Comment on Tompalski et al. Combining Multi-Date Airborne Laser Scanning and Digital Aerial Photogrammetric Data for Forest Growth and Yield Modelling. Remote Sens. 2018, 10, 347
Abstract
1. Introduction
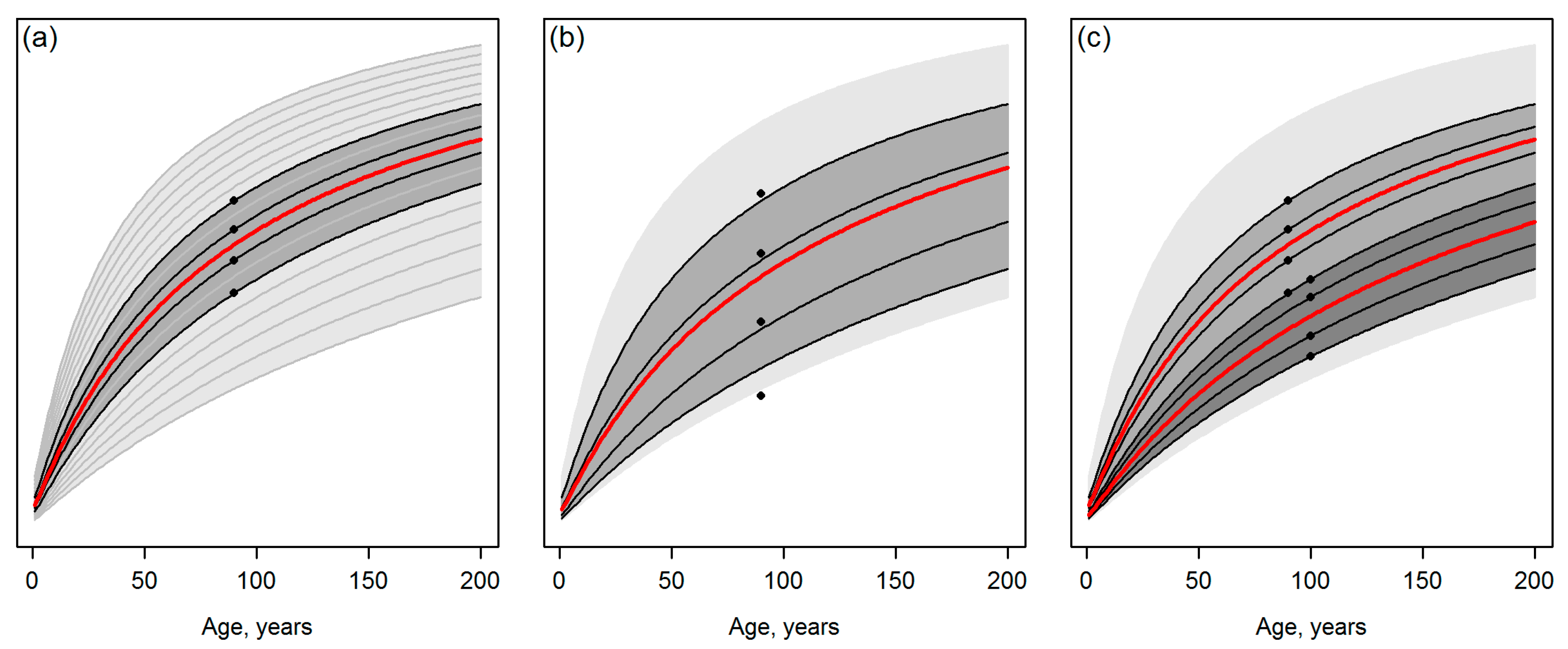
- Assume that Figure 1a represents an ideal case with error-free observations, whereas predicting the attributes of interest independently of each other for a grid cell hinders the curve matching as illustrated in Figure 1b. Tompalski et al. [1] do conclude that “the accuracy of curve matching was dependent on the ABA (area-based approach) prediction error”. However, what are the implications of a lower “accuracy of curve matching”? What is the sensitivity of the approach to produce such a severe incompatibility (e.g., [2] (Chapter 15)) between stand attributes or their future projections that causes problems for the further use of this information? Are there interactions in this sense between the composition of the observations and the template database (for example, what if observations or predictions are located on borders or slightly outside of the template space such as the lowest observation of Figure 1b)?
- Assume that multi-date data are available (Figure 1c) and observations or predictions for T2 systematically suggest a slower future development of the stand, compared to single-date observations or predictions for T1. Note that even if the training data for model fitting were filtered for recorded disturbances between the two time steps (as in Tompalski et al. [1]), situations corresponding to Figure 1c could occur due to artifacts in the remotely sensed data and ignoring the dependencies between T1 and T2 in the prediction models. Applying the workflow of Tompalski et al. [1] to wall-to-wall forest inventory grid data from multiple dates would thus produce situations that resemble real-world disturbances in the forest (cf. Figure 1c), but it is unclear if and how the underlying growth and yield modelling system logic can manage such situations.
- Overall, how well-reasoned is that with multi-date data, “the weighted means were calculated separately for each data set, and then averaged” [1] (Section 3.3), i.e., the observations for T1 and T2 were considered with an equal weight, even if the latter represent data acquired later and should therefore have a closer temporal match with the present forest state?
2. Experimental Implementation
2.1. The Growth and Yield Projection System (GYPSY) Models (gypsy_models.R)
- Above, the definition of “age” varies between models and tree species (groups): it is either total age (years since the point of germination) or breast height age (age at the height of 1.3 m above ground). Consequently, SI is the top height corresponding to the age definition that varies between the models and species. For my exercises, I assumed that all age and SI values inputted corresponded to the breast height age. Where necessary, I converted the breast height age to total age using average conversion factors [11] (p. 5). However, I did not convert SI, i.e., SI values between Equation (1) and the other models are not consistent regarding the definition, which causes a leveling difference with a magnitude that probably could be assessed by applying exact conversions [11] (Appendix A). This is further explored below.
- GYPSY provides a possibility to simulate growth and yield for both pure and mixed stands [11]. Tompalski et al. [1] do not indicate whether their stands were considered to be pure or mixtures of species. For the simulations below, I assumed pure stands. This choice affected the parameterization of Equations (1)–(4) as follows: The SC component of the BAINC model (Equation (3)) was always fixed to 1. The GYPSY models of BAINC additionally include term k, which is computed for the Aspen and Pine species as a transformation of the other model parameters. However, for the two Spruce species, the term k included the SDFs for each of the other three species. In the stands simulated here, where no other species occurred, the term k of the Spruce species received a value of zero.
2.2. Template Database (generate_templates.R)
“Following the approach of Tompalski et al. [21], we used the simulator to create a database of yield curve templates based on all possible input combinations (e.g., every combination of species groups, top height, total age, density, and basal area). The database represented all possible stand conditions in the study area and was based on the range of stand attributes in the existing forest inventory. The yield curves were generated for the four specified species groups, from 1 to 200 years, by 1 year increments. An individual yield curve template consisted of four sequences of values representing top height, basal area, volume, and stem density, between 1 and 200 years, and being estimated by the simulator based on species group, top height, total age, and basal area.”
- Site index (SI) as a sequence of values from 5 to 30 m with an interval of 1 m. The range was obtained as the one that included all observations for the species considered here, according to a graphical interpretation of the GYPSY Validation Summary report [14] (Figures 1 and 6).
- The initial density at the age of zero (Nini) as two alternative sequences: (1) from 700 to 2700 stems/ha with an interval of 100 stems/ha or (2) from 700 to 6700 stems/ha with an interval of 1000 stems/ha. The values above are derived from common tree spacing used for artificial regeneration in Alberta [15]. Even though a fixed density such as 2400 stems/ha could be assumed as the most typical tree planting density, the reference cited above also mentions natural regeneration as a possible method for Alberta [15], which probably results to more variations of Nini obviously impacting the GYPSY projections [11]. When neither regeneration type could be assumed, two alternative sequences of Nini values were used to derive two distinct template databases. The motivation was to assess how the alternative parameter ranges affect the growth trajectories and, subsequently, how they potentially propagate the template generation and matching.
- The GYPSY model documentation indicates that the BAINC model (Equation (3)) could be run with or without knowledge on the current basal area [11]. How to do it is, however, not trivial based on the model documentation [11] and Tompalski et al. [1] do not mention if the basal area predictions were explicitly used as the current basal area in the models. In the analyses below, a single basal area sequence corresponding to the age sequence was generated by running the BAINC model (Equation (3)) as follows. First, the BAINC model was run with age = 1, current basal area set to 0, and other parameters as each of the possible combinations of the sequences described above. Then, the obtained BAINC was added to the previous basal area and, incrementing the basal area similarly for each age value, the iteration was continued until a single basal area sequence was obtained for each of the parameter combinations above. The sequences generated this way are abbreviated as G to differentiate from BA used in Equations (3) and (4). To obtain Tvol (Equation (4)), G was used together with Htop obtained from Equation (1) as parameters of Equation (4).
2.3. Template Matching (matching.R)
and“First, candidate curves were selected from the database, based on stand age, species group and the minimal difference between the stand attribute and a value of a yield curve.”
“the final yield curve was derived by calculating a weighted mean of the candidate curves, with the percent of explained variance in the ABA model used as a weight.”
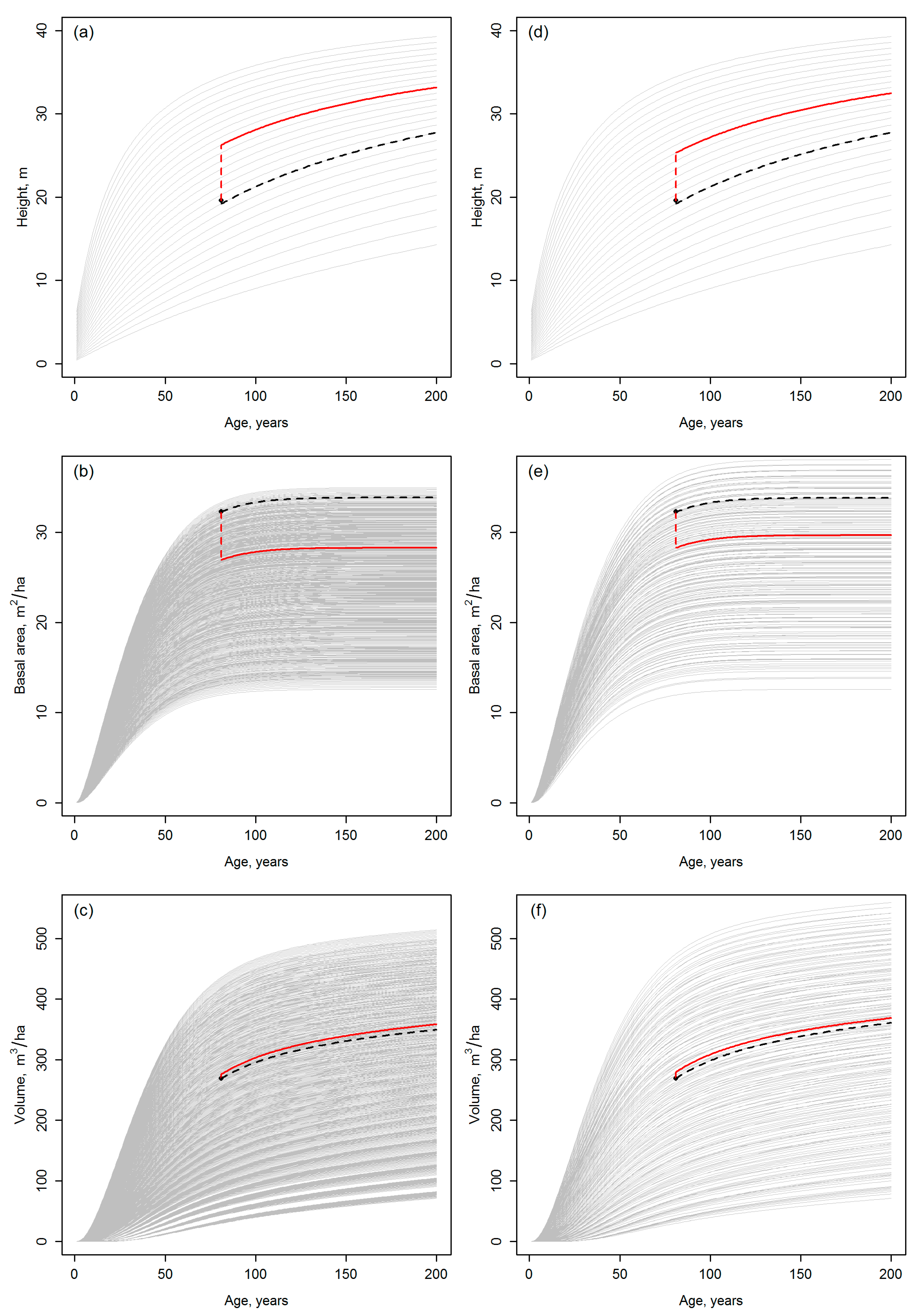
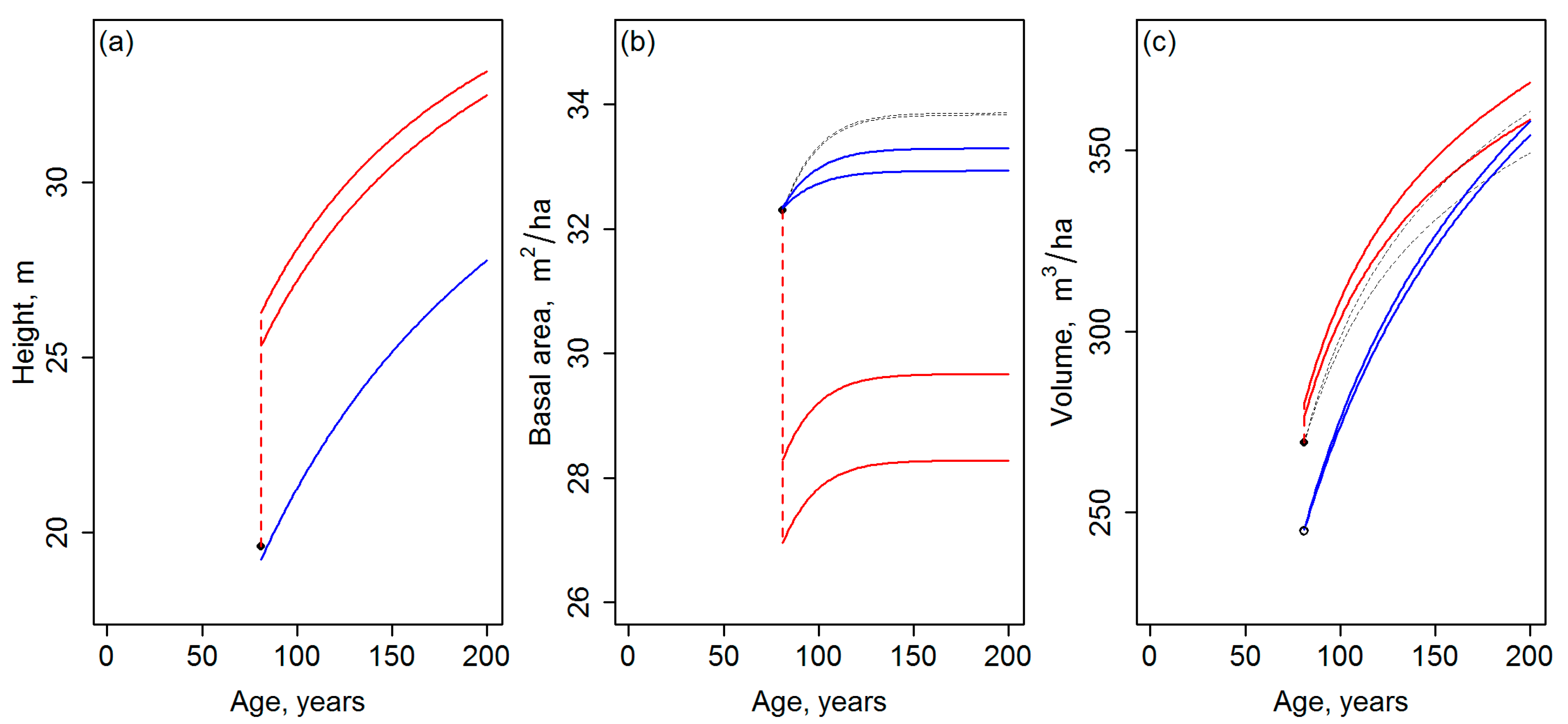
2.4. Evaluation (draw_fig2.R & draw_fig3.R)
3. Discussion
3.1. Implications of the Results from the Experimental Implementation
- The results of the template matching are very sensitive to the assumptions made when generating the template database, for which reason the results should be presented as a function of the applied parameters. With the GYPSY models, especially assumptions on the initial tree planting densities (Nini) had a considerable effect on the final development trajectories obtained. The template matching could even produce trajectories with different site indices, when carried out with template databases based on different assumptions on Nini. Whether the driver for the future development is either different planting density or productivity (site index) affects the future growth and yield estimates drastically, especially if the simulations are continued beyond the current rotation.
- Template matching may inherently produce discontinuities between the current state of the forest and its projected future state. Note that in Figure 2 and Figure 3, the discontinuities are probably magnified, because the site index differing between Equations (1) and (2) by its definition was not standardized (Section 2.1). Nevertheless, it is difficult to argue against the risk for the discontinuities, considering that observations or predictions are matched with the templates independently (cf. Figure 1). Tompalski et al. [1] do not seem to identify this problem, but to confirm the usability of the projections, those should be exposed to “model criticism and benchmarking” [2] (Chapter 15), with emphases on verifying the biological realism and compatibility of the resulting attributes (see also [3,8,9] (Chapter 18)) with respect to above.
3.2. Further Aspects Not Covered by Numeric Examples
4. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
Appendix A
References
- Tompalski, P.; Coops, N.C.; Marshall, P.L.; White, J.C.; Wulder, M.A.; Bailey, T. Combining multi-date airborne laser scanning and digital aerial photogrammetric data for forest growth and yield modelling. Remote Sens. 2018, 10, 347. [Google Scholar] [CrossRef]
- Weiskittel, A.R.; Hann, D.W.; Kershaw, J.A., Jr.; Vanclay, J.K. Forest Growth and Yield Modeling; John Wiley & Sons: Chichester, UK, 2011. [Google Scholar]
- Burkhart, H.E.; Tomé, M. Modeling Forest Trees and Stands; Springer Science & Business Media: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Bettinger, P.; Boston, K.; Siry, J.P.; Grebner, D.L. Forest Management and Planning; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Packalén, P.; Heinonen, T.; Pukkala, T.; Vauhkonen, J.; Maltamo, M. Dynamic treatment units in eucalyptus plantation. For. Sci. 2011, 57, 416–426. [Google Scholar]
- Pascual, A.; Pukkala, T.; Rodríguez, F.; de-Miguel, S. Using spatial optimization to create dynamic harvest blocks from LiDAR-based small interpretation units. Forests 2016, 7, 220. [Google Scholar] [CrossRef]
- Lamb, S.M.; MacLean, D.A.; Hennigar, C.R.; Pitt, D.G. Forecasting forest inventory using imputed tree lists for LiDAR grid cells and a tree-list growth model. Forests 2018, 9, 167. [Google Scholar] [CrossRef]
- Buchman, R.G.; Shifley, S.R. Guide to evaluating forest growth projection systems. J. For. 1983, 81, 232–254. [Google Scholar]
- Vanclay, J.K.; Skovsgaard, J.P. Evaluating forest growth models. Ecol. Model. 1995, 98, 1–12. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: https://www.R-project.org/ (accessed on 26 August 2018).
- Huang, S.; Meng, S.Y.; Yang, Y. A Growth and Yield Projection System (GYPSY) for Natural and Post-Harvest Stands in Alberta; Technical Report Pub. No.: T/216; Alberta Sustainable Resource Development: Edmonton, AB, Canada, 2009; ISBN 978-0-7785-8486-5. Available online: https://www1.agric.gov.ab.ca/$department/deptdocs.nsf/all/formain15784 (accessed on 10 March 2018).
- Biging, G.S.; Dobbertin, M. A comparison of distance-dependent competition measures for height and basal area growth of individual conifer trees. For. Sci. 1992, 38, 695–720. [Google Scholar]
- Biging, G.S.; Dobbertin, M. Evaluation of competition indices in individual tree growth models. For. Sci. 1995, 41, 360–377. [Google Scholar]
- Validation Summary of GYPSY Sub-Models (“Internal” Validation). Forest Resources Improvement Association of Alberta. Available online: https://www1.agric.gov.ab.ca/$department/deptdocs.nsf/all/formain15784 (accessed on 10 March 2018).
- Woodlot Regeneration. Alberta Department of Agriculture and Forestry. Available online: https://www1.agric.gov.ab.ca/$department/deptdocs.nsf/all/apa3318 (accessed on 27 June 2018).
- Ehlers, S.; Grafström, A.; Nyström, K.; Olsson, H.; Ståhl, G. Data assimilation in stand-level forest inventories. Can. J. For. Res. 2013, 43, 1104–1113. [Google Scholar] [CrossRef]
- Nyström, M.; Lindgren, N.; Wallerman, J.; Grafström, A.; Muszta, A.; Nyström, K.; Bohlin, J.; Willén, E.; Fransson, J.E.S.; Ehlers, S.; et al. Data assimilation in forest inventory: First empirical results. Forests 2015, 6, 4540–4557. [Google Scholar] [CrossRef]
- Babcock, C.; Finley, A.O.; Cook, B.D.; Weiskittel, A.; Woodall, C.W. Modeling forest biomass and growth: Coupling long-term inventory and LiDAR data. Remote Sens. Environ. 2016, 182, 1–12. [Google Scholar] [CrossRef]
- Saarela, S.; Grafström, A. DatAssim: Data Assimilation; R Package Version 1.0. Available online: https://CRAN.R-project.org/package=DatAssim (accessed on 27 June 2018).
- Publication Ethics Statement. Research and Publication Ethics. Remote Sensing—Instructions for Authors. Available online: https://www.mdpi.com/journal/remotesensing/instructions#ethics (accessed on 4 July 2018).
| Species Group | Age, Years | Htop, m | BA, m2/ha | Tvol, m3/ha |
|---|---|---|---|---|
| Aspen | 81 | 19.6 | 32.3 | 269.3 |
| Pine | 112 | 12.9 | 22.8 | 162.5 |
| Black spruce | 117 | 15.4 | 30.6 | 190.4 |
| White spruce | 120 | 21.8 | 36.1 | 304.4 |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vauhkonen, J. Comment on Tompalski et al. Combining Multi-Date Airborne Laser Scanning and Digital Aerial Photogrammetric Data for Forest Growth and Yield Modelling. Remote Sens. 2018, 10, 347. Remote Sens. 2018, 10, 1411. https://doi.org/10.3390/rs10091411
Vauhkonen J. Comment on Tompalski et al. Combining Multi-Date Airborne Laser Scanning and Digital Aerial Photogrammetric Data for Forest Growth and Yield Modelling. Remote Sens. 2018, 10, 347. Remote Sensing. 2018; 10(9):1411. https://doi.org/10.3390/rs10091411
Chicago/Turabian StyleVauhkonen, Jari. 2018. "Comment on Tompalski et al. Combining Multi-Date Airborne Laser Scanning and Digital Aerial Photogrammetric Data for Forest Growth and Yield Modelling. Remote Sens. 2018, 10, 347" Remote Sensing 10, no. 9: 1411. https://doi.org/10.3390/rs10091411
APA StyleVauhkonen, J. (2018). Comment on Tompalski et al. Combining Multi-Date Airborne Laser Scanning and Digital Aerial Photogrammetric Data for Forest Growth and Yield Modelling. Remote Sens. 2018, 10, 347. Remote Sensing, 10(9), 1411. https://doi.org/10.3390/rs10091411




