1. Introduction
Remote sensing hyperspectral images (HSIs) usually contain information about hundreds of spectral bands spanning from visible to infrared spectrum. Each pixel in HSIs is a high-dimensional vector whose entries correspond to the spectral reflectance in a specific wavelength, providing rich spectral information for distinguishing land covers of interest [
1]. Recently, HSI classification with the aim of identifying the land-cover type of each pixel has become one of the most active research fields in the remote sensing community, because it is an essential step in a wide variety of earth monitoring applications, such as environmental monitoring [
2] and precision agriculture [
3].
The spectral and the spatial information of HSIs are two major characteristics that can be exploited for classification [
4]. Traditional classification methods such as random forest [
5], support vector machine (SVM) [
6] and multinomial logistic regression [
7], mainly focus on making use of the abundant spectral information for classification. To improve classification performance, methods such as morphological profiles [
8], multiple kernel learning [
9], superpixel [
10] and sparse representation [
11] have been introduced to combine the spatial information with the spectral information for HSI classification [
12,
13]. For instance, Benediktsson et al. utilized extended morphological profiles (EMPs) to obtain spectral-spatial features of HSIs [
8]. Fang et al. proposed a multiscale adaptive sparse representation (MASR) model to exploit the multiscale spatial information of HSIs [
11]. Fauvel et al. proposed a morphological kernel based SVM classifier to jointly use the spatial and the spectral information for classification [
14]. Nevertheless, the common limitation of these methods is that they heavily rely on hand-crafted features, which require experts’ experiences and massive efforts in feature engineering, limiting their applicability in difficult scenarios.
Recently, deep learning-based methods have made great breakthroughs in many computer vision tasks, for example, image classification [
15,
16], semantic segmentation [
17], natural language processing [
18] and object detection [
19], for they can automatically extract robust and discriminative features from original data in a hierarchical way. Deep learning models have also been introduced for HSI classification and have achieved a remarkable progress [
20,
21]. In Reference [
22], a stacked auto-encoder (SAE) was first proposed for HSI spectral classification. Next, deep learning models, including deep belief network (DBN) [
23] and convolutional neural network (CNN) [
24,
25,
26], were introduced as deep spectral classifiers for HSI classification. To make use of both the spectral and spatial information of HSIs, a series of improved CNN-based spectral–spatial classifiers were then proposed [
27,
28,
29]. Zhao et al. proposed a spectral-spatial feature-based classification (SSFC) framework in which the CNN was used to extract spatial features and the balanced local discriminant embedding method to extract spectral features [
30]. To simultaneously extract the spectral-spatial features of HSIs, 3-D CNNs were proposed for HSI classification [
31,
32]. Due to the joint utilization of the spectral and spatial information of HSIs, spectral-spatial classifiers usually achieve better classification performance than spectral classifiers. To extract deeper discriminative spectral-spatial features, residual learning [
33], which helps to train CNNs up to thousands of layers without suffering gradient vanishing, was introduced for HSI classification [
34,
35,
36,
37,
38,
39]. For instance, a fully convolutional neural network was proposed for HSI classification in which multiscale filter bank was used to exploit both spectral and spatial information embedded in HSIs and residual learning to enhance the learning efficiency of the network [
34]. Song et al. proposed a deep feature fusion network in which multiple-layer features extracted from a deep CNN were fused for classification and residual learning was utilized to alleviate gradient vanishing problem [
35].
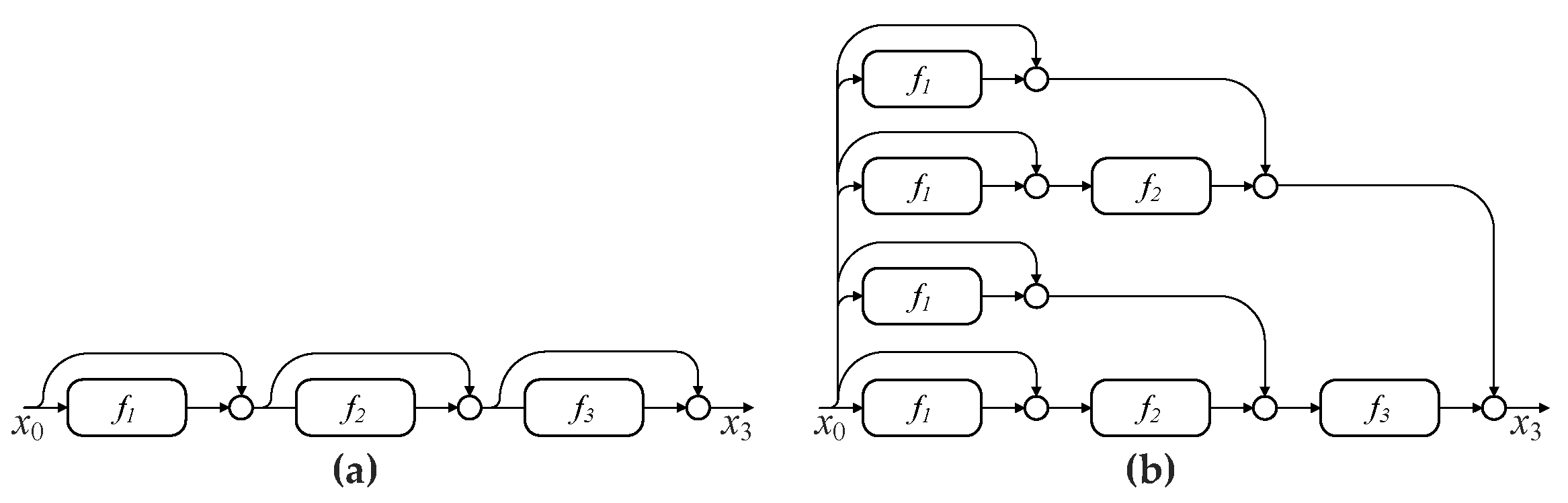
Although residual learning helps to extremely increase the network depth, recent studies pointed out that deep ResNets actually behave like a large ensemble of much shallower networks, instead of a ultra deep network [
40,
41]. By rewriting ResNets as an explicit collection of paths of different length, Veit et al. revealed that although these paths are trained together, they exhibit ensemble-like behavior, that is, different paths are not strongly dependent on each other [
40]. For example, the removal of a layer during the testing phase has a modest impact on the performance of a ResNet. Furthermore, deep paths do not contribute any gradient during training. For instance, most of the gradient in a 110-layer ResNet comes from paths between 10 to 34 layers deep, demonstrating that the effective paths are relatively shallow. Furthermore, a ResNet trained only on some effective paths can achieve a comparable performance to that of a full ResNet [
40].
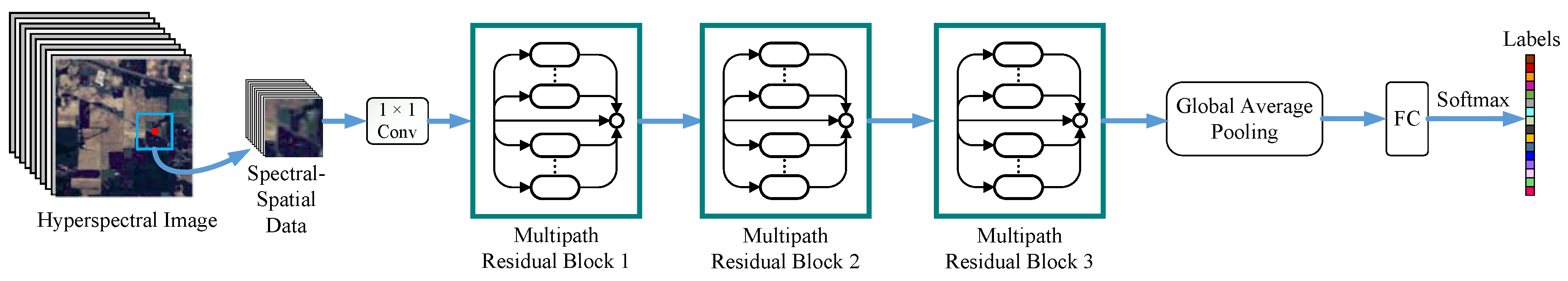
In this paper, inspired by the above observations, a novel multipath ResNet (MPRN) model that employs multiple residual functions in each residual block is proposed for HSI classification, utilizing both spectral and spatial information. Different from the previous networks used in HSI classification, the proposed network is wider and consists of shorter-medium paths for efficient gradient flow. The proposed network is more efficient than conventional ResNet, since deep paths, which do not contribute any gradient during training, are abandoned. The main contributions of this paper can be summarized as follows: (1) The increase of the number of residual functions in each residual block can enhance the performance of ResNet and can lead to a better performance than the increase of the network depth; (2) To the best of our knowledge, the idea of balancing network width and depth for accurate and efficient HSI classification is proposed for the first time in this paper; and (3) Experimental results on three real hyperspectral data sets demonstrate that the proposed method can achieve a better classification performance than several state-of-the-art approaches.
The remainder of this paper is organized as follows.
Section 2 introduces the general framework of CNN-based HSI classification and reviews the ResNet briefly. In
Section 3, the details of the proposed method are described. Experimental results conducted on three real hyperspectral data sets are then presented and discussed in
Section 4. Finally, some conclusions and suggestions are provided in
Section 5.
4. Experiments
4.1. Hyperspectral Data Sets
To demonstrate the effectiveness of our proposed method, now we consider three real hyperspectral data sets including Indian Pines, Houston University and Kennedy Space Center (KSC) data sets. These data sets are openly accessible online [
51,
52]. The number of samples per class of the three data sets are summarized in
Table 2.
The Indian Pines data set was collected by the Airborne Visible/Infrared Imaging Spectrometer (AVIRIS) sensor over the agricultural Indian Pines test area with a spatial resolution of 20 m. This HSI consists of 145 × 145 pixels with 224 spectral bands ranging from 400 to 2500 nm. After removing 20 water absorption bands and four null bands, 200 channels were used for the classification. Its ground reference map covered 16 classes of interest.
The Houston University data set was captured by the Compact Airborne Spectrographic Imager (CASI) sensor over the Houston University campus and its neighboring region with a spatial resolution of 2.5 m. It was used in the 2013 GRSS Data Fusion Contest. The image consists of 349 × 1905 pixels with 144 spectral bands ranging from 380 to 1050 nm. The ground reference map of this data set includes 15 classes of interest.
The KSC data set was captured by the AVIRIS sensor over KSC, Florida. This HSI is composed of 512 × 614 pixels with a spatial resolution of 18 m. After removing noisy bands, 176 spectral bands were used for the classification. Its ground reference map covered 13 classes of interest.
4.2. Experimental Setup
For each data set, the labeled samples were split into training, validation and testing sets. The training set was used to tune the model parameters. The validation set was utilized to evaluate the interim trained models created during training and the model with the highest validation accuracy was preserved. The testing set was employed to assess the classification performance of the saved model. For the Indian Pines and Houston University data sets, 10%, 10% and 80% of the labeled data per class were randomly selected to form the training, validation and testing sets, respectively. As for the KSC data set, the split ratio was 2%, 2% and 96%, respectively. Note that each data set was standardized to mean value with unit variance.
To assess the classification performance of the proposed method, the overall accuracy (OA), the average accuracy (AA), the Kappa coefficient, the F1-score and the Precision were adopted as evaluation metrics [
53]. To avoid biased estimation, the metrics obtained by averaging of five repeated experiments with randomly selected training samples were reported.
The proposed network was trained for 100 epochs with an L2 weight decay penalty of 0.0001. The batch size was set to 100 and a cosine shape learning rate was employed which starts from 0.001 and gradually reduces to 0 [
54]. In addition, our implementation was based on Pytorch framework [
55] and conducted on a PC with AMD Ryzen 7 2700X CPU, 16 GB of RAM and a NVIDIA RTX 2080 GPU.
4.3. Parameters Discussion
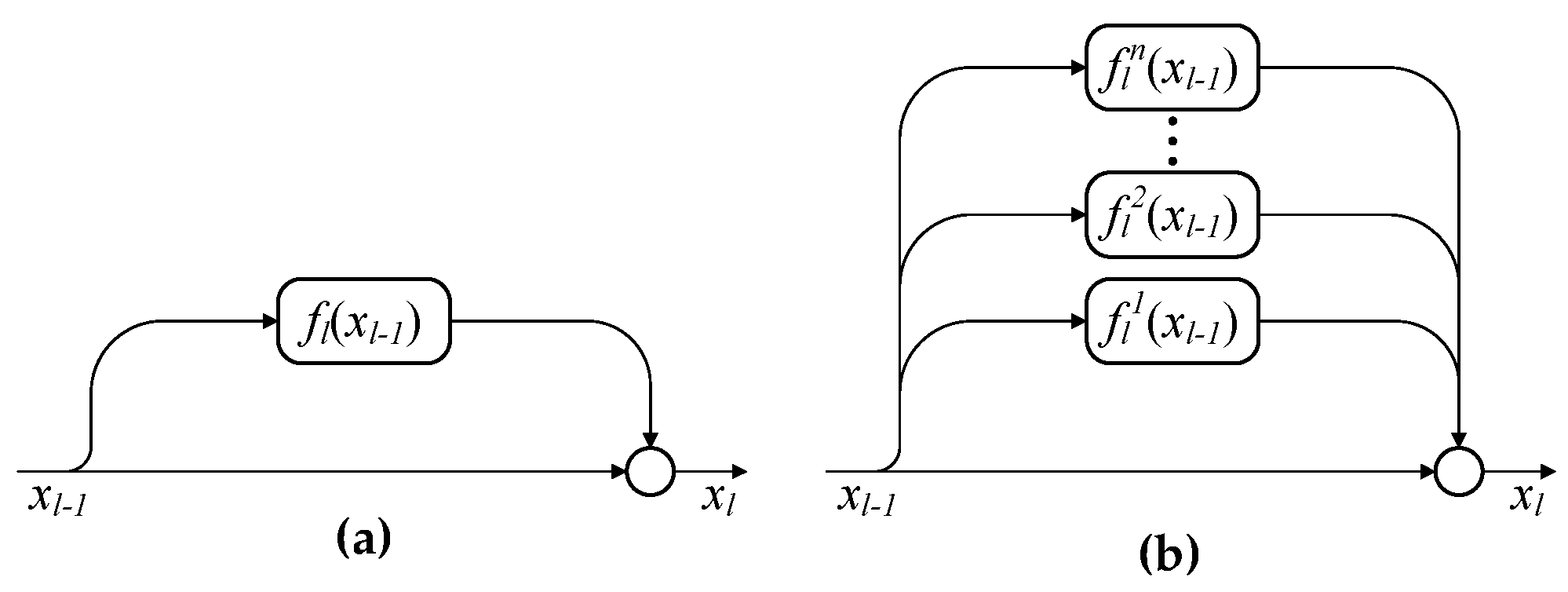
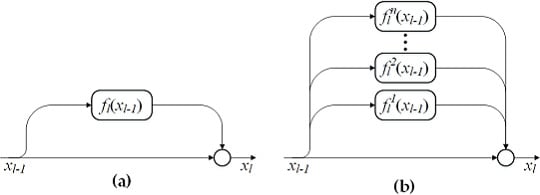
It is well known that increasing the network depth can enhance the model representation capability and lead to a better classification performance. In this section, we will show that depth is not the only factor for achieving high classification accuracy. In addition, the increase of network width is able to obtain a better performance than the increase of network depth. In the following experiments, network depth (i.e., m) is represented by the number of residual blocks and network width (i.e., n) is denoted by the number of residual functions in each block. Note that conventional ResNets have a single residual function in each block, that is, .
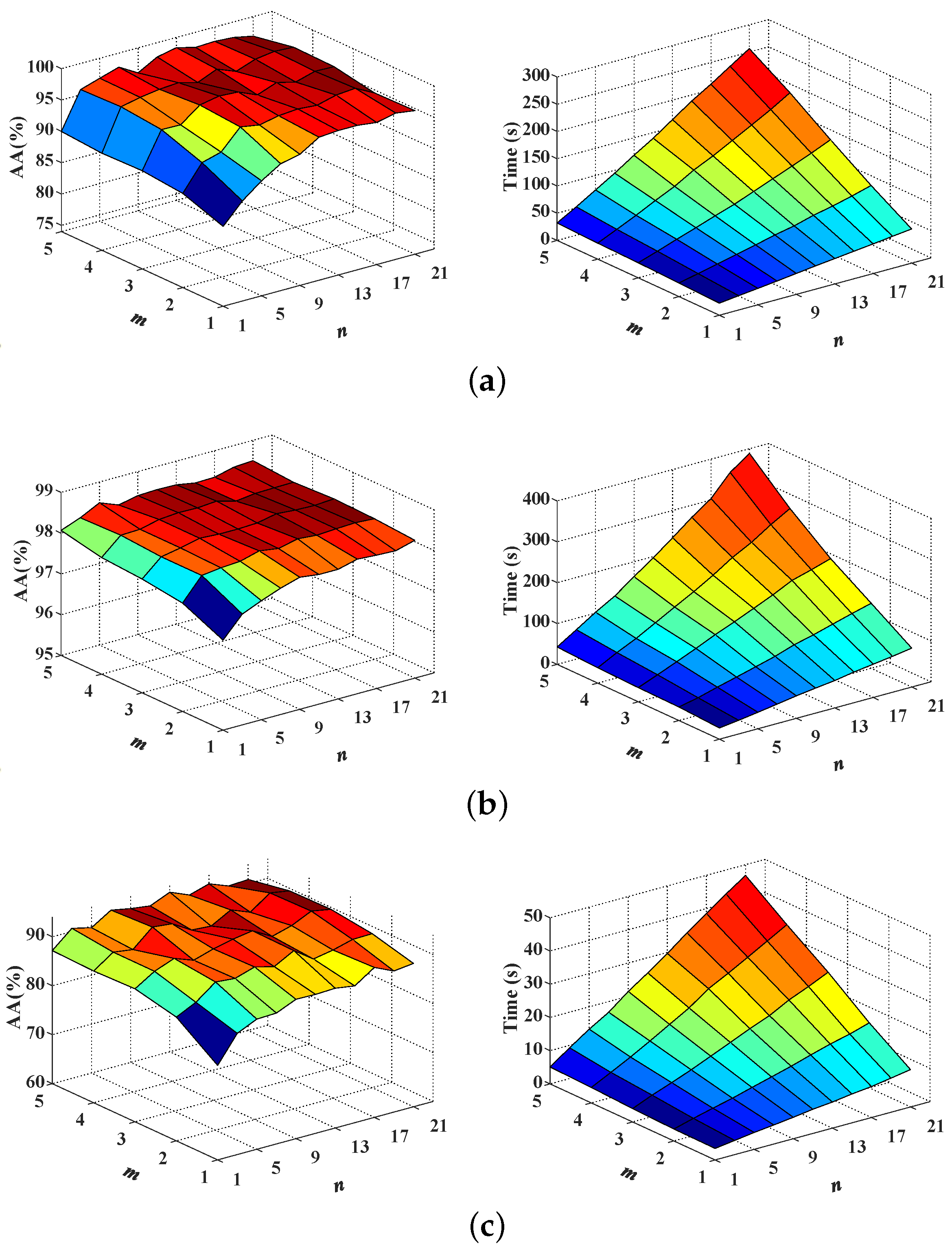
First, the network depth
m and width
n of MPRN are analyzed together. In our experiments, the
m ranges from 1 to 5 with step 1 and
n from 1 to 21 with step 2. Consider the fact that extremely shallow networks, compared to deep ones, tend to be difficult in capturing higher level features, which are beneficial for deep semantic feature extraction. However, over-deeper structure will spend great running time. Therefore, a proper network depth should be set to balance classification accuracy against timeliness. As can be observed from the left column of
Figure 5, when
m and
n are respectively larger than 3 and 7, MPRNs achieve relatively stable high accuracy for all data sets, demonstrating the robustness of MPRN to different
m and
n values. Meanwhile, with
m and
n values rise, the parameters of the corresponding models and thus the computing time will increase rapidly, as shown in the right column of
Figure 5. Therefore, to effectively leverage the overall performance, we set
m to 3 for all data sets.
To clearly show the impact of network width n on the classification performance of MPRN, we fix and show the effect of n with value ranging from 1 to 20 with step 1. In addition, we further give a contrastive evaluation of our method with ResNets () with different network depth. To make a fair comparison, for ResNet, the m ranges from 3 to 60 with step 3. In this way, each pair of the MPRN and ResNet have the same number of parameters, for example, MPRN with and has the same number of parameters as ResNet with . In addition, when and , MPRN and ResNet have the same network architecture.
Figure 6 shows the effects of network width
n on the performance (on AA) of the proposed MPRN method over the three data sets, while
Figure 7 demonstrates the impacts of network depth
m of the ResNet method. From
Figure 6 and
Figure 7, one can see that increasing any dimension of network, width or depth, will improve classification accuracy. Clearly, when the network depth goes beyond a certain level, increasing the depth become less effective. In contrast, increasing the width can further improve the classification performance.
Table 3 summarizes the optimal network architectures of MPRN and ResNet for each data set. Compared to the ResNet, our MPRN achieves better performance and with fewer parameters on the three data sets. For example, MPRN achieves 98.73% AA with 0.51 M parameters on the Indian Pines data set, while the ResNet achieves 98.60% with 1.10 M parameters. For the Houston University data set, MPRN achieves 98.36% AA with 0.39 M parameters, while the ResNet achieves 98.28% with 0.55 M parameters. For the KSC data set, MPRN achieves 92.30% AA with 0.45 M parameters, while the ResNet achieves 92.23% with 0.83 M parameters. In addition, with the increase of the model size, MPRN obtains better performance (98.48% and 93.27%) on the Houston University and KSC data sets, which further validate the effectiveness of our method. This is because paths in MPRN are relatively shallow, which have significant contribution towards the gradient updates during training. For ResNet, the increase of the depth will not only introduce more deeper paths that do not contribute significant gradient during training but also results in the overfitting phenomenon (see
Figure 7b,c). In the following experiments, the optimal architectures of ResNet and MPRN are employed for comparison (see
Table 3).
4.4. Comparison Results of Different Methods
The proposed method was compared with several state-of-the-art classification methods available in the literature: (1) 3-D CNN [
32]; (2) fully convolutional layer fusion network (FCLFN) [
56]; (3) deep feature fusion network (DFFN) [
35]; (4) DenseNet [
57]; and (5) ResNet [
46].
More specifically, 3-D CNN, FCLFN, DFFN, DenseNet, ResNet, together with the proposed MPRN, are spectral-spatial classifiers. 3-D CNN utilizes 3-D convolutional kernels to simultaneously extract the spatial and spectral features from HSIs. FCLFN combines features extracted from each Conv layer for classification. DFFN fuses multiple-layer features from a deep ResNet for classification. DenseNet employs shortcut connections between layers, in which the outputs of the previous layers are concatenated as inputs into all subsequent layers and hence can combine various spectral-spatial features across layers for HSI classification. ResNet is constructed by stacking multiple conventional residual blocks (with a single residual function in each block). In addition, some parameters of the compared methods had been set in advance. For the 3-D CNN, FCLFN, DFFN and DenseNet, the parameters were set according to the default values in the corresponding references. For ResNet and MPRN, the optimal architectures were used according to the
Table 3. In addition, they were trained under exactly the same experimental setting, for example, using the same optimizer and L2 weight decay penalty.
The first experiment was conducted on the Indian Pines data set and 10% of the labeled samples in each class were randomly selected for training. The quantitative classification results, that is, classification accuracy of each class, OA, AA, Kappa, F1-score and Precision values obtained by different approaches are reported in
Table 4. It can be seen that MPRN achieves the best results in terms of the five overall metrics, that is, OA, AA, Kappa, F1-score and Precision. From
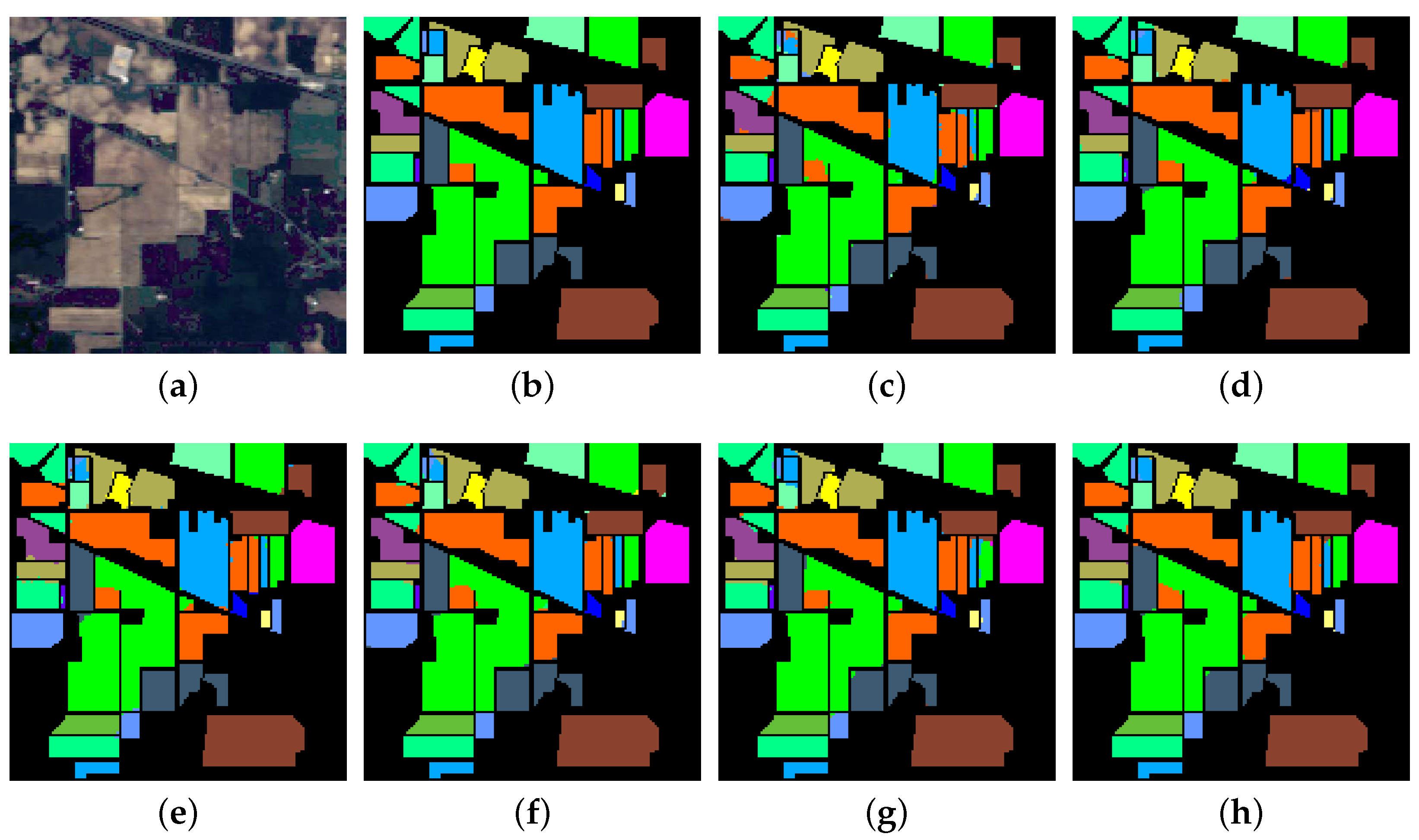
Table 4, one can observe that MPRN improves the performance of 11 classes out of 16 compared with ResNet, indicating that MPRN is more effective than ResNet. Moreover, the false-color composite image, ground reference map and the classification maps obtained by the six considered methods in a single experiment are shown in
Figure 8.
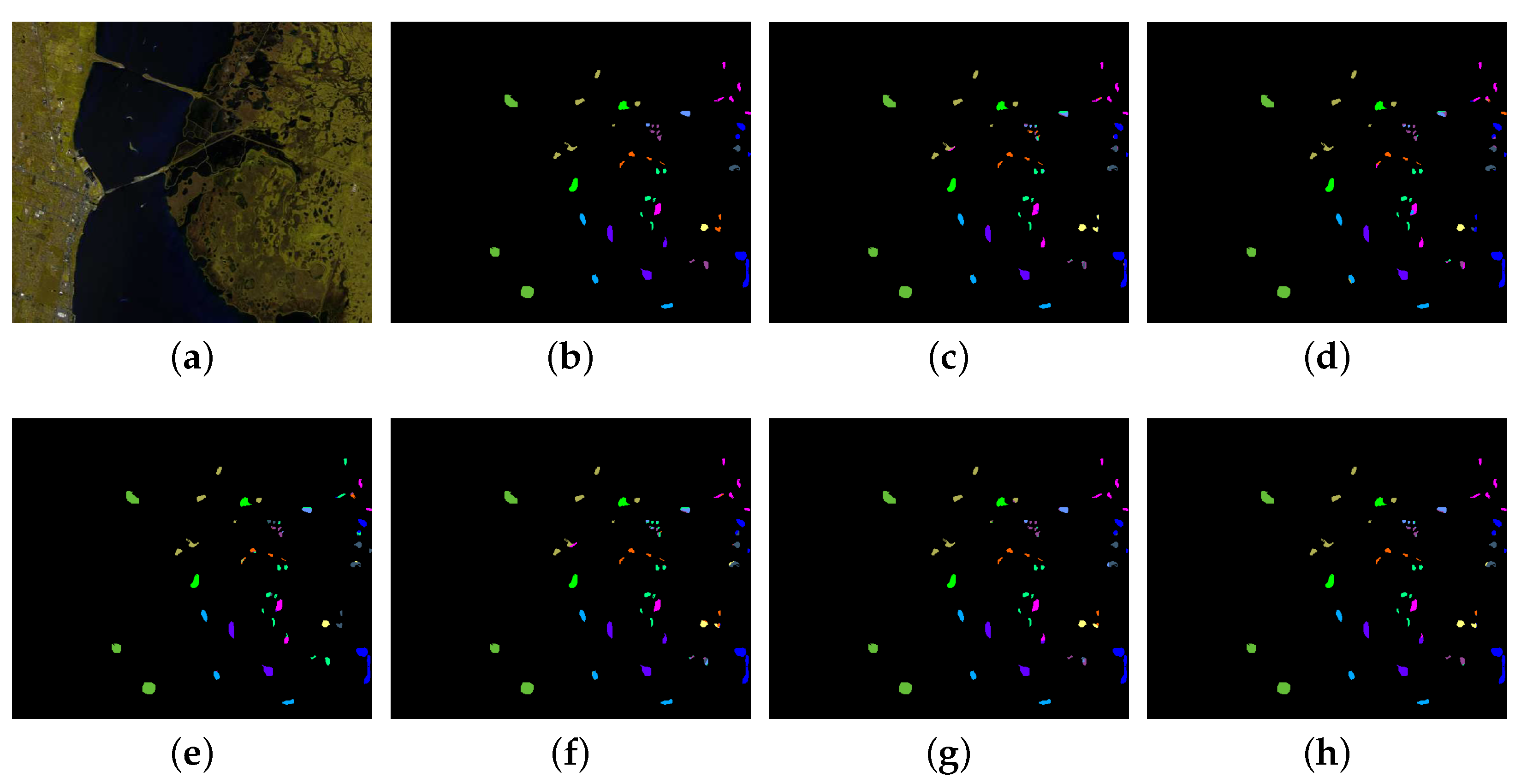
The second and third experiments were conducted on the Houston University and KSC data sets, respectively. For the Houston University data set, 10% of the labeled samples in each class were randomly selected for training. For the KSC data set, 2% of the labeled samples per class were randomly chosen for training.
Table 5 and
Table 6, respectively, show the quantitative classification results obtained by different approaches on the two data sets. It can be seen that the proposed MPRN improves the OA value from 98.53% to 98.88% for the Houston University data set and 95.24% to 96.00% for the KSC data set compared with the ResNet. In addition, the proposed method obtains the best classification performance in terms of the five overall metrics (the OA, AA, Kappa, F1-score and Precision) among all the six methods on the two data sets, which demonstrates the effectiveness of the proposed method. The corresponding classification maps are respectively illustrated in
Figure 9 and
Figure 10.
As shown in
Table 7, the standardized McNemar’s test [
58] was performed to demonstrate the statistical significance in accuracy improvement of the proposed MPRN. When the
Z value of McNemar’s test is larger than 1.96 and 2.58, it indicates that the difference in accuracy between classifiers 1 and 2 are statistically significant at the 95% and 99% confidence levels, respectively. The
Z value larger than 0 means that classifier 1 is more accurate than classifier 2 and vice versa. In this experiment, the proposed MPRN is compared with five other methods, that is, 3-D CNN, FCLFN, DFFN, DenseNet and ResNet. From
Table 7, one can see that all the
Z values are larger than 2.58, demonstrating that the proposed MPRN can significantly outperform the compared methods.
Finally, the total number of parameters and computing time of the six considered methods on the three data sets are reported in
Table 8 and
Table 9, respectively. From
Table 9, we can find that FCLFN achieves the lowest training times on the three data sets. In addition, MPRN spends less time than ResNet on the Indian Pines data set because it has fewer parameters compared with ResNet. For the Houston University and KSC data sets, the proposed method is the most time-consuming, which is attributed to the processing of a large number of Conv layers.
4.5. Effect of Input Spatial Patch Size
In this experiment, we compare our MPRN method with the spatial-spectral ResNet (SSRN) in Reference [
37]. In this case, the Indian Pines and KSC data sets are considered. Following Reference [
37], 20% of the available labeled samples are randomly selected to form the training set. In addition, input patches with four different spatial sizes {
,
,
and
} have been considered. Since a patch too large may contain pixels from multiple classes that detract from the target pixel. In addition, it results in the degradation of intersample diversity, increasing the possibility of overfitting and curse of dimensionality as well.
Table 10 shows the overall accuracies obtained in this experiment. From
Table 10, one can see that MPRN achieves remarkable improvements in terms of OA regardless of the sizes of the considered image patches. For example, the proposed MPRN reach 6.58 percent higher OA than the SSRN with the same amount of spatial information (
patch size) on the Indian Pines data set. Furthermore, all the OAs, obtained by MPRN with different patch sizes on the two data sets, are higher than 99%, indicating the robustness of our MPRN method to input patch size.
4.6. Effect of Limited Training Samples
Since manual labeling of hyperspectral data is expensive and time demanding, labeled samples are usually limited in practice. Therefore, it is necessary to assess the performance of the proposed method when limited training data is available.
Figure 11 illustrates the overall classification accuracies achieved by different methods on the three data sets using limited numbers of training samples (ranging from 0.1% to 0.5%, with a step of 0.1% per class). As can be seen in
Figure 11, for each data set, the proposed MPRN consistently performs the best among all methods under all different training samples, demonstrating the effectiveness and robustness of the proposed approach.
In the face of limited training data, deep networks with a large number of parameters tend to overfit the training set and thus obtain poor accuracy on the testing set. However, MPRN can be interpreted as an ensemble of exponential relatively shallow networks, each of which has a small number of parameters to be optimized and thus avoids the overfitting problem naturally. Therefore, the proposed method is able to provide superior performance when facing limited training data.