, L.S. Tropical Montane Cloud Forests: Science for Conservation and Management; Cambridge University Press: London, UK, 2010; p. 740. ISBN 9780521760355. [Google Scholar]
- Bubb, P.; May, I.; Miles, L.; Sayer, J. Cloud Forest Agenda; PNUMA-CMVC: Cambridge, UK, 2014; p. 32. [Google Scholar]
- Bertoncello, R.; Yamamoto, K.; Meireles, L.D.; Shepherd, G.J. A phytogeographic analysis of cloud forests and other forest subtypes amidst the Atlantic forests in south and southeast Brazil. Biodivers. Conserv. 2011, 20, 3413–3433. [Google Scholar] [CrossRef]
- Rahbek, C. The role of spatial scale and the perception of large-scale species—Richness patterns. Ecol. Lett. 2005, 8, 224–239. [Google Scholar] [CrossRef]
- Grytnes, J.A.; McCain, C.M. Elevational trends in biodiversity. Encycl. Biodivers. 2007, 2, 1–8. [Google Scholar]
- Slik, J.W.; Shin-Ichiro, A.S.I.; Brearley, F.Q.; Cannon, C.H.; Forshed, O.; Kitayama, K. Environmental correlates of tree biomass, basal area, wood specific gravity and stem density gradients in Borneo’s tropical forests. Glob. Ecol. Biogeogr. 2010, 19, 50–60. [Google Scholar] [CrossRef]
- Webster, G.L. The panorama of Neotropical Cloud Forests. In Biodiversity and Conservation of Neotropical Montane Forests. In Proceedings of the Neotropical Montane Forest Biodiversity and Conservation Symposium, New York, NY, USA, 21–26 June 1993; Churchill, S.P., Balslev, H., Forero, E., Luteyn, J.L., Eds.; The New York Botanical Garden: New York, NY, USA, 1995; pp. 53–77. [Google Scholar]
- Santos, M.F.; Serafim, H.; Sano, P.T. Fisionomia e composição da vegetação florestal na Serra do Cipó, MG, Brasil. Acta Bot. Bras. 2011, 25, 793–814. [Google Scholar] [CrossRef]
- Streher, A.S.; Silva, T.S.F. Geração de imagens sintéticas Landsat TM para a avaliação da fenologia de diferentes fitofisonomias na região do Espinhaço Meridional, MG. In Anais XVII Simpósio Brasileiro de Sensoriamento Remoto—SBSR. In Proceedings of the XVII Simpósio Brasileiro de Sensoriamento Remoto—SBSR, João Pessoa-PB, Brazil, 25–29 April 2015; INPE: São Paulo, Brazil, 2015; pp. 6343–6350. [Google Scholar]
- Morellato, L.P.C.; Talora, D.C.; Takahashi, A.; Bencke, C.S.C.; Romera, E.C.; Ziparro, V. Phenology of atlantic rain forest trees: A comparative study. Biotropica 2000, 32, 811–823. [Google Scholar] [CrossRef]
- Staggemeier, V.G.; Morellato, L.P.C. Reproductive phenology of coastal plain Atlantic forest vegetation: Comparisons from seashore to foothills. Int. J. Biometeorol. 2011, 55, 843–854. [Google Scholar] [CrossRef]
- Pereira, C.R.; Barbosa, R.S. Quantificação da chuva oculta na Serra do Mar, Estado do Rio de Janeiro. Cienc. Florest. 2016, 26, 1061–1073. [Google Scholar] [CrossRef]
- BRASIL. Portaria Instituto Estadual de Florestas (IEF), nº 22 de 17 de Maio de 2018; Diário do Executivo; Diário do Executivo: Minas Gerais, Brazil, 2018; p. 29. Available online: http://www.siam.mg.gov.br/sla/download.pdf?idNorma=46324 (accessed on 19 October 2019).
- Nummer, A.R. Mapeamento Geológico e Tectônico Experimental do Grupo Andrelândia na Região de Santa Rita do Ibitipoca—Lima Duarte, Sul de Minas Gerais. Master’s Thesis, Geology Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil, 1991. [Google Scholar]
- Sousa, A.M.O.; Mesquita, P.; Gonçalves, A.C.; Silva, J.R.M. Segmentação e classificação de tipologias florestais a partir de imagens QUICKBIRD. Ambiência 2010, 6, 57–66. [Google Scholar]
- Disperati, A.A.; Santos, J.R.; Oliveira Filho, P.C.; Neeff, T. Aplicação da técnica “filtragem de locais máximas” em fotografia aérea digital para a contagem de copas em reflorestamento de Pinus elliottii. Sci. For. 2007, 76, 45–55. [Google Scholar]
- Silva, S.T.; Mello, J.M.; Acerbi Junior, F.W.; Reis, A.A.; Raimundo, M.G.; Silva, I.L.G.; Scolforo, J.R.S. Using remote sensing images for stratification of the cerrado in forest inventories. Pesqui. Florest. Bras. 2014, 34, 337–343. [Google Scholar] [CrossRef]
- Cambule, A.H.; Rossiter, D.G.; Stoorvogel, J.J. A methodology for digital soil mapping in poorly-accessible areas. Geoderma 2013, 192, 341–353. [Google Scholar] [CrossRef]
- Minasny, B.; McBratney, A.B. A conditioned Latin hypercube method for sampling in the presence of ancillary information. Comput. Geosci. 2006, 32, 1378–1388. [Google Scholar] [CrossRef]
- Roudier, P.; Hewitt, A.E.; Beaudette, D.E. A conditioned latin hypercube sampling algorithm incorporating operational constraints. In Digital Soil Assessments and Beyond; Minasny, B., Malone, B.P., McBratney, A.B., Eds.; CRC Press/Balkema: London, UK, 2012; pp. 227–232. [Google Scholar]
- Carvalho Júnior, W.; Chagas, C.S.; Muselli, A.; Pinheiro, H.S.K.; Pereira, N.R.; Bhering, S.B. Método do hipercubo latino condicionado para a amostragem de solos na presença de covariáveis ambientais visando o mapeamento digital de solos. Rev. Bras. Cienc. Solo 2014, 38, 386–396. [Google Scholar] [CrossRef]
- Costa, E.M.; Tassinari, W.S.; Pinheiro, H.S.K.; Beutler, S.J.; Anjos, L.H.C. Mapping Soil Organic Carbon and Organic Matter Fractions by Geographically Weighted Regression. J. Environ. Qual. 2018, 47, 718–725. [Google Scholar] [CrossRef]
- Fernández-Manso, A.; Fernández-Manso, O.; Quitano, C. SENTINEL-2A red-edge spectral indices suitability for discriminating burn severity. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 170–175. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, E.J. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Gitelson, A.A. Remote estimation of crop and grass chlorophyll and nitrogen content using red-edge bands on Sentinel-2 and 3. Int. J. Appl. Earth Obs. Geoinf. 2012, 23, 344–351. [Google Scholar] [CrossRef]
- Holdridge, L.R. Life Zone Ecology; Tropical Science Center: San Jose, CA, USA, 1967; p. 206. [Google Scholar]
- Arruda, D.M.; Fernandes-Filho, E.I.; Solar, R.R.C.; Schaefer, C.E.G.R. Combining climatic and soil properties better predicts covers of Brazilian biomes. Sci. Nat. 2017, 104, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wang, P.; Masek, J. Integrating remote sensing data from multiple optical sensors for ecological and crop condition monitoring. In Proceedings of the SPIE—The International Society for Optical Engineering, San Diego, CA, USA, 24 September 2013. [Google Scholar] [CrossRef]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Seitz, N.; White, J.C.; Gao, F.; Masek, J.G.; Stenhouse, G. Generation of dense time series synthetic Landsat data through data blending with MODIS using a spatial and temporal adaptive reflectance fusion model. Remote Sens. Environ. 2009, 113, 1988–1999. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Gao, F.; Chen, X.; Masek, J.G. An enhanced spatial and temporal adaptive reflectance fusion model for complex heterogeneous regions. Remote Sens. Environ. 2010, 114, 2610–2623. [Google Scholar] [CrossRef]
- Rocha, G.C. O meio físico da região de Ibitipoca: Características e Fragilidade. In Flora do Parque Estadual do Ibitipoca e seu Entorno, 1st ed.; Forzza, R.C., Menini Neto, L., Salimena, F.R.G., Zappi, D., Eds.; Editora UFJF: Juiz de Fora, Brazil, 2013; pp. 27–52. ISBN 978857672187-2. [Google Scholar]
- Rodela, L.G.; Tarifa, J.R. O clima da serra do Ibitipoca, sudeste de Minas Gerais. GEOUSP Espaço Tempo 2002, 101–113. [Google Scholar] [CrossRef]
- System for Automated Geoscientific Analyses. SAGA. Version: 2.1.4. Copyrights (c) 2002–2014 by Olaf Conrad. GNU General Public License Version 2.0. 1999. Available online: http://www.saga-gis.org (accessed on 2 April 2013).
- Grass Development Team. Geographic Resources Analysis Support System (GRASS). Copyright, 1999–2013. GRASS Development Team, and Licensed under Terms of the GNU General Public License—GPL. Available online: http://grass.osgeo.org/home/copyright (accessed on 28 April 2013).
- Jasiewicz, J.; Stepinski, T.F. Geomorphons: A pattern recognition approach to classification and mapping of landforms. Geomorphology 2013, 182, 147–156. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources; World Soil Resources Reports, No. 106; FAO: Rome, Italy, 2014; p. 191. ISBN 978-92-5-108369-7. [Google Scholar]
- Dias, H.C.T.; Fernandes Filho, E.I.; Schaefer, C.E.G.R.; Fontes, L.E.F.; Ventorim, L.B. Geoambientes do parque estadual do ibitipoca, município de Lima Duarte-MG. Rev. Árvore 2002, 26, 777–786. [Google Scholar] [CrossRef]
- Pinto, C.P. Lima Duarte: Folha SF.23-X-C-VI: Estado de Minas Gerais; Programa Levantamentos Geológicos Básicos do Brasil—PLGB; DNPM: Rio de Janeiro, Brizal, 1991; p. 201. [Google Scholar]
- USGS. United States Geological Survey Website. Sentinel Data and Specifications. Available online: https://www.usgs.gov/centers/eros/science/usgs-eros-archive-sentinel-2?qt-science_center_objects=0#qt-science_center_objects (accessed on 25 June 2019).
- European Space Agency—ESA. User Guides. Sentinel Online. Available online: https://sentinel.esa.int/web/sentinel/user-guides (accessed on 11 July 2019).
- Antunes, M.A.H.; Bolpato, I.F. Atmospheric Correction and Cirrus Clouds Removal from MSI Sentinel 2A Images. In Proceedings of the IGARSS IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; pp. 9145–9148. [Google Scholar] [CrossRef]
- Vermonte, E.F.; Tanre, D.; Deuze, J.L.; Herman, M.; Mocrette, J.J. Second simulation of the satellite signal in the solar spectrum, 6S: An overview. IEEE Trans. Geosci. Remote Sens. 1997, 35, 675–686. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Shell, J.A.; Deering, D.W.; Harlan, J.C. Monitoring the Vernal Advancement of Retrogradation of Natural Vegetation; NASA/GSFC: Greenbelt, MD, USA, 1973; p. 37. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/isbn 3-900051-07-0 (accessed on 5 August 2015).
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Breiman, L. Random Forests. Technical Report for Version 3. 2001. Available online: http://oz.berkeley.edu/users/breiman/randomforest2001.pdf (accessed on 28 December 2014).
- Grimm, R.; Behrens, T.; Märker, M.; Elsenbeer, H. Soil organic carbon concentrations and stocks on Barro Colorado Island—Digital soil mapping using Random Forests analysis. Geoderma 2008, 146, 102–113. [Google Scholar] [CrossRef]
- Young, P.; Parkinson, S.; Lees, M. Simplicity out of complexity in environmental modelling: Occam’s razor revisited. J. Appl. Stat. 1996, 23, 165–210. [Google Scholar] [CrossRef]
- Bonate, P.L. Effect of correlation on covariate selection in linear and nonlinear mixed effect models. Pharm. Stat. 2017, 16, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Monserud, R.A.; Leemans, R. Comparing global vegetation maps with the Kappa statistic. Ecol. Model. 1992, 62, 275–293. [Google Scholar] [CrossRef]
- Chagas, C.S.; Carvalho Júnior, W.; Bhering, S.B. Integration of quickbird data and terrain attributes for digital soil mapping by artificial neural networks. Rev. Bras. Cienc. Solo 2011, 35, 693–704. [Google Scholar] [CrossRef]
- Chagas, C.S.; Vieira, C.A.O.; Fernandes Filho, E.I. Comparison between artificial neural networks and maximum likelihood classification in digital soil mapping. Rev. Bras. Cienc. Solo 2013, 37, 339–351. [Google Scholar] [CrossRef]
- Pinheiro, H.S.K.; Owens, P.R.; Anjos, L.H.C.; Carvalho Júnior, W.; Chagas, C.S. Tree-based techniques to predict soil units. Soil Res. 2017, 55, 788–798. [Google Scholar] [CrossRef]
- Pinheiro, H.S.K.; Owens, P.R.; Chagas, C.S.; Carvalho Júnior, W.; Anjos, L.H.C. Applying Artificial Neural Networks Utilizing Geomorphons to Predict Soil Classes in a Brazilian Watershed. In Digital Soil Mapping across Paradigms, Scales and Boundaries; Zhang, G.L., Brus, D., Liu, F., Song, X.D., Lagacherie, P., Eds.; Springer Environmental Science and Engineering: Singapore, 2016; pp. 89–102. ISBN 978-981-10-0415-5. [Google Scholar]
- Reece, N.; Wingard, G.; Mandakh, B.; Reading, R.P. Using random forest to classify vegetation communities in the southern area of Ikh Nart Nature Reserve in Mongolia. Mong. J. Biol. Sci. 2019, 17, 31–39. [Google Scholar] [CrossRef]
- Ayala-Izurieta, J.E.; Márquez, C.O.; García, V.J.; Recalde-Moreno, C.G.; Rodríguez-Llerena, M.V.; Damián-Carrión, D.A. Land Cover Classification in an Ecuadorian Mountain Geosystem Using a Random Forest Classifier, Spectral Vegetation Indices, and Ancillary Geographic Data. Geosciences 2017, 7, 34. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First experience with Sentinel-2 data for crop and tree species classifications in central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Ramoelo, A.; Cho, M.; Mathieu, R.; Skidmore, A.K. Potential of Sentinel-2 spectral configuration to assess rangeland quality. J. Appl. Remote Sens. 2015, 9, 094096. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Odindi, J.; Abdel-Rahman, E.M. Land-use/cover classification in a heterogeneous coastal landscape using Rapid Eye imagery: Evaluating the performance of random forest and support vector machines classifiers. Int. J. Remote Sens. 2014, 35, 3440–3458. [Google Scholar] [CrossRef]
- Trisasongko, B.H.; Panuju, D.R.; Paull, D.J.; Jia, X.; Griffin, A.L. Comparing six pixel-wise classifiers for tropical rural land cover mapping using four forms of fully polarimetric SAR data. Int. J. Remote Sens. 2017, 38, 3274–3293. [Google Scholar] [CrossRef]
- De Luca, G.; Silva, J.M.N.; Cerasoli, S.; Araújo, J.; Campos, J.; Di Fazio, S.; Modica, G. Object-Based Land Cover Classification of Cork Oak Woodlands using UAV Imagery and Orfeo ToolBox. Remote Sens. 2019, 11, 1238. [Google Scholar] [CrossRef]
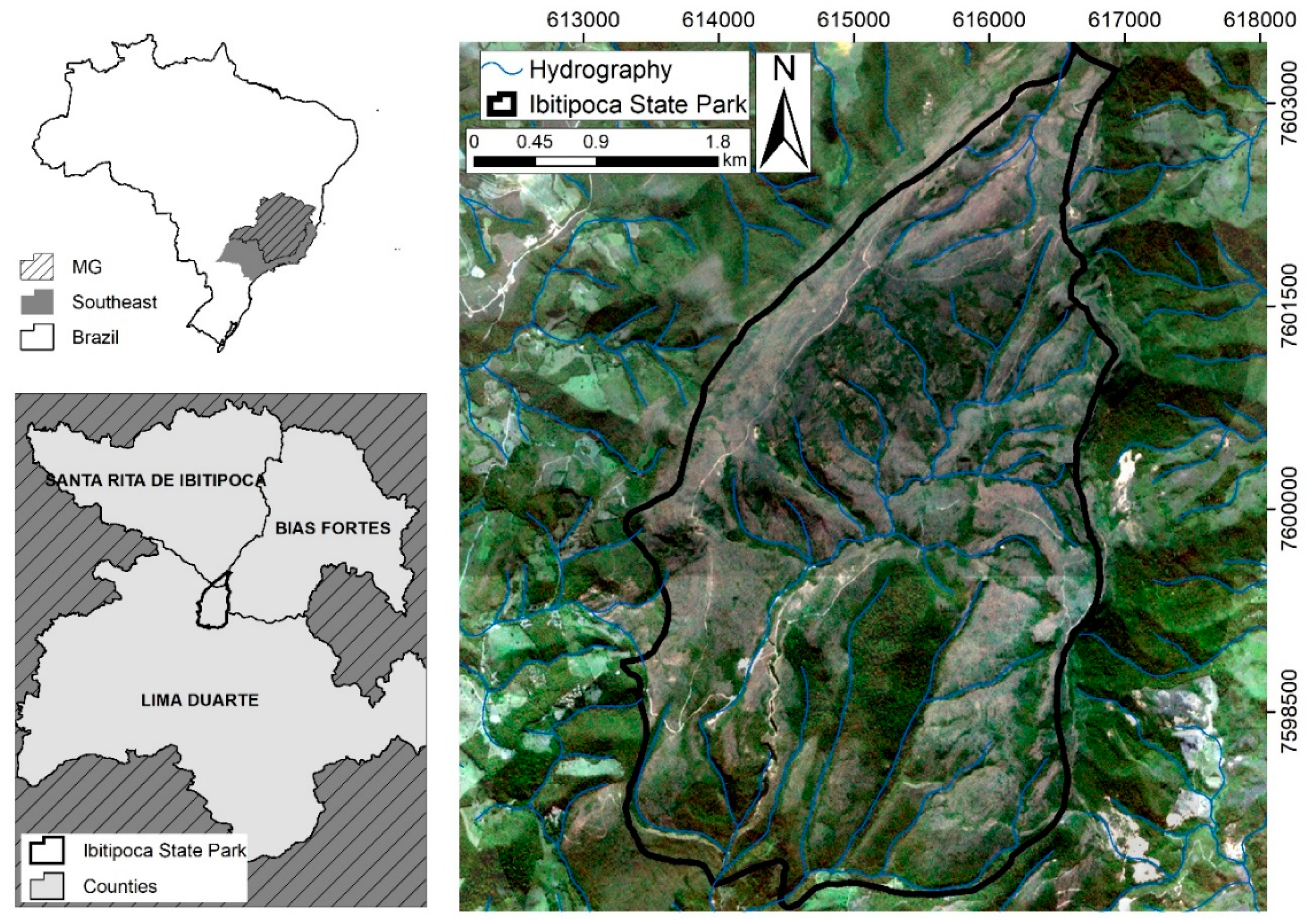

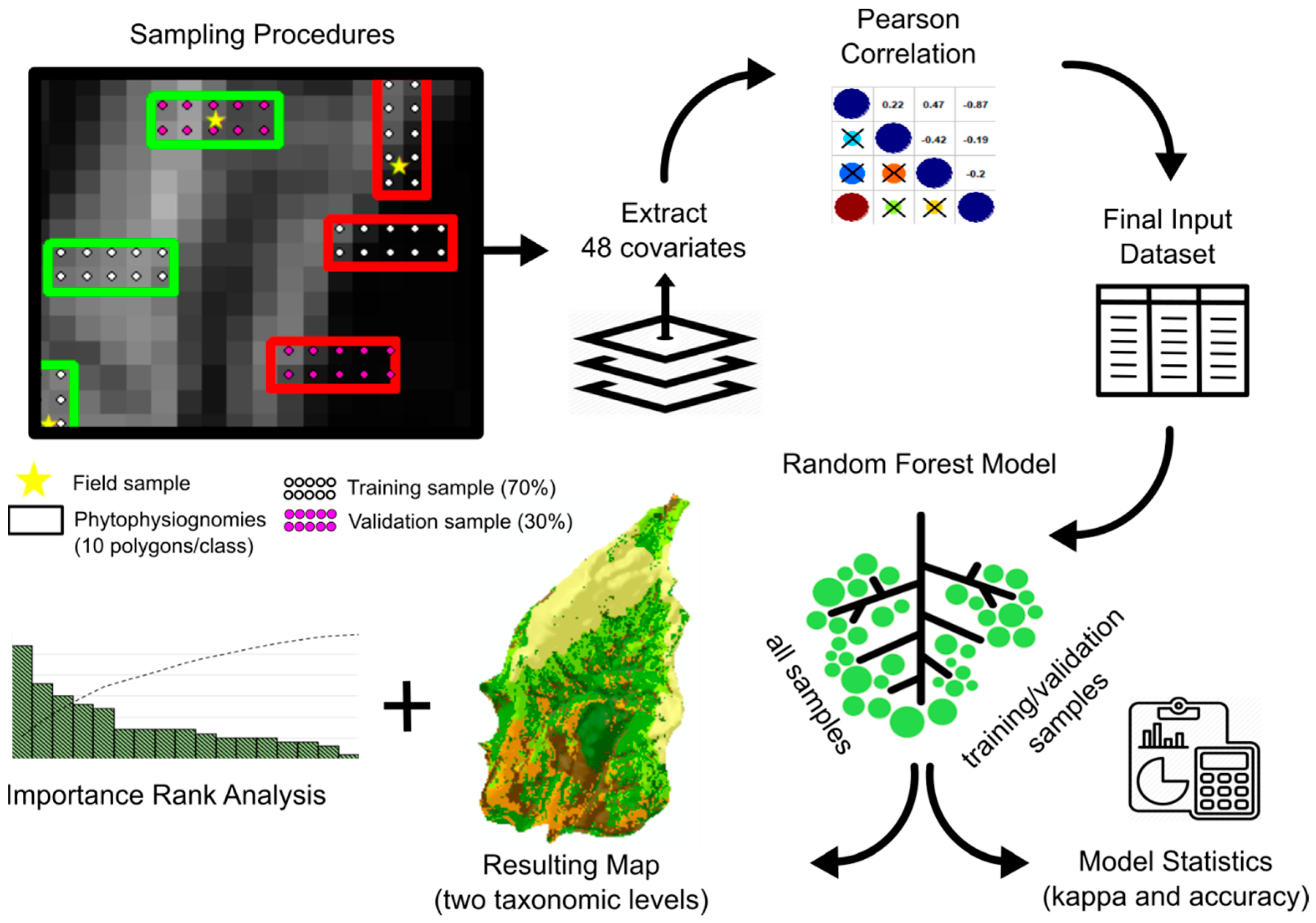
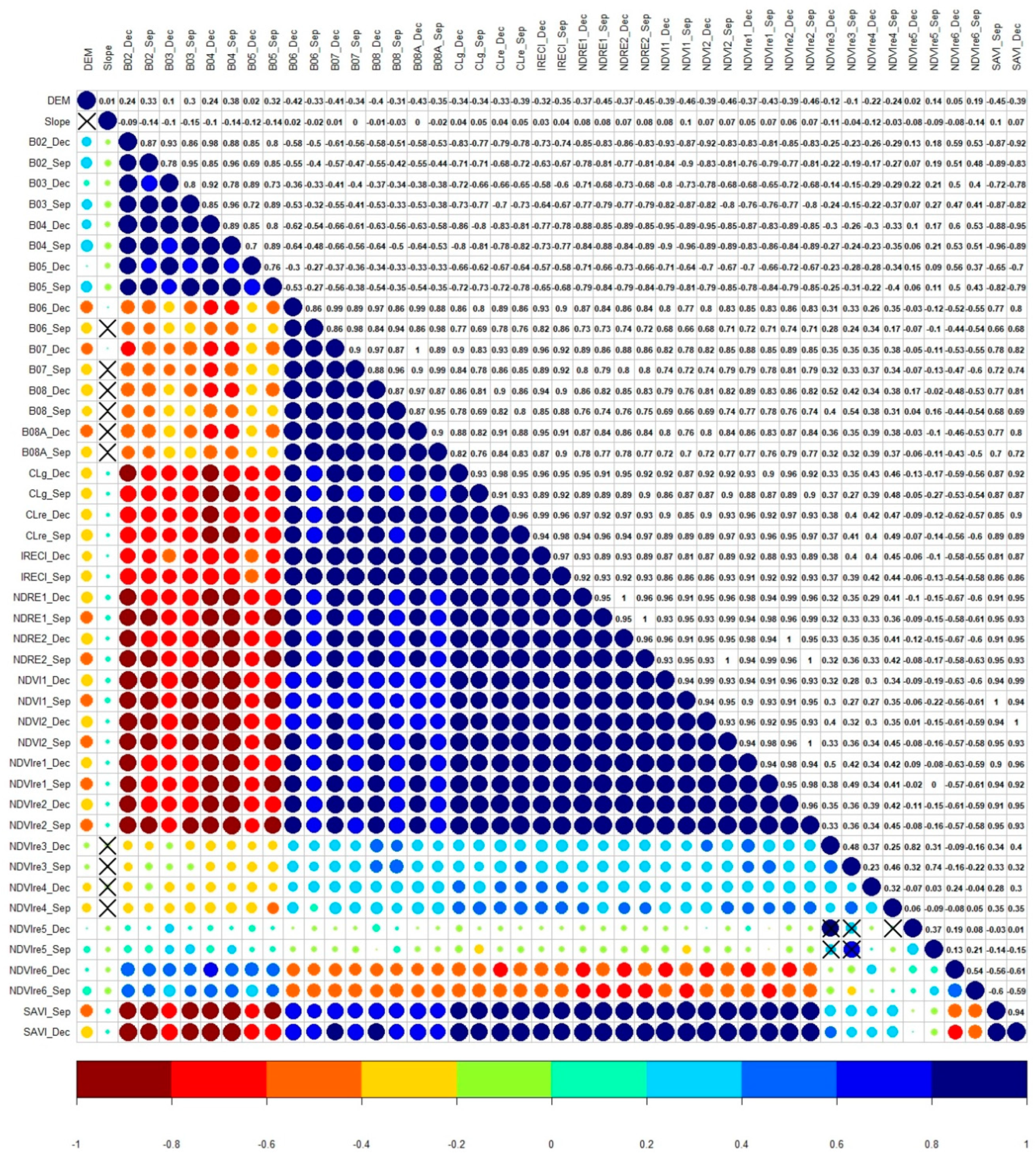



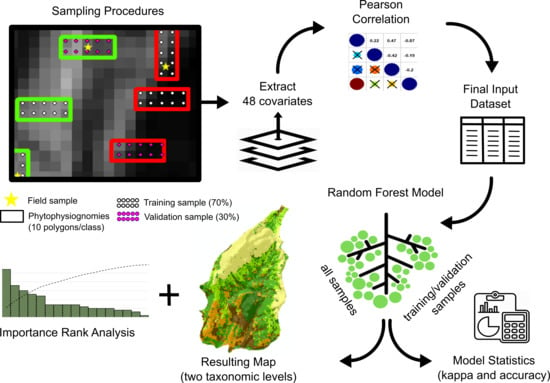
| Physiognomies (1st) | Description of General Taxonomic Level | Attribute (5th) * | N ** |
|---|---|---|---|
| Broadleaved forest (FL) | The trees usually have broad leaves and make up a canopy between 5 and 30 m in height, although the emergent trees may be sparsely populated. Climbers and epiphytes are very frequent in this forest formation. | Broadleaved forest over deep soils (FLd) | 1 |
| Broadleaved forest over shallow soils (FLs) | 1 | ||
| Broadleaved dwarf-forest (NL)t | Nearly all trees are broadleaved and form a low canopy, between 3 and 5 m in height. Scattered taller trees may emerge from the canopy. Climbers, epiphytes, and shrubs may be relevant. | Broadleaved dwarf-forest over deep soils (NLd) | 11 |
| Parkland savanna (SL) | The woody biomass is predominant, but the trees do not form a continuous canopy. The shrubs are abundant, and the bush forms an almost continuous vegetation cover. | Parkland savanna over shallow soils (SLs) | 3 |
| Shrubland savanna (SA) | The woody component is mainly composed of shrubs, while the trees are very rare. The shrub component is also significant and forms an almost continuous vegetation cover. | Shrubland savanna over shallow soils (SAs) | 5 |
| Shrubland savanna and rocky outcrop (SAr) | 2 | ||
| Broadleaved scrub (AL) | Subshrubs and broadleaved shrubs form a nearly uniform vegetable mass, but without herbaceous plants coating the soil. There may be an expressive biomass of climbers and epiphytes. | Broadleaved scrub over shallow soils (ALs) | 3 |
| Bushy grassland (CL) | Broadleaved subshrubs, short-lived or perennial herbs make up discontinuous vegetable formation. Scattered shrubs and isolated trees may also occur. | Bushy grassland over shallow soils (CLs) | 10 |
| Vegetation Index | Sentinel-2 MSI Bands | Reference |
|---|---|---|
| Normalized difference vegetation index (NDVI1) | ||
| Normalized difference vegetation index (NDVI2) | [27,46] | |
| Soil-adjusted vegetation index (SAVI) | [45,47] | |
| Inverted Red-Edge Chlorophyll Index (IRECI) | [46] | |
| Chlorophyll index ( | [26,28] | |
| Green chlorophyll index | [28] | |
| Normalized difference vegetation index red-edge (NDVIre1) | [26] | |
| Normalized difference vegetation index red-edge (NDVIre2) | [26] | |
| Normalized difference vegetation index red-edge (NDVIre3) | [26] | |
| Normalized difference vegetation index red-edge (NDVIre4) | [26] | |
| Normalized difference vegetation index red-edge (NDVIre5) | [26] | |
| Normalized difference vegetation index red-edge (NDVIre6) | [26] | |
| Normalized Difference Red-edge (NDRE1) | [26,28] | |
| Normalized Difference Red-edge (NDRE2) | [26,28] |
| Training Dataset (n = 560) | Validation Dataset (n = 240) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Max. | Min. | Median | Avg. | SD | Max. | Min. | Median | Avg. | SD | |
| DEM | 1761 | 1300 | 1440 | 1468 | 38.31 | 1696 | 1352 | 1455 | 1471 | 38.35 |
| Slope | 70.559 | 2.418 | 18.737 | 22.37 | 4.73 | 50.928 | 1.953 | 27.971 | 27.25 | 5.22 |
| B2_Dec | 0.0759 | 0.0091 | 0.0284 | 0.0291 | 0.1706 | 0.0849 | 0.0086 | 0.0269 | 0.0844 | 0.2905 |
| B3_Sep | 0.1072 | 0.0083 | 0.0339 | 0.0357 | 0.1889 | 0.0915 | 0.0090 | 0.0295 | 0.0332 | 0.1823 |
| B3_Dec | 0.1093 | 0.0147 | 0.0493 | 0.0500 | 0.2237 | 0.1188 | 0.0301 | 0.0470 | 0.0494 | 0.2222 |
| B5_Sep | 0.1612 | 0.0404 | 0.0788 | 0.0787 | 0.2805 | 0.1302 | 0.0414 | 0.0720 | 0.0751 | 0.2740 |
| B5_Dec | 0.1457 | 0.0403 | 0.0894 | 0.0898 | 0.2997 | 0.1409 | 0.0648 | 0.0856 | 0.0880 | 0.2967 |
| B6_Sep | 0.2823 | 0.1093 | 0.1629 | 0.1693 | 0.4115 | 0.2621 | 0.1030 | 0.1617 | 0.1647 | 0.4058 |
| B6_Dec | 0.3762 | 0.0714 | 0.1925 | 0.2007 | 0.4480 | 0.3160 | 0.1325 | 0.1976 | 0.2049 | 0.4527 |
| B8_Sep | 0.4298 | 0.1102 | 0.2022 | 0.2135 | 0.4621 | 0.3426 | 0.1093 | 0.2032 | 0.2096 | 0.4578 |
| CLg_Sep | 29.1347 | 0.4948 | 4.2525 | 5.7762 | 2.4034 | 24.3490 | 1.2140 | 4.9070 | 6.3850 | 2.5269 |
| NDVIre1_Sep | 0.7524 | 0.0914 | 0.4253 | 0.4487 | 0.6698 | 0.7395 | 0.2192 | 0.4441 | 0.4653 | 0.6821 |
| NDVIre3_Sep | 0.2597 | –0.0476 | 0.1119 | 0.1119 | 0.3345 | 0.2352 | 0.0158 | 0.1115 | 0.1169 | 0.3419 |
| NDVIre4_Sep | 0.1918 | 0.0768 | 0.1242 | 0.1263 | 0.3553 | 0.2033 | 0.0196 | 0.1279 | 0.1285 | 0.3585 |
| NDVIre5_Sep | 0.2243 | –0.1227 | 0.0467 | 0.0450 | 0.2120 | 0.1532 | –0.1136 | 0.4996 | 0.0474 | 0.2177 |
| NDVIre6_Sep | 0.1102 | 0.0134 | 0.0581 | 0.0594 | 0.2438 | 0.1029 | 0.0127 | 0.0587 | 0.0591 | 0.2431 |
| NDVIre3_Dec | 0.2405 | –0.1681 | 0.1051 | 0.1065 | 0.3264 | 0.2111 | –0.0092 | 0.1043 | 0.1098 | 0.3314 |
| NDVIre4_Dec | 0.1878 | 0.0891 | 0.1377 | 0.1398 | 0.3739 | 0.1872 | 0.0946 | 0.1372 | 0.1393 | 0.3732 |
| NDVIre5_Dec | 0.1299 | –0.2267 | 0.0299 | 0.0247 | 0.1570 | 0.1214 | –0.0943 | 0.0269 | 0.0256 | 0.1599 |
| NDVIre6_Dec | 0.1069 | 0.0235 | 0.0574 | 0.0583 | 0.2414 | 0.0331 | 0.0331 | 0.0540 | 0.0554 | 0.2354 |
| SAVI_Dec | 2.4500 | 1.7890 | 2.1450 | 2.1710 | 1.4734 | 2.4580 | 1.8650 | 2.1960 | 2.1950 | 1.4816 |
| First Categorical Level—Phytophysiognomies (OA: 76.7%; Kappa: 0.72) | |||||||||
| AL | CL | FL | NL | SA | SL | UA (%) | |||
| Broadleaved scrub (AL) | 26 | 0 | 3 | 2 | 7 | 0 | 68.4 | ||
| Bushy grassland (CL) | 0 | 26 | 0 | 9 | 0 | 0 | 74.3 | ||
| Broadleaved forest (FL) | 0 | 26 | 1 | 0 | 4 | 83.9 | |||
| Broadleaved dwarf-forest (NL) | 0 | 4 | 1 | 15 | 0 | 1 | 71.4 | ||
| Shrubland savanna (SA) | 3 | 0 | 0 | 0 | 23 | 3 | 79.3 | ||
| Parkland savanna (SL) | 1 | 0 | 0 | 3 | 0 | 22 | 84.6 | ||
| Producer’s accuracy—PA (%) | 86.7 | 86.7 | 86.7 | 50.0 | 76.7 | 73.3 | |||
| Fifth Categorical Level—Phytophysiognomies + Soil Depth (OA: 61.7%; Kappa: 0.56) | |||||||||
| ALs | CLs | FLd | FLs | NLd | SAr | SAs | SLs | UA (%) | |
| Broadleaved scrub shallow (ALs) | 24 | 0 | 0 | 4 | 3 | 0 | 1 | 0 | 75.0 |
| Bushy grassland shallow (CLs) | 0 | 17 | 0 | 0 | 10 | 0 | 0 | 0 | 63.0 |
| Broadleaved forest deep FLd) | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | 100.0 |
| Broadleaved forest shallow (FLs) | 1 | 0 | 0 | 10 | 0 | 0 | 4 | 6 | 47.6 |
| Broadleaved dwarf-forest deep (NLd) | 0 | 10 | 0 | 0 | 4 | 9 | 1 | 0 | 16.7 |
| Shrubland savanna rocky (SAr) | 3 | 0 | 0 | 0 | 0 | 21 | 0 | 2 | 80.8 |
| Shrubland savanna shallow (SAs) | 0 | 3 | 0 | 0 | 10 | 0 | 20 | 0 | 60.6 |
| Parkland savanna (SLs) | 2 | 0 | 0 | 16 | 3 | 0 | 4 | 22 | 46.8 |
| PA (%) | 80.0 | 56.7 | 100.0 | 33.3 | 13.3 | 70.0 | 66.7 | 73.3 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinheiro, H.S.K.; Barbosa, T.P.R.; Antunes, M.A.H.; Carvalho, D.C.d.; Nummer, A.R.; Carvalho Junior, W.d.; Chagas, C.d.S.; Fernandes-Filho, E.I.; Pereira, M.G. Assessment of Phytoecological Variability by Red-Edge Spectral Indices and Soil-Landscape Relationships. Remote Sens. 2019, 11, 2448. https://doi.org/10.3390/rs11202448
Pinheiro HSK, Barbosa TPR, Antunes MAH, Carvalho DCd, Nummer AR, Carvalho Junior Wd, Chagas CdS, Fernandes-Filho EI, Pereira MG. Assessment of Phytoecological Variability by Red-Edge Spectral Indices and Soil-Landscape Relationships. Remote Sensing. 2019; 11(20):2448. https://doi.org/10.3390/rs11202448
Chicago/Turabian StylePinheiro, Helena S. K., Theresa P. R. Barbosa, Mauro A. H. Antunes, Daniel Costa de Carvalho, Alexis R. Nummer, Waldir de Carvalho Junior, Cesar da Silva Chagas, Elpídio I. Fernandes-Filho, and Marcos Gervasio Pereira. 2019. "Assessment of Phytoecological Variability by Red-Edge Spectral Indices and Soil-Landscape Relationships" Remote Sensing 11, no. 20: 2448. https://doi.org/10.3390/rs11202448
APA StylePinheiro, H. S. K., Barbosa, T. P. R., Antunes, M. A. H., Carvalho, D. C. d., Nummer, A. R., Carvalho Junior, W. d., Chagas, C. d. S., Fernandes-Filho, E. I., & Pereira, M. G. (2019). Assessment of Phytoecological Variability by Red-Edge Spectral Indices and Soil-Landscape Relationships. Remote Sensing, 11(20), 2448. https://doi.org/10.3390/rs11202448