Discovering the Representative Subset with Low Redundancy for Hyperspectral Feature Selection
Abstract
1. Introduction
2. Related Works
- (1)
- When compared with the the LCMV-based methods; although both the LCMV-based methods and the MRMR method evaluate the representativeness of bands, their explicit selection criteria are totally different. The LCMV-based methods measure one band’s representativeness relative to the whole dataset by using a finite impulse response (FIR) filter [10]. The MRMR method evaluates the representativeness of a band subset relative to the remaining bands by using OP. Moreover, LCMV cannot consider redundancy among selected bands [10,19], but the MRMR method can achieve it.
- (2)
- When compared with the existing OP-based methods like OPBS, OSP-BSVD and VGBS; although both these similar methods and the MRMR method use OP to measure the relationship among bands, their objectives are totally different. For the OPBS, OSP-BSVD and VGBS methods, OP is used to evaluate the redundancy or the dissimilarity between a candidate band and the currently selected bands [19,20,21]; while for the MRMR method, OP is used to measure the representativeness of a band subset relative to the remaining unselected bands. The existing OP-based mainly consider the redundancy among selected bands but do not pay sufficient attention on the selected bands’ representativeness [19], in contrast, the MRMR method can well consider both the redundancy and the representativeness of the selected band subset.
- (3)
- Finally, all the LCMV, OPBS, OSP-BSVD and VGBS methods are point-wise band selection methods, namely, the desired bands are obtained individually [10,19,20,21]; whereas the MRMR method is a group-wise method, in which the desired bands are obtained simultaneously. Because the selected bands actually works together in the applications like pixel classification, the effect of the selected bands should be considered jointly. The group-wise methods are usually more effective than the point-wise methods, since the group searching strategy is more suitable for evaluating the joint effect of multiple bands.
3. The Proposed Method
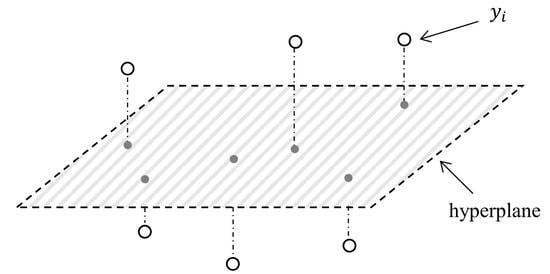
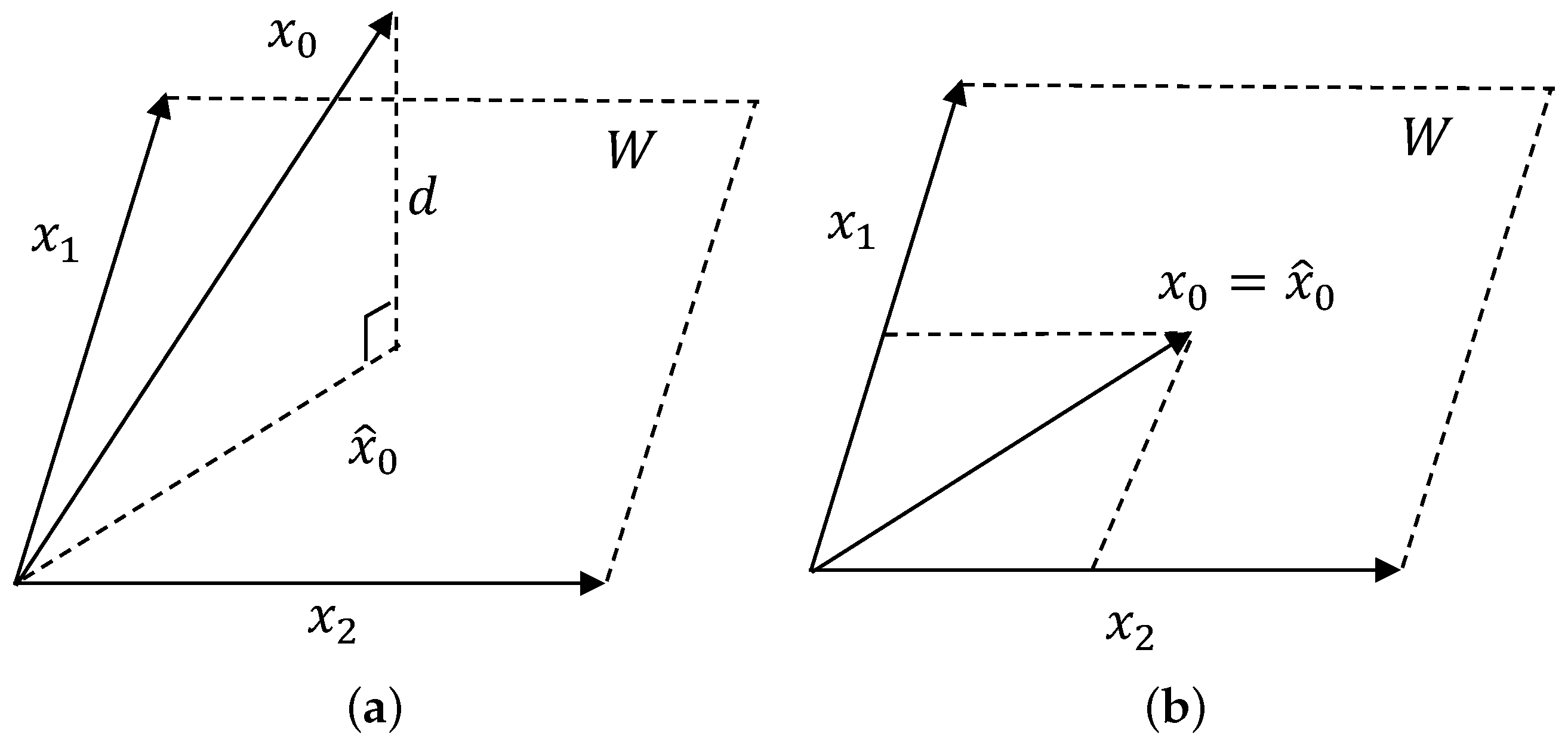
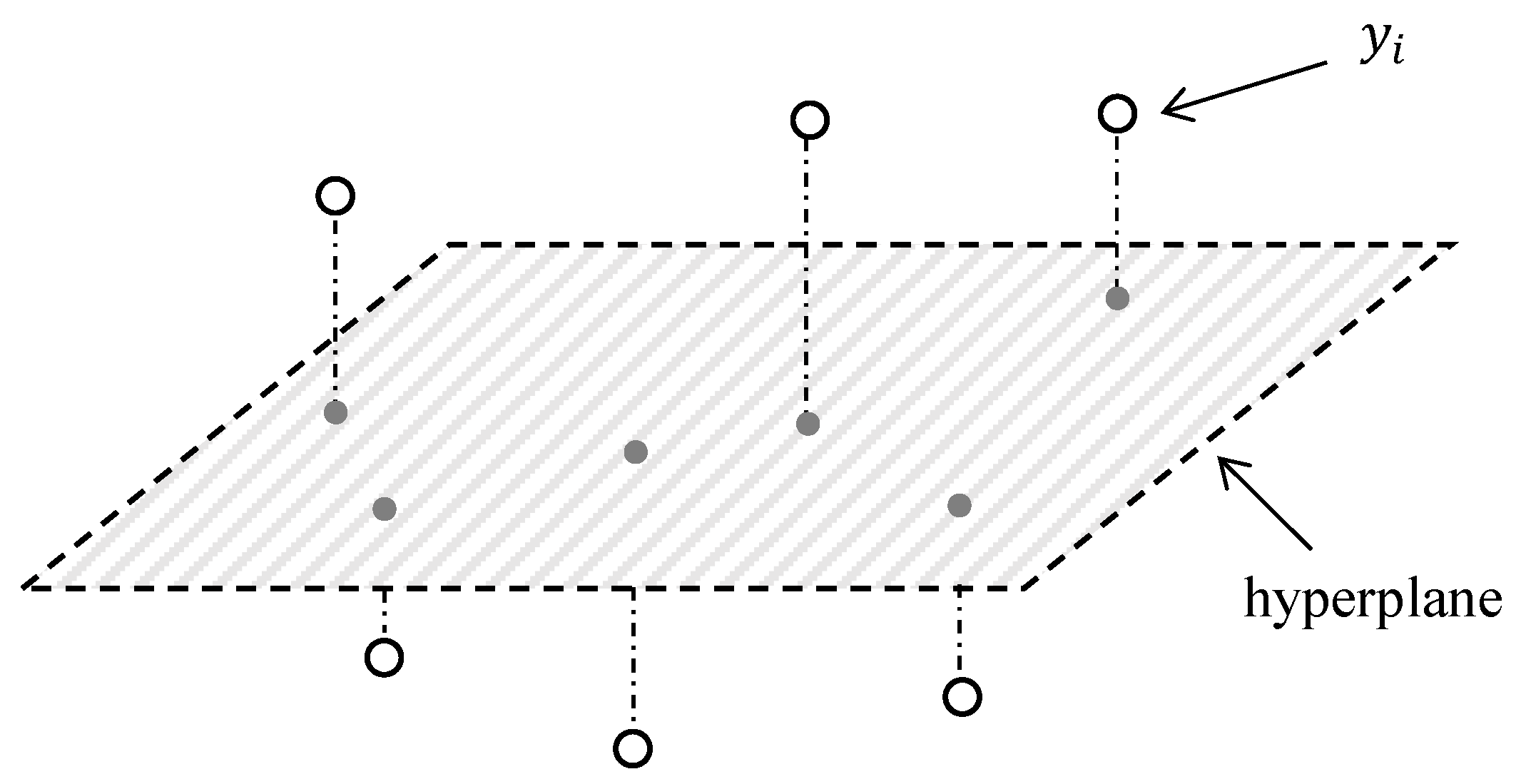
3.1. Background of OP
3.2. MRMR Selection Criterion
3.3. Subset Searching Strategy
3.4. Practical Considerations
3.4.1. Adaptive Determination of
3.4.2. Accelerating Tricks of Computing
3.4.3. The Number of Selected Bands
| Algorithm 1: The MRMR Algorithm |
| Input: Observations , the number of selected bands n. |
| Initialize: m, , and . |
| Step1: Compute the Gram matrix , then use it to compute subsets’ representativeness (using (20)) in the following processes. |
| Step2: Compute the correlation coefficient matrix of D, then use it to compute subsets’s redundancy (using (8)) in the following processes. |
| Step3: Establish the initial set of the antibody population, i.e., . |
| Step4: |
| while the stop criterion is not met do |
| 1: Copy the antibodies according to their affinities. |
| 2: According to the clone selection strategy, randomly select some bands from each copied antibody and replace them with other candidate bands. |
| 3: Select the m antibodies that have the highest affinities to construct the new antibody population. |
| end while |
| Step5: The antibody that has the largest affinity is regarded as the final selected band subset. |
| Output:n selected bands. |
4. Experiments
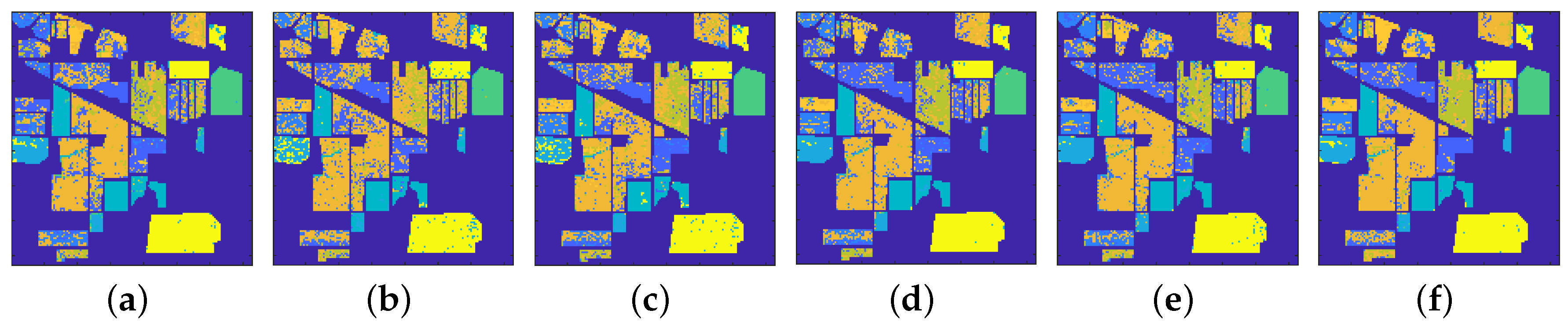
4.1. Indian Pine Dataset
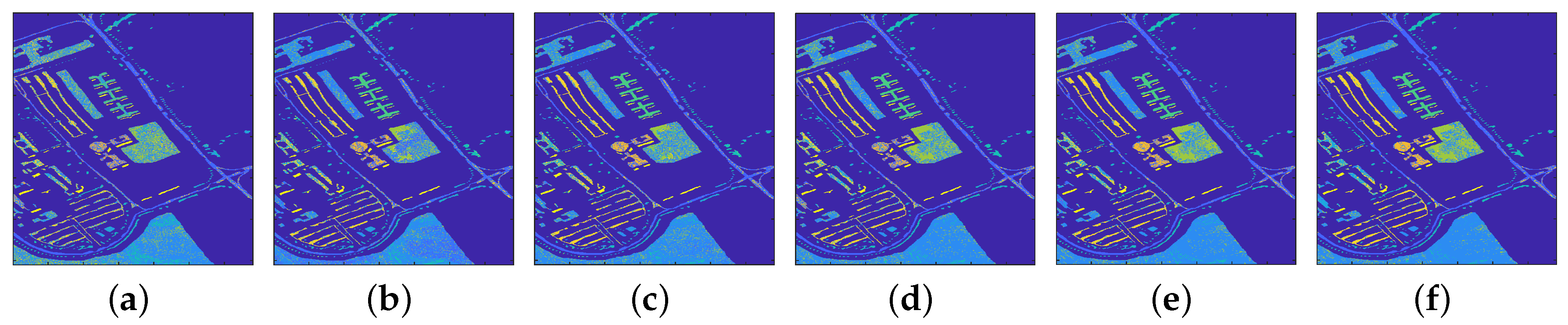
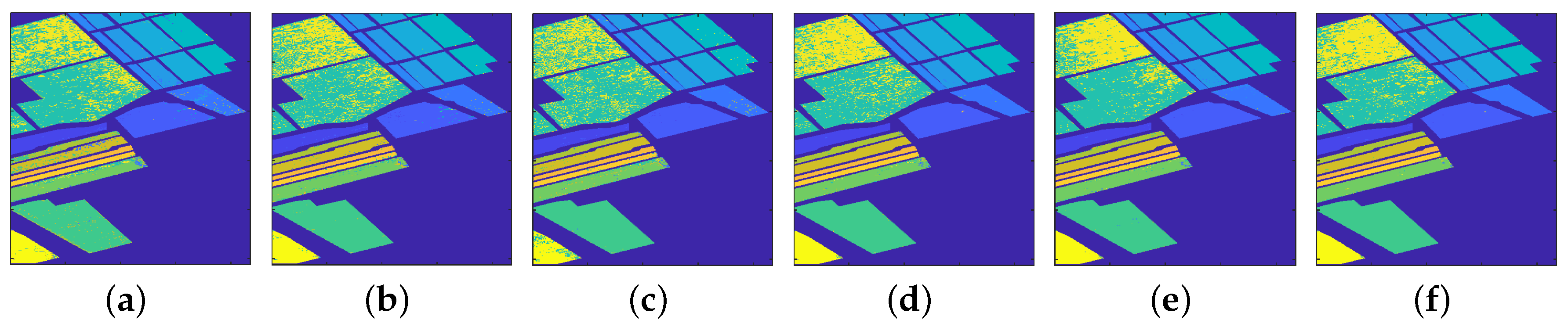
4.1.1. Classification Results
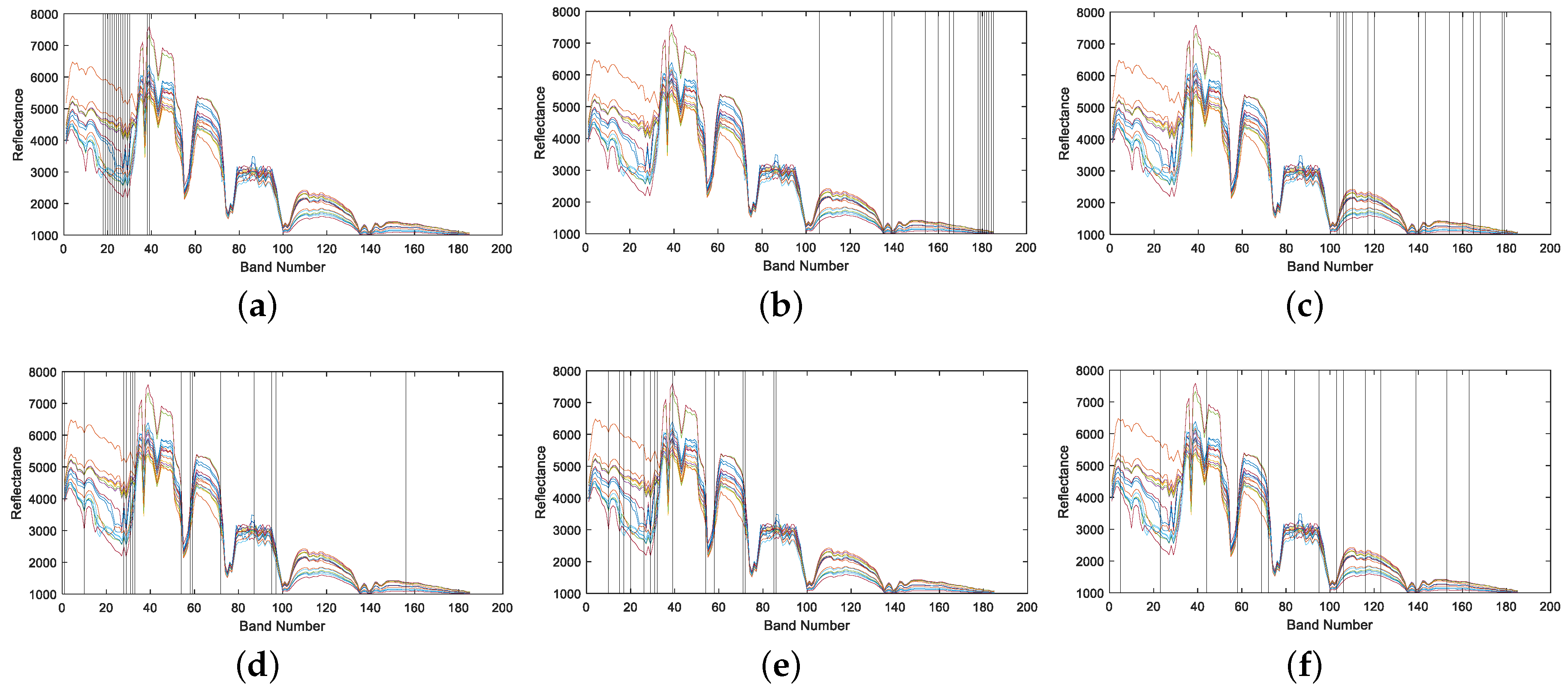
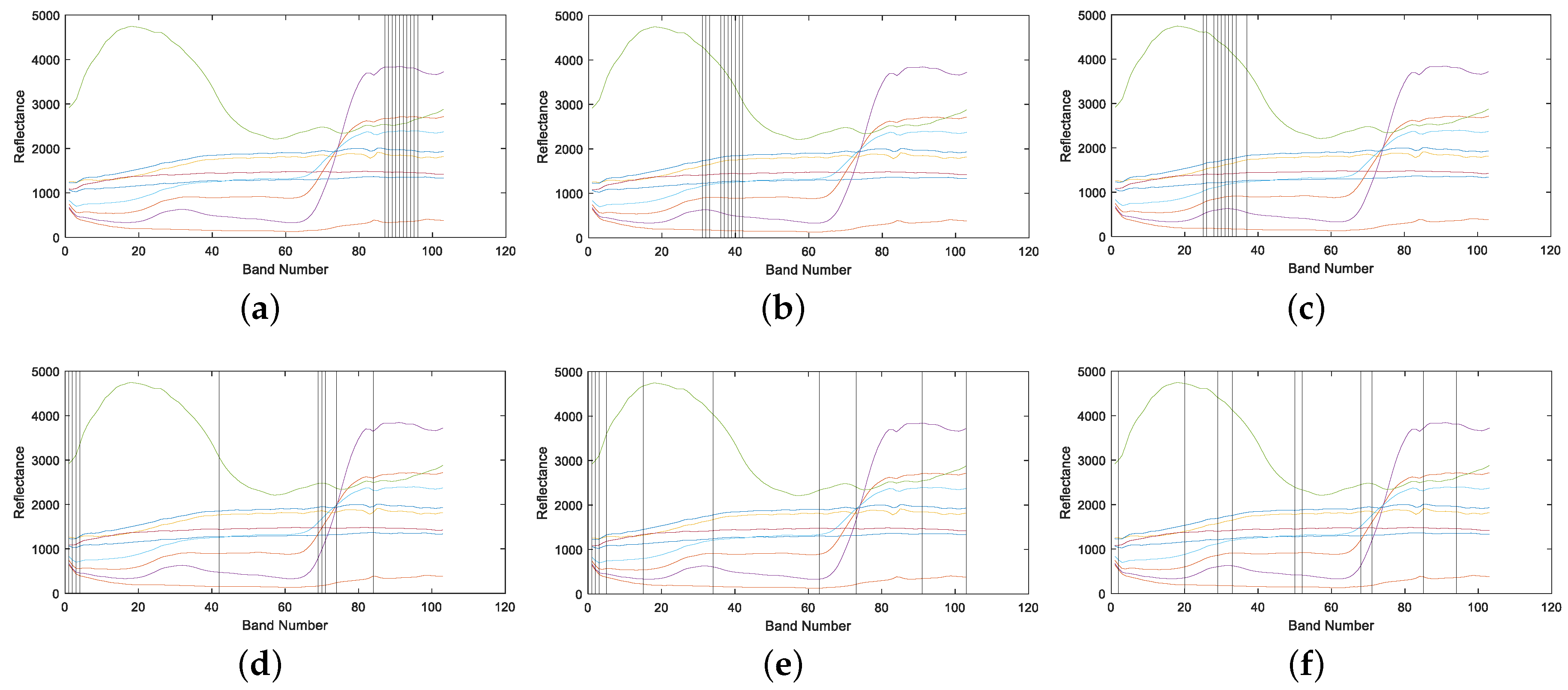
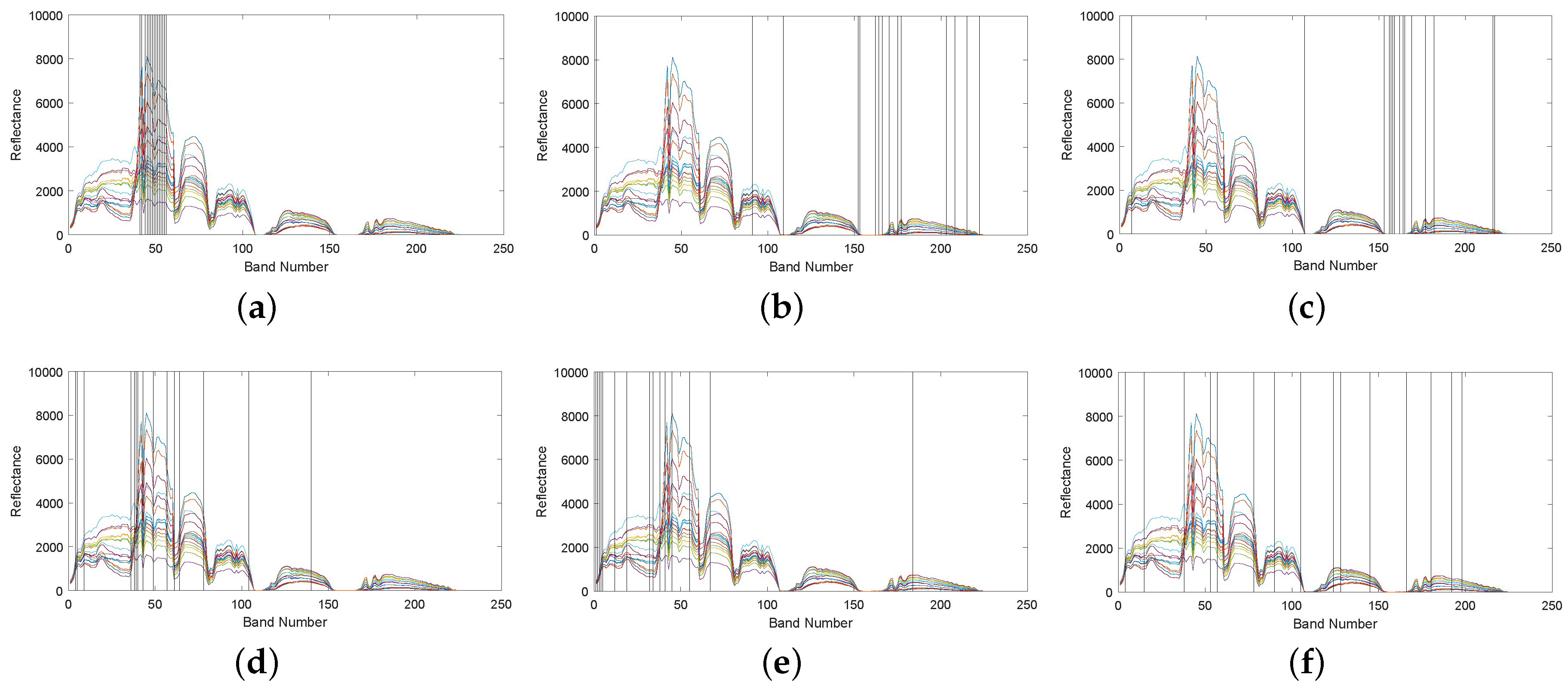
4.1.2. Band Correlation Comparison
4.1.3. Computing Time Comparison
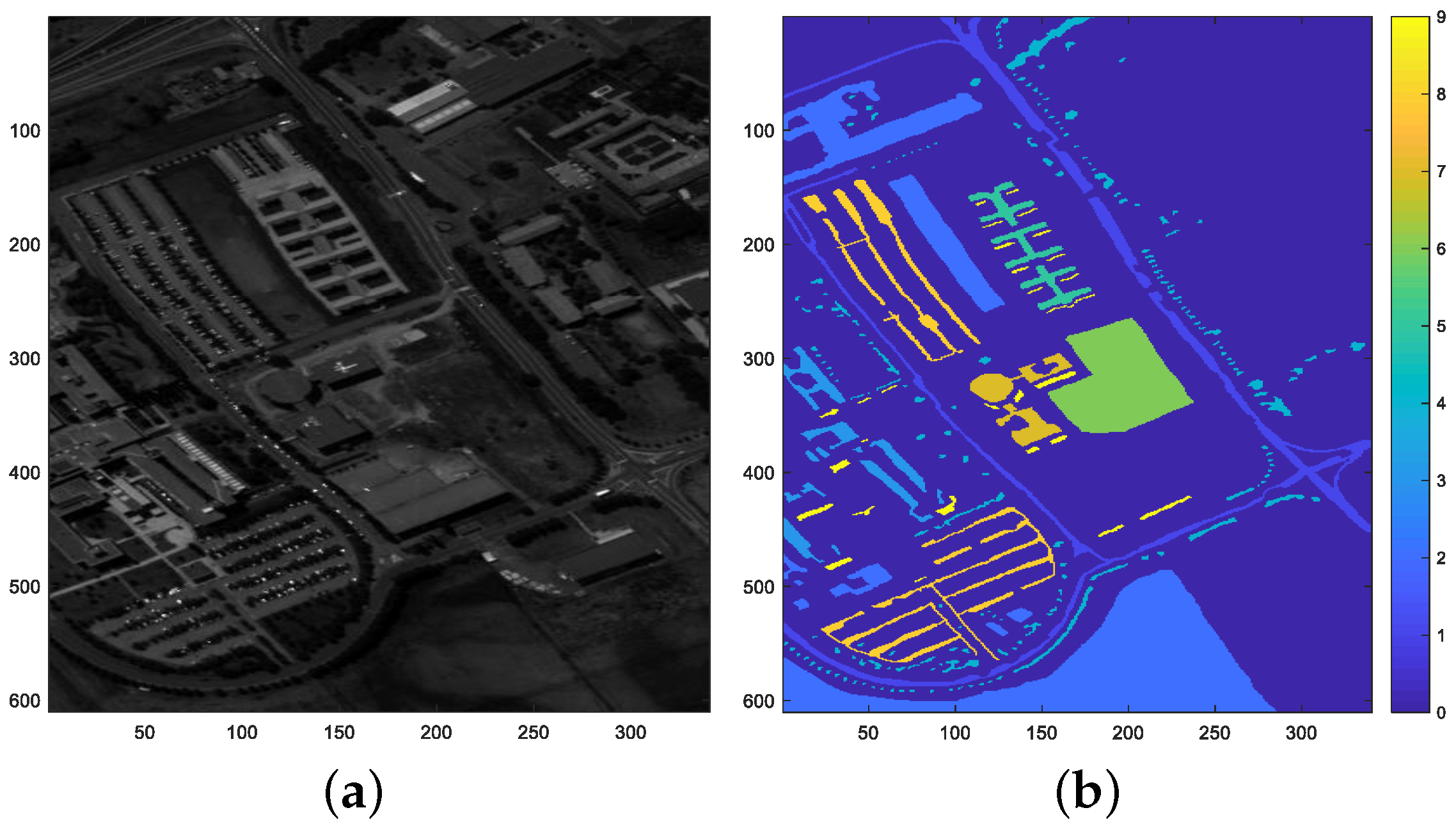
4.2. Pavia University Image
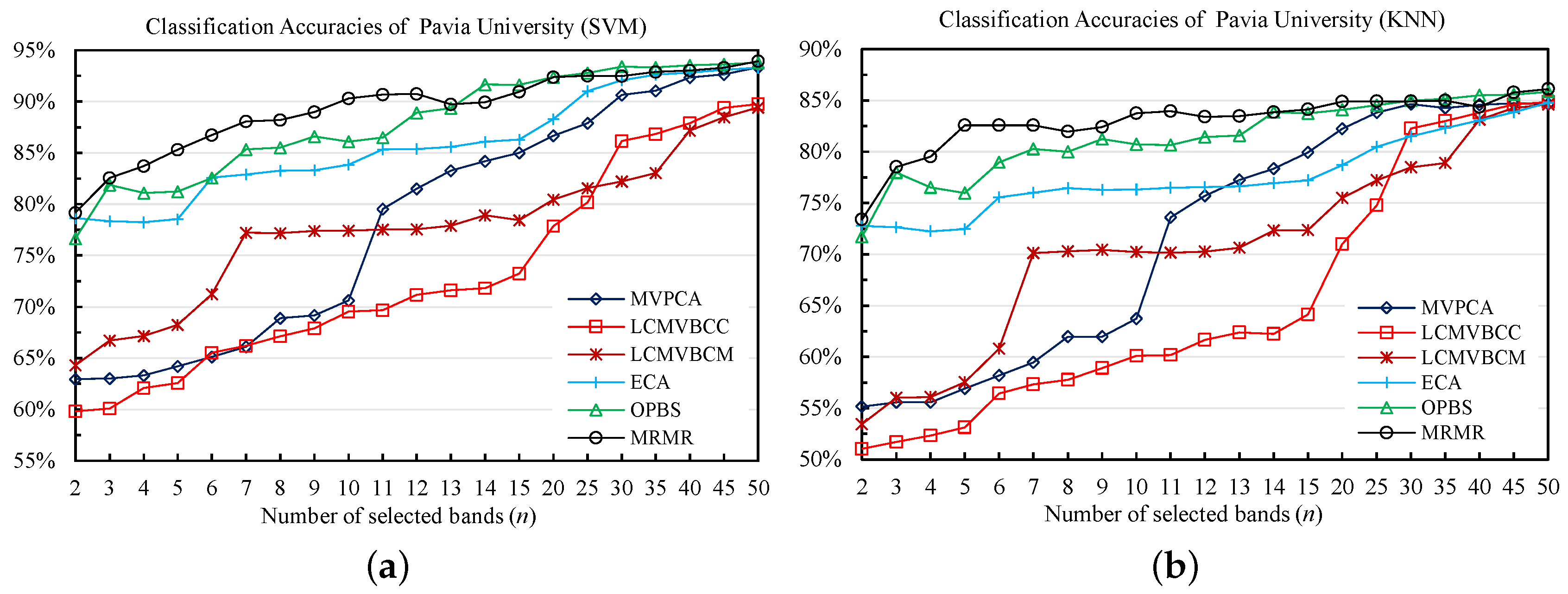
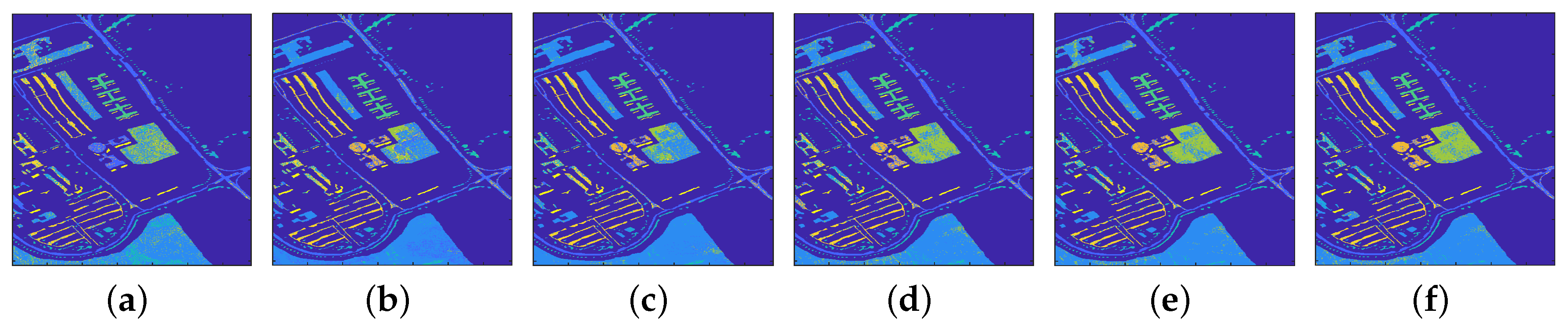
4.2.1. Classification Results
4.2.2. Band Correlation Comparison
4.2.3. Computing Time Comparison
4.3. Salinas Dataset
4.3.1. Classification Results
4.3.2. Band Correlation Comparison
4.3.3. Computing Time Comparison
4.4. Summary
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jolliffe, I.T. Principal Component Analysis. In Principal Component Analysis; Springer: Berlin/Heidelberg, Germany, 1986; pp. 115–128. [Google Scholar]
- Khan, Z.; Shafait, F.; Mian, A. Joint Group Sparse PCA for Compressed Hyperspectral Imaging. IEEE Trans. Image Process. 2015, 24, 4934–4942. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, A.; Hurri, J.; Hoyer, P.O. Independent component analysis. Natural Image Statistics; Springer Science & Business Media: New York, NY, USA, 2009; pp. 151–175. [Google Scholar]
- Roweis, S.T.; Saul, L.K. Nonlinear dimensionality reduction by locally linear embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef] [PubMed]
- Prabukumar, M.; Shrutika, S. Band clustering using expectation–maximization algorithm and weighted average fusion-based feature extraction for hyperspectral image classification. J. Appl. Remote Sens. 2018, 12, 046015. [Google Scholar] [CrossRef]
- Prabukumar, M.; Sawant, S.; Samiappan, S.; Agilandeeswari, L. Three-dimensional discrete cosine transform-based feature extraction for hyperspectral image classification. J. Appl. Remote Sens. 2018, 12, 046010. [Google Scholar] [CrossRef]
- Taşkın, G.; Kaya, H.; Bruzzone, L. Feature Selection Based on High Dimensional Model Representation for Hyperspectral Images. IEEE Trans. Image Process. 2017, 26, 2918–2928. [Google Scholar] [CrossRef] [PubMed]
- Persello, C.; Bruzzone, L. Kernel-Based Domain-Invariant Feature Selection in Hyperspectral Images for Transfer Learning. IEEE Trans. Geosci. Remote Sens. 2016, 54, 2615–2626. [Google Scholar] [CrossRef]
- Chang, C.I.; Wang, S. Constrained band selection for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1575–1585. [Google Scholar] [CrossRef]
- Yuan, Y.; Zheng, X.; Lu, X. Discovering diverse subset for unsupervised hyperspectral band selection. IEEE Trans. Image Process. 2017, 26, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Gunn, S.R.; Damper, R.I.; Nelson, J.D. Band selection for hyperspectral image classification using mutual information. IEEE Geosci. Remote Sens. Lett. 2006, 3, 522–526. [Google Scholar] [CrossRef]
- Chang, C.I.; Du, Q.; Sun, T.L.; Althouse, M.L. A joint band prioritization and band-decorrelation approach to band selection for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 1999, 37, 2631–2641. [Google Scholar] [CrossRef]
- Liu, X.S.; Ge, L.; Wang, B.; Zhang, L.M. An unsupervised band selection algorithm for hyperspectral imagery based on maximal information. J. Infrared Millim. Waves 2012, 31, 166–176. [Google Scholar] [CrossRef]
- Sheffield, C. Selecting band combinations from multispectral data. Photogramm. Eng. Remote Sens. 1985, 51, 681–687. [Google Scholar]
- Zhang, W.; Li, X.; Zhao, L. Hyperspectral band selection based on triangular factorization. J. Appl. Remote Sens. 2017, 11, 025007. [Google Scholar] [CrossRef]
- Zare, A.; Gader, P. Hyperspectral band selection and endmember detection using sparsity promoting priors. IEEE Geosci. Remote Sens. Lett. 2008, 5, 256–260. [Google Scholar] [CrossRef]
- Zhang, W.; Li, X.; Dou, Y.; Zhao, L. Fast linear-prediction-based band selection method for hyperspectral image analysis. J. Appl. Remote Sens. 2018, 12, 016027. [Google Scholar] [CrossRef]
- Zhang, W.; Li, X.; Dou, Y.; Zhao, L. A Geometry-Based Band Selection Approach for Hyperspectral Image Analysis. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4318–4333. [Google Scholar] [CrossRef]
- Geng, X.; Sun, K.; Ji, L.; Zhao, Y. A fast volume-gradient-based band selection method for hyperspectral image. IEEE Trans. Geosci. Remote Sens. 2014, 52, 7111–7119. [Google Scholar] [CrossRef]
- Yu, C.; Lee, L.; Chang, C.; Xue, B.; Song, M.; Chen, J. Band-Specified Virtual Dimensionality for Band Selection: An Orthogonal Subspace Projection Approach. IEEE Trans. Geosci. Remote Sens. 2018, 56, 2822–2832. [Google Scholar] [CrossRef]
- Wang, L.; Jia, X.; Zhang, Y. A novel geometry-based feature-selection technique for hyperspectral imagery. IEEE Geosci. Remote Sens. Lett. 2007, 4, 171–175. [Google Scholar] [CrossRef]
- Sun, K.; Geng, X.; Ji, L. Exemplar component analysis: A fast band selection method for hyperspectral imagery. IEEE Geosci. Remote Sens. Lett. 2015, 12, 998–1002. [Google Scholar]
- Ahmad, M.; Haq, D.I.U.; Mushtaq, Q.; Sohaib, M. A new statistical approach for band clustering and band selection using K-means clustering. IACSIT Int. J. Eng. Technol. 2011, 3, 606–614. [Google Scholar]
- Jia, S.; Tang, G.; Zhu, J.; Li, Q. A Novel Ranking-Based Clustering Approach for Hyperspectral Band Selection. IEEE Trans. Geosci. Remote Sens. 2016, 54, 88–102. [Google Scholar] [CrossRef]
- Li, H.; Xiang, S.; Zhong, Z.; Ding, K.; Pan, C. Multicluster Spatial-Spectral Unsupervised Feature Selection for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1660–1664. [Google Scholar] [CrossRef]
- Qian, Y.; Yao, F.; Jia, S. Band selection for hyperspectral imagery using affinity propagation. IET Comput. Vis. 2009, 3, 213–222. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, J.; Yuan, Y. Salient band selection for hyperspectral image classification via manifold ranking. IEEE Trans. Neural Netw. Learn. Syst. 2016, 27, 1279–1289. [Google Scholar] [CrossRef]
- Sun, K.; Geng, X.; Ji, L. A new sparsity-based band selection method for target detection of hyperspectral image. IEEE Geosci. Remote Sens. Lett. 2015, 12, 329–333. [Google Scholar]
- Xenaki, S.D.; Koutroumbas, K.D.; Rontogiannis, A.A.; Sykioti, O.A. A new sparsity-aware feature selection method for hyperspectral image clustering. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Milan, Italy, 26–31 July 2015; pp. 445–448. [Google Scholar]
- De Castro, L.N.; Von Zuben, F.J. Learning and optimization using the clonal selection principle. IEEE Trans. Evol. Comput. 2002, 6, 239–251. [Google Scholar] [CrossRef]
- Chang, C.I. Hyperspectral Imaging: Techniques for Spectral Detection and Classification; Springer Science & Business Media: New York, NY, USA, 2003; Volume 1. [Google Scholar]
- Pudil, P.; Ferri, F.J.; Novovicova, J.; Kittler, J. Floating search methods for feature selection with nonmonotonic criterion functions. In Proceedings of the 12th IAPR International Conference on Pattern Recognition, Jerusalem, Israel, 9–13 October 1994; Volume 2, pp. 279–283. [Google Scholar]
- Zhang, X.D. Matrix Analysis and Applications; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar]
- Jain, A.; Zongker, D. Feature selection: Evaluation, application, and small sample performance. IEEE Trans. Pattern Anal. Mach. Intell. 1997, 19, 153–158. [Google Scholar] [CrossRef]
- Somol, P.; Pudil, P.; Kittler, J. Fast branch & bound algorithms for optimal feature selection. IEEE Trans. Pattern Anal. Mach. Intell. 2004, 26, 900–912. [Google Scholar] [CrossRef]
- Gen, M.; Cheng, R. Genetic Algorithms and Engineering Optimization; John Wiley & Sons: Hoboken, NJ, USA, 2000; Volume 7. [Google Scholar]
- Serpico, S.B.; Bruzzone, L. A new search algorithm for feature selection in hyperspectral remote sensing images. IEEE Trans. Geosci. Remote Sens. 2001, 39, 1360–1367. [Google Scholar] [CrossRef]
- Furcy, D.; Koenig, S. Limited discrepancy beam search. In Proceedings of the 19th International Joint Conference on Artificial Intelligence (IJCAI’05), Edinburgh, Scotland, UK, 30 July–5 August 2005; pp. 125–131. [Google Scholar]
- Kennedy, J. Particle swarm optimization. In Encyclopedia of Machine Learning; Springer: Berlin/Heidelberg, Germany, 2011; pp. 760–766. [Google Scholar]
- Chang, C.I.; Du, Q. Estimation of number of spectrally distinct signal sources in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 608–619. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Devroye, L.; Györfi, L.; Lugosi, G. A Probabilistic Theory of Pattern Recognition; Springer Science & Business Media: New York, NY, USA, 2013; Volume 31. [Google Scholar]
- Rifkin, R.; Klautau, A. In defense of one-vs-all classification. J. Mach. Learn. Res. 2004, 5, 101–141. [Google Scholar]
- Dópido, I.; Li, J.; Gamba, P.; Plaza, A. A new hybrid strategy combining semisupervised classification and unmixing of hyperspectral data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3619–3629. [Google Scholar] [CrossRef]






| SVM | KNN | |||
|---|---|---|---|---|
| OA (100%) | AA (100%) | OA (100%) | AA (100%) | |
| 1.MVPCA | 67.74 | 67.99 | 59.95 | 61.19 |
| 2.LCMVBCC | 64.39 | 63.82 | 54.25 | 54.90 |
| 3.LCMVBCM | 70.48 | 72.02 | 62.60 | 63.52 |
| 4.ECA | 77.45 | 76.99 | 70.62 | 69.41 |
| 5.OPBS | 75.90 | 76.15 | 68.70 | 67.86 |
| 6.MRMR | 81.32 | 82.70 | 72.94 | 72.86 |
| Band Correlation (ACC) | Computing Time (s) | |
|---|---|---|
| 1.MVPCA | 0.5950 | 0.2825 |
| 2.LCMVBCC | 0.9816 | 3.0452 |
| 3.LCMVBCM | 0.9882 | 2.5048 |
| 4.ECA | 0.2988 | 1.6770 |
| 5.OPBS | 0.1815 | 0.6376 |
| 6.MRMR | 0.2179 | 1.7101 |
| SVM | KNN | |||
|---|---|---|---|---|
| OA (100%) | AA (100%) | OA (100%) | AA (100%) | |
| 1.MVPCA | 70.96 | 62.83 | 63.87 | 59.09 |
| 2.LCMVBCC | 69.08 | 64.36 | 60.69 | 62.52 |
| 3.LCMVBCM | 77.19 | 70.20 | 68.27 | 68.27 |
| 4.ECA | 83.89 | 79.96 | 76.62 | 72.65 |
| 5.OPBS | 86.58 | 83.62 | 80.87 | 78.38 |
| 6.MRMR | 90.15 | 87.78 | 83.76 | 82.57 |
| Band Correlation (ACC) | Computing Time (s) | |
|---|---|---|
| 1.MVPCA | 0.9981 | 1.1549 |
| 2.LCMVBCC | 0.9880 | 5.7662 |
| 3.LCMVBCM | 0.9934 | 4.3998 |
| 4.ECA | 0.6223 | 15.2934 |
| 5.OPBS | 0.5267 | 2.5998 |
| 6.MRMR | 0.5788 | 2.3671 |
| SVM | KNN | |||
|---|---|---|---|---|
| OA (100%) | AA (100%) | OA (100%) | AA (100%) | |
| 1.MVPCA | 84.75 | 87.78 | 80.16 | 83.56 |
| 2.LCMVBCC | 88.06 | 90.91 | 84.08 | 86.57 |
| 3.LCMVBCM | 86.36 | 87.02 | 82.65 | 85.44 |
| 4.ECA | 92.16 | 95.67 | 88.58 | 92.87 |
| 5.OPBS | 91.72 | 95.34 | 84.54 | 88.77 |
| 6.MRMR | 93.02 | 96.31 | 88.83 | 93.13 |
| Band Correlation (ACC) | Computing Time (s) | |
|---|---|---|
| 1.MVPCA | 0.9976 | 1.1549 |
| 2.LCMVBCC | 0.6282 | 5.7662 |
| 3.LCMVBCM | 0.7001 | 4.3998 |
| 4.ECA | 0.4509 | 15.2934 |
| 5.OPBS | 0.3728 | 2.5998 |
| 6.MRMR | 0.3039 | 2.3671 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Li, X.; Zhao, L. Discovering the Representative Subset with Low Redundancy for Hyperspectral Feature Selection. Remote Sens. 2019, 11, 1341. https://doi.org/10.3390/rs11111341
Zhang W, Li X, Zhao L. Discovering the Representative Subset with Low Redundancy for Hyperspectral Feature Selection. Remote Sensing. 2019; 11(11):1341. https://doi.org/10.3390/rs11111341
Chicago/Turabian StyleZhang, Wenqiang, Xiaorun Li, and Liaoying Zhao. 2019. "Discovering the Representative Subset with Low Redundancy for Hyperspectral Feature Selection" Remote Sensing 11, no. 11: 1341. https://doi.org/10.3390/rs11111341
APA StyleZhang, W., Li, X., & Zhao, L. (2019). Discovering the Representative Subset with Low Redundancy for Hyperspectral Feature Selection. Remote Sensing, 11(11), 1341. https://doi.org/10.3390/rs11111341