Abstract
Terrestrial laser scanning (TLS) can produce precise and detailed point clouds of forest environment, thus enabling quantitative structure modeling (QSM) for accurate tree morphology and wood volume allocation. Applying QSM to plot-scale wood delineation is highly dependent on wood visibility from forest scans. A common problem is to filter wood point from noisy leafy points in the crowns and understory. This study proposed a deep 3-D fully convolution network (FCN) to filter both stem and branch points from complex plot scans. To train the 3-D FCN, reference stem and branch points were delineated semi-automatically for 14 sampled areas and three common species. Among seven testing areas, agreements between reference and model prediction, measured by intersection over union (IoU) and overall accuracy (OA), were 0.89 (stem IoU), 0.54 (branch IoU), 0.79 (mean IoU), and 0.94 (OA). Wood filtering results were further incorporated to a plot-scale QSM to extract individual tree forms, isolated wood, and understory wood from three plot scans with visual assessment. The wood filtering experiment provides evidence that deep learning is a powerful tool in 3-D point cloud processing and parsing.
1. Introduction
Modern wood management is a balance of economic activities and environmental stewardships. It is a dynamic system integrating chains of wood production, marketing, conservation, energy, biodiversity, carbon cycling, and climate consequences. It is therefore important to inventory storage and change of wood resources in a timely and comprehensive manner. A typical indicator of wood resources is wood biomass. Estimating wood biomass directly from harvesting or destruction is infeasible given a forest’s massive extent. In practice, regional or national wood biomass is estimated from plot-level surveys of inventory attributes (e.g., diameter and height) extrapolated with remote sensing imagery [1,2]. Plot-level surveys can provide accurate inventory attributes, but most are gross dimension attributes (e.g., the diameter-at-breast-height (DBH) and tree height). Regional or national wood biomass is usually modeled in simple allometric equations using these gross dimension attributes. Using such simple allometric equations, relative root mean squared error (RMSE) of biomass is around 30% [3,4] and absolute error has 50% variation with different tree forms [5]. Large errors are found in branch biomass estimation [6]. With the advent of three-dimensional (3-D) point clouds from Terrestrial laser scanning (TLS), detailed stem and plot-level structural attributes become extractable [7]. Kankare et al. [6] proposes an allometric equation with 20 TLS features for Scots pine and Norway spruce and reported a total relative biomass RMSE of 11.90%. Further efforts, particularly quantitative structure modeling (QSM), present lower relative biomass RMSE (<10%) by modeling biomass simply as a product of wood volume, basic density, and biomass expansion factor (BEF), with volume extracted automatically from TLS point clouds [8,9].
Compared to allometric approaches, the success of QSM depends on whether stems and branches can be delineated. Apparently, QSM is challenged by leafy crown and understory where wood points are hardly separable. A compromise is to limit analyses to deciduous trees in leaf-off condition [8,10,11], or simply ignore the leaf issue because leaf points can be partially filtered out in wood reconstruction or branch skeletonizing models [12,13,1415,16]. The side effect of ignoring leaf points in QSM is overestimation of branch volume [12]. In contrast, many studies recognize the importance of independent filtering algorithm prior to QSM. Several wood filtering algorithms have been proposed, which can be categorized into seeking high laser intensity [9,17,18,19], seeking high point density [9,20,21] or comparing dual-wavelength difference [22,23]. Nevertheless, all existing filtering algorithms rely on ‘hard’ thresholds in filter configuration. Their filtering performance are susceptible to varying scanning scenarios, for example, species and sensor variation, point density attenuation, surrounding shadowing, wind sway, or environmental noise. A robust and versatile wood filtering algorithm is demanded in terms of dataset imperfection.
The complexity of scanning scenarios is a constraint to developing successful wood filtering algorithms. Fortunately, artificial intelligence, especially the revolutionary deep learning network, is tailored to handle complex recognition and filtering problems [24]. In general, deep learning networks comprise multiple layers with millions of parameters that explain complex data structures and achieve high intelligence. Initial parameter values need to be trained in a gradual optimization process where gradients between the predicted and the desired are backpropagated and minimized. The configuration of layers, however, has various forms and depends on application type. A close application to our wood filtering case is image classification, which is also termed image semantic segmentation, under the context of computer vision and machine learning. Correspondingly, the most popular deep learning network is the Fully Convolutional Network (FCN). As the name implies, FCN typically has hierarchical convolutional layers that extract abstract image features in hyperspace, then fully-connected layers that define wall-to-wall mappings between the abstract features and classes, and finally transposed convolution layers that interpolate class predictions to fine resolution. The convolutional layers are also termed Convolutional Neural Network (CNN) layers. Such a layer configuration of FCN has had significant success in practice for the capability of automatic feature selection and high-degree nonlinearity. Different FCN realizations and variations are reviewed in Garcia-Garcia et al. [25].
Applying FCN to 3-D wood filtering has several practical difficulties. First, training an FCN structure with 3-D point clouds is time and storage challenging. So far, the convolution step of FCN from most deep learning programs does not support a discrete point format, and 3-D point clouds need to be voxelized first. However, inputting the FCN structure with all voxels in fine resolution easily leads to computation overflow. This tradeoff between computation load and 3-D information abundance calls for additional transformation of 3-D point data or FCN structure. Existing transformation methods involve projecting 3-D points to multi-view or multi-feature 2-D images [26,27,28], subdividing point clouds into blocks [29,30], or enabling sparse computation of CNN layers [31,32]. Second, a gradient vanishing problem arises. Most voxels are empty voxels because point clouds are sparse. Non-empty voxel values have little effect on network computing, and their influence decreases or even disappears when passed on to deeper layers. Similarly, backpropagation of gradients to shallower layers hardly affects non-empty voxels. This phenomenon is a symptom of gradient vanishing [33], which significantly slow the training process or even lead to training failure. The gradient vanishing problem in 3-D has no sound solution yet, albeit compromises can be replacing CNN with a multi-layer perceptron structure that weakens spatial relationships [34,35]. Third, it is difficult to have either ground truth or manual reference of wood points. The training process of FCN requires a considerable number of samples with correct labels. Yet it is laborious to either obtain ground truth by repetitively scanning a plot with and without leaves or create a manual reference by picking and labeling individual points from dense and noisy scans at branch level.
A 3-D FCN structure was customized in this study, aiming to filter stems and branches from TLS point clouds in a robust and flexible manner. TLS scans were collected with varying scanning and geometric conditions from three common species. An automatic region-growing method was developed in aid of manual reference generation. The filtering results from 3-D FCN were finally input to a QSM model to test application possibility for plot-level wood delineation. From this experimental study, we hope to unveil the potential of applying deep learning methods in complicated 3-D processing tasks, particularly in parsing forest plot scans, in pursuit of automatic and intelligent wood resource management.
2. Data and Sampling
In 2015 and 2016 summers, we collected plot scans for three widespread species in Canada: sugar maple (Acer saccharum) from Vivian Forest (Ontario), trembling aspen (Populus tremuloides) from West Castle (Alberta), and lodgepole pine (Pinus contorta) from Cypress Hills (Saskatchewan). Maple, aspen, and pine plots have an approximate stem density of 400, 200, and 700 ha−1, and average tree height of 17, 12, and 20 m, respectively. All sites were scanned with an Optech ILRIS LR (1064 nm) from plot center and three-to-five corners. The mean point spacing was between 10–20 mm and range accuracy was ±2 mm at a distance of 20 m from the scanner. Laser intensity values ranged from 0–255. For the sampling and evaluating purpose, all our scans were not trimmed in size.
From corner scans, we manually clipped typical areas with at least one tree to be our sample areas. Our training sample included six maple areas, six aspen areas, and two pine areas, whereas our testing sample includes three maple areas, two aspen areas, and two pine areas. We intentionally selected sample with varying tree numbers and scanning conditions, with a quantitative summary in Table 1. We also created a sample quality ranking between ‘1’ and ‘10’, explained in Table 2, indicating how difficult it is to infer inventory details from human interpretation. Point sparsity and occlusion are important reference to our subjective quality ranking, which can be reflected by the high Pearson’s correlation (r) of 0.72 between quality ranking and point spacing, and by the low p value of 0.0003. It is noteworthy that a quality ranking of ‘1’ does not mean perfect quality. All our samples encounter a certain degree of point occlusion, especially in upper crown. Such scan quality imperfection and inconsistency are notorious characteristics of natural forest scans and are where the challenge is. Great quality variation can help develop a robust or universal filtering model to maximize plot data usage.

Table 1.
Quantitative characteristics of each training and testing sample. DBH min-max means minimum and maximum DBH extracted from sample point clouds at a height of 1.3 m. A DBH with 0 value means unmeasurable. Height means vertical distance between lowest and highest points. Point spacing means median nearest neighbor distance. Quality ranking is described in Table 2.

Table 2.
Criteria for sample quality ranking.
All sample areas were labeled with three classes: ‘stem’, ‘branch’, and ‘other’ components. The labeling step was semi-automatic, to be described in Section 3.1. For completeness, stems and branches of understory saplings (height < 4 m) were also labeled in the sample areas. Finally, center scans and corner scans were manually co-registered in preparation for plot-level filtering visualization and tree reconstruction.
3. Methodology
Our experimental design of training, evaluating, and applying a 3-D FCN filtering model in wood reconstruction is presented in Figure 1. The process of generating training and testing reference is described in Section 3.1, specification of our 3-D FCN in Section 3.2, and schemes of plot-level QSM to retrieve tree structure and volume in Section 3.3.
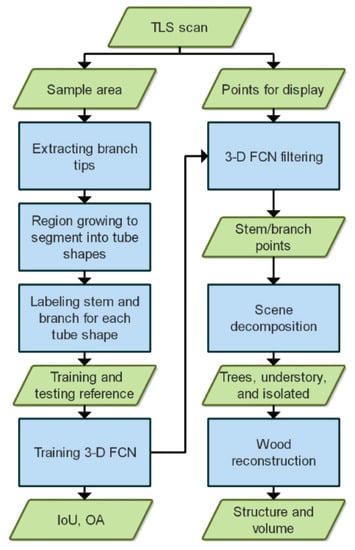
Figure 1.
A framework of wood filtering and reconstruction from terrestrial laser scanning (TLS) plot scans. IoU stands for intersection over union and OA stands for overall accuracy.
3.1. Labeling Reference Points
Per-point labeling of subtle branches, especially in crown and understory areas, can be difficult. This study relied on an automatic segmentation algorithm to help locate tube-shaped branches and filter out noise [36]. Generally, the algorithm started from selecting a few core points from point clouds, which iteratively incorporated neighboring points and eventually grew to be tube-shaped regions. Major requirements of the region-growing routine were to constrain growth direction and tube width changes. As a result, most points were segmented into tube-shaped regions, while other remaining points especially from leaves and ground had bulky shapes. The tube-shaped regions were extracted based on their three-dimensional features from principal components analysis (PCA) [37]: any regions with linearity >0.4 and sphericity <0.3. The extracted regions were then exported to CloudCompare Open Source Software for further editing. Specifically, the tube-shaped regions were manually trimmed and refined to represent clean wood segments. Wood segments, with thick long shape, near-vertical direction, tight connection to base part, or high laser intensity, were preferably recognized as stems. The remaining wood segments were thus intended to be branches. We labeled stems as class 2, branches as class 3, and other points as class 1.
The above-stated region-growing algorithm was an iterative process, meaning that later region-growing steps were affected by errors in earlier steps. It was therefore critical to ensure correctness of initial region-growing steps. For example, locating initial region-growing points should avoid areas with ambiguous shapes, such as joints between branches. Our experiments suggested that region-growing initialized from branch tip points would yield most correct shapes and cause less conflict with following region-growing steps. We then proposed a simple method of locating branch tips, to improve the initialization step in Xi et al. [36]. First, for operational convenience, all points were converted to voxels at a resolution of 5 × 5 × 5 cm3 (xyz). A metric termed mean curvature was used to describe the spatial local sharpness (Figure 2). Specifically, for each voxel, we calculated its unit vectors directing to all neighboring voxels. The average of those unit vectors was a vector (red arrow in Figure 2), the direction and amplitude of which represented surface normal and sharpness degree, respectively. This amplitude value was indeed a discrete expression of surface mean curvature [38]. Using surface mean curvature, branch tips were coarsely located where a voxel’s mean curvature exceeded 0.75. These voxels representing branch tips were regarded as initial region-growing points. Each branch tip was searched in turn to grow initial regions, until no tips were available. Then any arbitrary point was selected to continue region-growing until no points were selectable. Both region-growing and initialization algorithms were programmed in MATLAB.
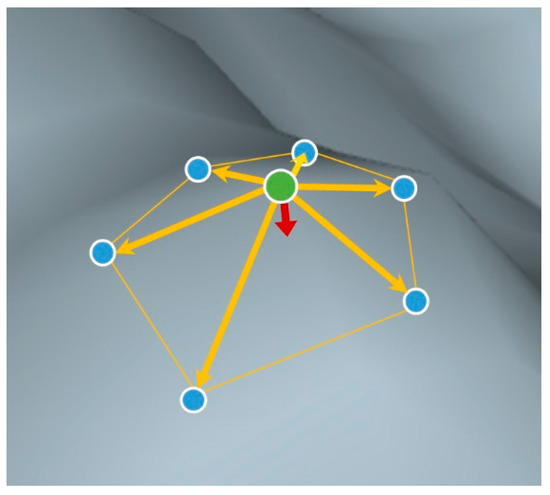
Figure 2.
A sketch of surface normal and surface mean curvature (red arrow), calculated as average of unit vectors (yellow arrow) pointing from center point (green dot) to neighboring points (blue dots).
3.2. Configuring 3-D FCN
Our 3-D FCN model and evaluation steps were programmed in Tensorflow (GPU edition). We adopted the point cloud subdivision approach [29,30] and limited the model input to be a block of 128 × 128 × 128 voxels, with a voxel size of 5 × 5 × 5 cm3. Larger block size and smaller voxel size were desired but could cause computing overflow problems. We cut raw point clouds evenly into blocks and then shuffled blocks randomly before the training process. Each voxel was assigned with three attributes, derived from summary statistics of all points within that voxel. The first attribute was binary, with 1 meaning point occupancy and 0 otherwise. The second attribute was average of laser intensity values, whereas the third attribute was average of point heights (z value). The final FCN input was a 128 × 128 × 128 × 3 block. In addition, the class label of a voxel was calculated as the dominant label from all points within the voxel. If a voxel was empty, its class was set to be 0. Hence, the total class number was four.
Our FCN structure (Figure 3) was customized based on the FCN-VGG-16 structure described in Long et al. [39]. In addition to expanding 2-D layers to a 3-D version, we changed the fusion strategy between transposed convolution (TC) and low-level pooling, shown by the plus sign in Figure 3. The initial method was to fuse 1st TC results with 4th pooling results, 2nd TC with 3rd pooling, and so on. Our strategy was fusing 1st TC directly with 3rd pooling results, and 2nd TC with 2nd pooling. The omission of fusing 4th pooling results was intended to reduce blurring degree on final prediction. The final prediction output a 128 × 128 × 128 × 4 block, with last dimension denoting the probability of each class. The final class label was predicted as the maximum value in the last dimension of the probability block.
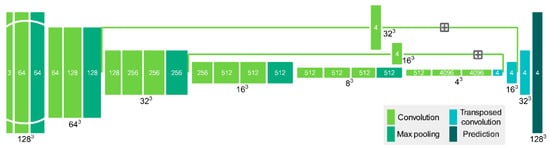
Figure 3.
Configuration of 3-D FCN structure. Colored bars denote different types of network layers, black numbers outside bar denote input size in 3-D domain, white numbers inside bar denote input size in feature domain, and gray plus sign denote addition of layer outputs.
We trained our model from scratch, because existing 2-D pretrained models were hardly adaptable to our scenes and data structure. Our convolution filter size was 3 × 3 × 3, and dropout ratio was 0.5. The weights of convolution and fully-connected layers were initialized using default values in Tensorflow, and the weights of transposed convolution layer were based on trilinear interpolation. To train our 3-D FCN model, errors (or disagreement) between probability block and reference, namely loss function, were measured using a mean softmax cross entropy metric [40]. Nevertheless, directly minimizing this loss function could encounter the gradient vanishing issue. Our solution was to calculate loss function only based on non-empty voxels in the probability block. We used an Adam optimizer [41] with an initial step of 10−4 to iteratively minimize the loss function. It was observed that loss function from only non-empty voxels could always descend to a minimum level, albeit the convergence process was slow and variable.
Prediction accuracy (or filtering accuracy) was defined as an agreement between predicted and referenced labels for this study. Three accuracy metrics were adopted: Overall Accuracy (OA), Intersection over Union (IoU) per class, and its mean value (mIoU) [25]. The OA divided the total number of matched labels between prediction and reference by the total number of points. It represented an overall matching degree. The IoU was class-specific, which divided the total number of matched labels by the total number of labels for one class. The mIoU was an average of IoUs among all classes. Comparing to OA, mIoU weighed per-class accuracy equally, and therefore less influenced by the accuracy of dominant classes. The metrics of OA, IoU, or mIoU all ranged from 0–1, with higher value indicating greater agreement.
3.3. Reconstructing 3-D Tree Geometries
We designed a follow-up wood reconstruction model (or QSM) at plot-level to showcase a potential application value of our 3-D FCN. The results of FCN filtering were labelled voxels. We extracted stem points from stem voxels, and branch points from branch voxels. The non-wood points were discarded, as well as the height and intensity attributes from wood points. Only 3-D coordinates of wood points were reserved for the reconstruction model. The wood reconstruction modeling (or QSM) was a top-to-bottom and bottom-to-top process: decomposing the plot to individual trees, extracting nodes from branch or stem points, connecting nodes along separate branches or stems, and finally connecting branches to stems. Success of all reconstruction steps relied significantly on clean filtering of stems and branches from the 3-D FCN. All wood points were assumed to be tube-shaped.
At the plot level, we used a simple region-growing algorithm to decompose plots into individual trees. Region growing process started from an arbitrary point, kept incorporating its buffer points within 0.1 m, and terminated when no buffer points were found. Results were isolated components of branches and stems. We applied a simple rule to merge isolated stem components into complete individual stems. Any two stem components were identified as being from the same stem if one stem component was completely above the other, and the upper components fell within five times stem radius of the lower component. Then, with individual stems identified, branch components were assigned to their nearest stem, together to form an individual tree. We limited the nearest distance between branch components and stem to be no greater than 2 m, otherwise the branch components would be identified as isolated branches. We also identified any branches not higher than 4 m from the ground as understory wood. Those branches were not assigned to their nearest stem. Consequently, a plot of wood points was decomposed into individual trees, isolated branches, and understory wood.
At the individual-tree level, stem and branch points were segmented respectively using the same region-growing algorithm as described in Section 3.1, except for filtering tubes based on linearity and sphericity criteria. The resulting grown regions were evenly sliced along their main axis at 1 cm intervals, to form a sequence of nodes representing the tube. The main axis direction was estimated from PCA [36], and each node location was at the center of the slice. At each node location, we applied a circle-fitting algorithm for its neighboring points [42]. The search radius of neighboring points was set to be three times the tube width retrieved from the region-growing process. The fitted circle center became the new node location, the circle radius became node radius, and the circle plane normal becomes the node direction.
At the individual-stem or branch level, nodes were connected based on a weighted similarity score (Equation (1)) utilizing distance , angle , and radius . Figure 4 has intuitive illustration of these geometric terms, with meaning distance between two nodes, included angle between node direction and node connection, and node radius difference. Any large , or can lead to a low similarity score. The weight of each term was customizable, and this study empirically set and to approximate consistent and smooth branch curves. The process of node connection started with an arbitrary node, searched its neighboring nodes within 0.1 m, connected nodes with highest similarity scores, and repeated search and connection, until no node is available. Nodes with were discarded to avoid turn-around of connection lines. We then reversed the direction of the starting node and performed the same connection process, so that both sides of starting nodes could be searched and connected. The entire connection process produced individual stems and branches.
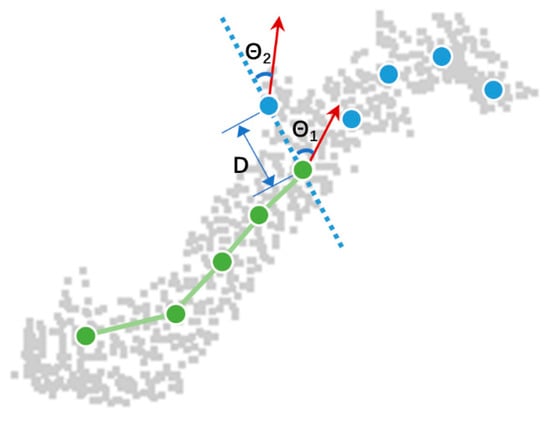
Figure 4.
An illustration of similarity score composition. Gray points are example of scanned branch points, green dots are connected nodes, blue dots are candidate nodes to a target node, is distance between target node and one of the candidate nodes, and is included angle between their connection (dashed line) and node direction (red arrow). For clarity, fitted circles around each node are not shown here.
At the individual-tree level, each branch, only represented by its starting and ending nodes, was connected to the stem or another branch based on the similarity score shown in Equation (2). Comparing to Equation (1), no penalty was required for radius difference or direction difference between target and connection line. The connection process was iterative. First, branches nearest to the stem were connected to a stem node with highest similarity score. This branch-to-stem connection shaped a general 3-D tree form. Then, remaining branches nearest to the tree form were connected to it based on a similarity score, and so forth, until no branches were available. The resulting tree form was further refined by combining similar branches (Figure 5). Any two branches with similar branch directions (<50° difference) would join at their shared node, if the joint branch were to be longer than both branches.
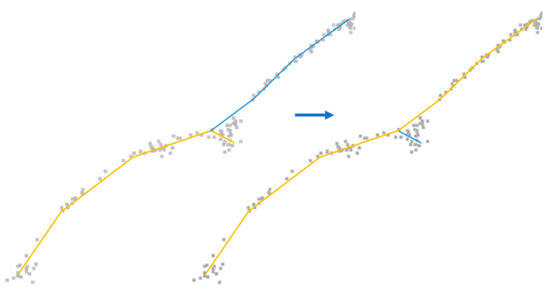
Figure 5.
Example of branch connection refinement. Gray dots are scanned branch points, yellow and blue lines are connection lines. The right panel shows a more natural way of connection than the left.
Returning to the plot level, in addition to individual trees, isolated branches and understory wood were simply reconstructed following the steps of node connection. We did not apply branch-to-branch connection to avoid an excessive number of plausible connection lines. Wood volumes of individual trees, isolated branches and understory were all computed. All the QSM parameter settings were the same for different plots.
4. Results and Discussion
4.1. Reference Creation
Results of branch tip extraction from ‘aspen 3’ are shown as white dots in Figure 6a. Initialized with these branch tips, our region-growing algorithm can segment point clouds mostly into tube shapes. Those segments are assigned random colours in Figure 6b to visualize differences. After segment-wise PCA filtering, manual selection and minor trimming, the resultant wood reference is displayed in Figure 6c. The stem and branch points after splitting are shown in green and red, respectively, whereas other points are in blue (Figure 6d). Two trees are covered in this sample area, with only one tree exposing its main stem. Both trees contain many ghost points, particularly near the crown area, due to the difficulty of interpretation and trimming. These are typical quality issues in preparing reference data which hinder deep learning algorithms from detecting branches correctly. Yet eliminating reference errors simply from human interference is limited and costly. It is anticipated in the future that a benchmarking reference dataset could be developed for an algorithm optimizing purpose where scene complexity and data quality could be finely controlled.
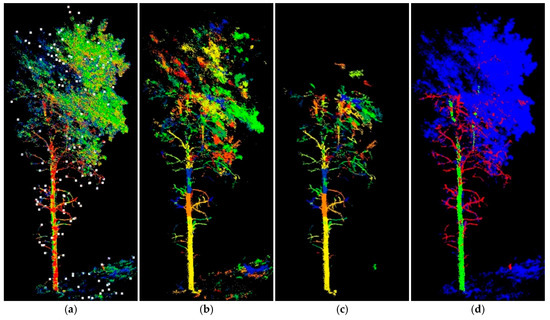
Figure 6.
Example of preparing aspen reference sample (‘aspen 3’): (a) clipped point clouds colored by laser intensity, overlaid with branch tip extraction in white dots; (b) region-growing segments in random colors; (c) segments with only stem and branches; and (d) reference tree with stem, branch, and other components classified in green, red, and blue colors, respectively. This sample area covers two trees: one exhibiting detailed stem form and the other showing only partial upper stem due to occlusion.
4.2. 3-D FCN Filtering and Evaluation
Figure 7a,b tracks accuracy changes during the training process for a training sample (aspen) and a testing sample (maple). A total of 60,000 training iterations are presented, corresponding to a running time of 30.19 h using a computer with Intel® Core™ i7-6700K 8 × 4.00 GHz, 32 GB RAM and NVIDIA GeForce GTX 1070. As iteration increases, both training sample and testing sample show an increasing trend for mIoU and OA, with only one or two abrupt slumps, indicating our 3-D FCN is a global optimization process. Note that an IoU or OA of 1.0 means perfect prediction. After 30,000 iterations, mIoU and OA of training sample converges to 0.98, indicating the strong fitting ability of the 3-D FCN. On the other hand, mIoU and OA of testing sample levels off to a limit of 0.76 and 0.96, respectively, after 10,000 iterations. This result indicates that the generalizability of FCN would saturate and that the later learning phase after 10,000 iterations may only focus on classifying marginal features.
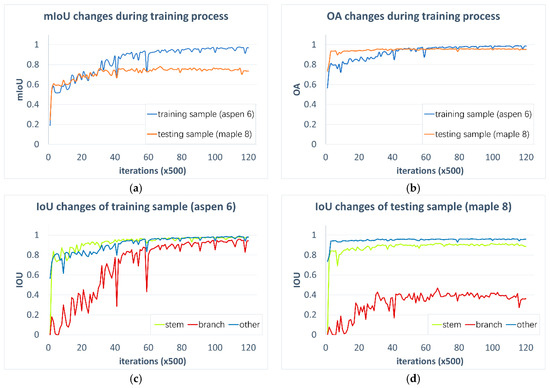
Figure 7.
Relationship between accuracy changes and training iterations, for ‘aspen 6’ from training dataset and ‘maple 8’ from testing dataset: (a,b) fluctuation of mIoU and OA; (c,d) fluctuation of stem IoU, branch IoU, and other components’ IoU.
The testing sample has a high OA limit but relatively lower mIoU limit. This result indicates that the majority of wood points are filtered accurately, but some classes may be predicted incorrectly. Class-wise IoU changes for the training and testing samples are thus examined in Figure 7c,d. An increasing trend is shown for all classes of both training and testing samples. Apparently, branch points, due to geometric complexity, have slower and lower convergence values than stem and other points. Therefore, the FCN model can identify simple classes in the early phase, but learning difficult classes takes much longer and may not achieve the same high accuracy. The testing sample has a low branch IoU convergence of 0.4, which also explains the low mIoU in Figure 7a. The branch IoU issue can be caused by imperfect branch reference and complex branch structure, which should be mitigated by adding training samples, or refining the deep learning model with higher spatial resolution and stronger feature extraction ability.
Filtered wood points and reference can be visually contrasted using examples in Figure 8a–c. The main wood points are recognized and most difference occur in crown branches in Figure 8a,c, understory stems in Figure 8a and ghost points in Figure 8b,c. The classified wood appears slightly fatter than reference wood partly due to the setting of 5 cm voxel resolution.

Figure 8.
Visualization of reference trees (left of each panel) and filtered trees (right of each panel) from testing dataset, including (a) ‘maple 7’, (b) ‘aspen 8’, and (c) ‘pine 4’. Per-class IoU is denoted on top of each panel. Stem, branch, and other points are in green, red, and blue colors, respectively.
The final accuracies of all training and testing samples are provided in Table 3. An average of stem IoU, branch IoU, ‘other’ IoU, mIoU, and OA is 0.90, 0.72, 0.96, 0.86, and 0.97 for the training sample, respectively, and 0.89, 0.54, 0.93, 0.79, and 0.94 for the testing sample, respectively. The mIoU shows no significant difference (p = 0.15) between training and testing samples based on a two-sample t test, implying that deep learning model is generalizable. The mIoU difference between the three species is also insignificant (p = 0.16) based on an ANOVA test using all samples. This result presents a possibility of having a universal wood filtering solution without need for exhaustive sampling of species.

Table 3.
3-D FCN filtering accuracies (IoU, mIoU, and OA) among training and testing samples.
Not all IoU results from Table 3 are satisfactory. Many factors could contribute to the low IoU situation, e.g. geometrical complexity and details. Results from a left-tailed t test show that branch IoU is significantly lower than stem IoU (p < 0.001), probably due to complex forms of branches. More details, denoted by a smaller point spacing, would lead to a lower mIoU. This statement can be indicated by a high Pearson’s r of 0.51 between point spacing from Table 1 and mIoU from Table 3. Similarly, worse quality ranking (Table 1) tends to produce higher branch IoU (r = 0.39) and lower stem IoU (r = −0.21). As quality ranking descends, stem form becomes fragmentary or blank at a rising risk of misclassification, whereas branches become simplified and conducive to classification. However, a clear understanding of IoU variation requires systematic investigation of model configuration, reference uncertainty and sample sufficiency, which is beyond the scope of this study.
Attribution of input voxels plays a vital role in our 3-D FCN filtering accuracy. As Table 4 shows, with only point occupancy, the 3-D FCN model can still achieve an impressive accuracy, for example, a stem IoU of 0.922 for the ‘aspen 6’. It is clear that local spatial pattern is mostly relied by the 3-D FCN filter. Adding the intensity attribute shows a slight improvement over accuracy. By contrast, introducing the height attribute provides leads to a greater accuracy increase, because branch is height-dependent.

Table 4.
Effect of voxel attribution on 3-D FCN filtering accuracies (per-class IoU, mIoU, and OA) at the 25,000th iteration.
Traditionally, wood points (branch + stem) can be filtered efficiently based on simple metrics such as intensity threshold. The accuracy of applying an intensity threshold filter to our sample is examined in Figure 9. We represent wood filtering accuracy with the ‘other’ IoU metric. For each sample, ‘other’ IoU is plotted against a range of intensity thresholds, and the highest ‘other’ IoU is marked with a red triangle. It is obvious that no universal threshold exists that can promise the highest wood filtering accuracy for all the samples. The upper boundary of ‘other’ IoU from intensity filter also vary drastically between 0.5 and 0.9. In comparison, ‘other’ IoU from our FCN filter, denoted by the blue starred line, is drawn in Figure 9. An overwhelming advantage of using FCN filter is presented, considering the accuracy level and stability among all the samples.
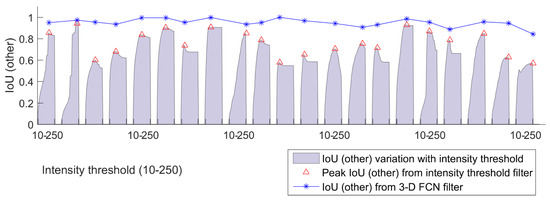
Figure 9.
IoU (other) of all the 21 training and testing samples from two filters. The area plot in lavender grey shows IoU (other) variation from an intensity filter based on ‘hard’ thresholds (10–250), with peak IoU (other) in red triangle. The blue starred line shows IoU (other) from a 3-D FCN filter.
After the training process, the 3-D FCN model can be directly applied to filter stem and branch points from entire plots, based on a moving block of 128 × 128 × 128 voxels. Our plot scan has an average file size of about 1 GB. Filtering an aspen plot of about 500 trees requires 3.12 hrs, a maple plot of 1000 trees 1.95 hrs, and a pine plot of 2000 trees 2.23 hrs.
4.3. Wood Reconstruction
Individual wood reconstruction results are illustrated in Figure 10 with ‘aspen 7’ provided as an example. Branch and stem curves are reconstructed separately from FCN-filtered branch and stem points, and branch curves are finally connected to the stem curve. Shown in Figure 10, our node connection can overcome a certain degree of stem occlusion in the crown area, and the slice-based circle fitting algorithm is tolerant to some branch noise in the top of the canopy. The extracted stem volume is 0.45 m3, and the branch volume 0.22 m3. Replacing FCN prediction with reference labels and using the same QSM settings produces a stem volume of 0.38 m3, and a branch volume of 0.23 m3. Based on a wood density of 0.35 g cm−3 [43], the resulting stem biomass and branch biomass from FCN-QSM are 159 kg and 77 kg, respectively, and from reference-QSM are 133 kg and 80 kg, respectively. The ground truth of biomass is not available in this study. A referable biomass can be allometric biomass [44] based on inventory information of species, sites, DBH, and height. The allometric stem biomass (including bark) is 149 kg and allometric branch biomass 26 kg. In this case, the branch biomass from QSM is probably overestimated. The causes can be noise and ghost points (as indicated in Figure 10) occurring during scanning collection, reference preparation and voxelization. However, considering all 21 sample areas, the mean branch biomass from FCN-QSM is only 15% of allometric branch biomass, due to high degree of branch occlusion in most sample areas. For branch biomass, the relative root-mean-square-error (RMSE%) and r2 between FCN-QSM and allometric approach are 130% and 0.05, respectively, comparing to the RMSE% of 19.6% and r2 of 0.95 for stem biomass.
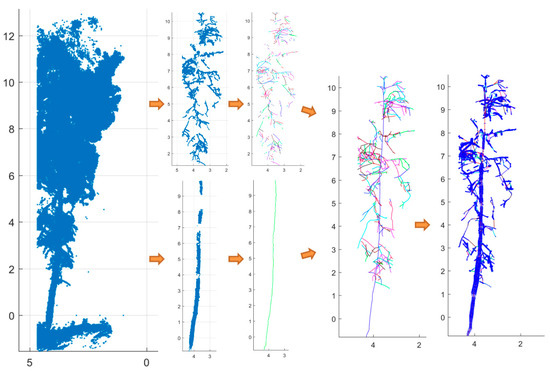
Figure 10.
Illustration of individual-tree QSM process for ‘aspen 7’. From left to right are raw points, stem and branch points filtered from the 3-D FCN model, stem and branch curve in random colors, tree curve connection in random colors, and tree curve overlaid with cross-section circles.
An example of plot-level wood reconstruction results is visualized in Figure 10. It is the maple plot, with FCN-filtered components (stem, branch and other) in different colours in Figure 11a. Its wood reconstructed stem, branch, isolated, and understory are also assigned with different colours in Figure 11b. The plot scan comprises five corner scans and one center scan, stretching over 100 m distance from plot center. In our maple plot, the farthest reconstructed tree is 148 m away from the plot center. However, a problem is also apparent in that reconstructing farther woods is strongly affected by a lower signal-noise ratio. Availability of both stem and branch points descends rapidly with the distance from the plot center. The reconstructed maple plot has 80% of stem volumes within a distance of 44 m, and 80% of branch volumes within 26 m. The individual tree branch-to-stem volume ratio also descends, from 0.15 within 10 m, to 0.13 within 25 m, to 0.05 within the entire plot. To mitigate the weak wood signal problem, we consider 25 m as an effective plot size for evaluating plot-level wood reconstruction. The reconstructed wood volume of maple plot has a composition of 92.3% stem, 6.5% branch, 0.3% isolated, and 0.9% understory. Correspondingly, the reconstructed aspen plot has 81.2% stem, 17.6% branch, 0.05% isolated, and 1.2% understory, and the pine plot has 91.5% stem, 8.4% branch, 0.03% isolated, and 0.1% understory. The average branch-to-stem ratio of individual tree with branch detected is 0.15 for maple, 0.24 for aspen, and 0.13 for pine. According to Ung and colleagues’ [45] biomass database, the average branch-to-stem ratio and its standard deviation should be 0.27 ± 0.24 for sugar maple, 0.14 ± 0.14 for trembling aspen, and 0.15 ± 0.12 for lodgepole pine. Comparing to maple and aspen in leaf-on conditions, lodgepole pine has better visibility of wood points, leading to less bias of branch-to-stem ratio. A future study should involve rigid evaluation and calibration of occlusion effect on QSM’s volume and branch-to-stem ratio calculation.
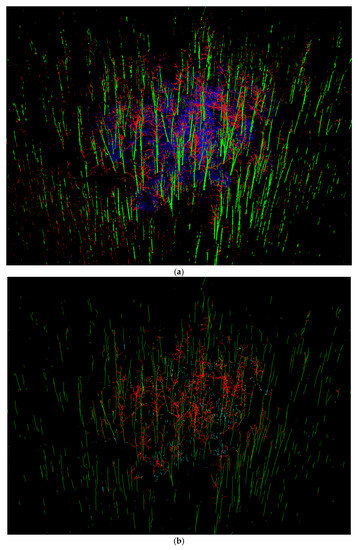
Figure 11.
Example wood reconstruction from a maple plot (1 ha): (a) a 3-D FCN filtered plot scan, with stem in green, branch in red, and other points in blue; (b) QSM reconstructed wood curves, with stem in green, branch in red, understory in cyan, and isolated wood in yellow. To balance the level of graphic details, blue points in (a) are assigned with transparency degree of 0.99, and stem curves are set four times thicker than all other curves.
Finally, it should be clarified that reconstruction results were not validated, due to lack of ground truth measurement of branch curve and volume. The parameters of reconstruction model are also not optimized, for the same reason of lacking accurate reference. Yet one function of this QSM model can be exposing detailed wood components, and reflecting any issues of integrating filtering and QSM for natural forest scans. The delineated tree curves can also provide high-level geometric features, in terms of raw point metrics, to help develop more accurate allometric models of wood volume or biomass.
5. Conclusions
Delineating wood components in complex natural forest environments has attained wide research interest, represented by QSM development, due to its importance to modern wood management in pursuit of correct and complete biomass, carbon budget, and tree physiology. At the plot level, the wood amount in dense and noisy areas such as crown and understory are however, uncharted or gross in common QSM models. Recent QSMs incorporate simple wood filtering methods aimed to reduce noise. In contrast, this study introduces the use of a deep learning model FCN in 3-D space to filter both stem and branch points from complex forest scans, with an average testing accuracy of 0.89 (stem IoU), 0.54 (branch IoU), 0.79 (mIoU), and 0.94 (OA) over three plots. We further explored the potential application of wood filtering in constructing a plot-level QSM and extracting the wood volume component. From visual inspection, wood filtering generally produces tube shapes beneficial to QSM modeling.
Some challenges remain, however. Creating reference data, although assisted with a semi-automation algorithm, inevitably contains interpretation errors and further induces low branch filtering accuracy. Besides, point cloud is a discrete form of 3D data, but continuous voxel form is the only supported choice in deep learning tools. As a result, coarse voxel resolution was fixed to tradeoff deep learning accuracy with sufficient computation efficiency. Last but not least, wood filtering cannot address the most severe problem of wood delineation, which is occlusion. Improving TLS scan coverage and QSM modeling, although sophisticated, are still the most reliable solutions to overcome occlusion.
Deep learning modeling offers significant potential to intellectualize processing and analysis in various data-driven applications. This study only intends to probe the feasibility of filtering 3-D wood points of interest, without activating diverse options of refinement. For example, the instability issue of branch accuracy can be mitigated by larger sample sizes, more classes, and broader species. Filtering low-quality scans, particularly from needle-leaf species (e.g., pine and spruce), remains a challenge. Tentative refinement can be introducing more complex network structures such as ResNet [46], Inception [47,48], and CRF [49], sparse point network such as PointNet [34,35], instance segmentation network such as R-CNN [50,51]. However, refinement and surgery of the all-purpose deep networks are not optimal solutions to many practical 3-D problems. It is strongly hoped that deep network mechanisms can be rationalized systematically in the future, with regard to how data structure, network architecture, sample property, and problem dimension are interrelated and delimited, before optimal practice is achieved.
Author Contributions
Conceptualization, C.H.; Methodology, Z.X.; Formal Analysis, Z.X.; Investigation, C.H., L.C., and Z.X.; Data Curation, C.H., L.C., and Z.X.; Writing-Original Draft Preparation, Z.X.; Writing-Review & Editing, C.H.; Supervision, C.H.
Funding
This research was supported by the S.G.S. International Tuition Award and the Dean’s Scholarship from the University of Lethbridge, Campus Alberta Innovates Program (CAIP), and NSERC Discovery Grants Program.
Acknowledgments
The authors sincerely value advice from Stewart Rood and Derek Peddle at the University of Lethbridge. Zhouxin Xi would appreciate inspiration and motivation from Yuchu Qin at the Institute of Remote Sensing and Digital Earth (RADI), Chinese Academy of Sciences (CAS), and an additional proofreading from Andrew Boyd at the Cambridge Proofreading.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Blackard, J.A.; Finco, M.V.; Helmer, E.H.; Holden, G.R.; Hoppus, M.L.; Jacobs, D.M.; Lister, A.J.; Moisen, G.G.; Nelson, M.D.; Riemann, R.; et al. Mapping U.S. forest biomass using nationwide forest inventory data and moderate resolution information. Remote Sens. Environ. 2008, 112, 1658–1677. [Google Scholar] [CrossRef]
- Beaudoin, A.; Bernier, P.; Guindon, L.; Villemaire, P.; Guo, X.; Stinson, G.; Bergeron, T.; Magnussen, S.; Hall, R. Mapping attributes of canada’s forests at moderate resolution through k nn and modis imagery. Can. J. For. Res. 2014, 44, 521–532. [Google Scholar] [CrossRef]
- Hauglin, M.; Gobakken, T.; Astrup, R.; Ene, L.; Næsset, E. Estimating single-tree crown biomass of norway spruce by airborne laser scanning: A comparison of methods with and without the use of terrestrial laser scanning to obtain the ground reference data. Forests 2014, 5, 384–403. [Google Scholar] [CrossRef]
- Lindberg, E.; Holmgren, J.; Olofsson, K.; Olsson, H. Estimation of stem attributes using a combination of terrestrial and airborne laser scanning. Eur. J. For. Res. 2012, 131, 1917–1931. [Google Scholar] [CrossRef]
- Melson, S.L.; Harmon, M.E.; Fried, J.S.; Domingo, J.B. Estimates of live-tree carbon stores in the pacific northwest are sensitive to model selection. Carbon Balance Manag. 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Kankare, V.; Holopainen, M.; Vastaranta, M.; Puttonen, E.; Yu, X.; Hyyppä, J.; Vaaja, M.; Hyyppä, H.; Alho, P. Individual tree biomass estimation using terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2013, 75, 64–75. [Google Scholar] [CrossRef]
- Hopkinson, C.; Chasmer, L.; Young-Pow, C.; Treitz, P. Assessing forest metrics with a ground-based scanning lidar. Can. J. For. Res. 2004, 34, 573–583. [Google Scholar] [CrossRef]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Hackenberg, J.; Wassenberg, M.; Spiecker, H.; Sun, D. Non destructive method for biomass prediction combining tls derived tree volume and wood density. Forests 2015, 6, 1274–1300. [Google Scholar] [CrossRef]
- Raumonen, P.; Casella, E.; Calders, K.; Murphy, S.; Åkerblom, M.; Kaasalainen, M. Massive-scale tree modelling from TLS data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, 2, 189–196. [Google Scholar] [CrossRef]
- Yan, D.-M.; Wintz, J.; Mourrain, B.; Wang, W.; Boudon, F.; Godin, C. Efficient and robust reconstruction of botanical branching structure from laser scanned points. In Proceedings of the 2009 11th IEEE International Conference on Computer-Aided Design and Computer Graphics, Huangshan, China, 19–21 August 2009; pp. 572–575. [Google Scholar]
- Burt, A.; Disney, M.; Raumonen, P.; Armston, J.; Calders, K.; Lewis, P. Rapid characterisation of forest structure from tls and 3d modelling. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS 2013), Melbourne, Australia, 21–26 July 2013; pp. 3387–3390. [Google Scholar]
- Bayer, D.; Seifert, S.; Pretzsch, H. Structural crown properties of Norway spruce (Picea abies [L.] Karst.) and European beech (Fagus sylvatica [L.]) in mixed versus pure stands revealed by terrestrial laser scanning. Trees 2013, 27, 1035–1047. [Google Scholar] [CrossRef]
- Livny, Y.; Yan, F.; Olson, M.; Chen, B.; Zhang, H.; El-Sana, J. Automatic reconstruction of tree skeletal structures from point clouds. ACM Trans. Graph. 2010, 29. [Google Scholar] [CrossRef]
- Xu, H.; Gossett, N.; Chen, B. Knowledge and heuristic-based modeling of laser-scanned trees. ACM Trans. Graph. 2007, 26. [Google Scholar] [CrossRef]
- Pfeifer, N.; Gorte, B.; Winterhalder, D. Automatic reconstruction of single trees from terrestrial laser scanner data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2004, 35, 114–119. [Google Scholar]
- Côté, J.-F.; Widlowski, J.-L.; Fournier, R.A.; Verstraete, M.M. The structural and radiative consistency of three-dimensional tree reconstructions from terrestrial lidar. Remote Sens. Environ. 2009, 113, 1067–1081. [Google Scholar] [CrossRef]
- Béland, M.; Baldocchi, D.D.; Widlowski, J.L.; Fournier, R.A.; Verstraete, M.M. On seeing the wood from the leaves and the role of voxel size in determining leaf area distribution of forests with terrestrial lidar. Agric. For. Meteorol. 2014, 184, 82–97. [Google Scholar] [CrossRef]
- Côté, J.F.; Fournier, R.A.; Egli, R. An architectural model of trees to estimate forest structural attributes using terrestrial lidar. Environ. Model. Softw. 2011, 26, 761–777. [Google Scholar] [CrossRef]
- Hackenberg, J.; Morhart, C.; Sheppard, J.; Spiecker, H.; Disney, M. Highly accurate tree models derived from terrestrial laser scan data: A method description. Forests 2014, 5, 1069–1105. [Google Scholar] [CrossRef]
- Raumonen, P.; Kaasalainen, M.; Akerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast automatic precision tree models from terrestrial laser scanner data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef]
- Danson, F.M.; Gaulton, R.; Armitage, R.P.; Disney, M.; Gunawan, O.; Lewis, P.; Pearson, G.; Ramirez, A.F. Developing a dual-wavelength full-waveform terrestrial laser scanner to characterize forest canopy structure. Agric. For. Meteorol. 2014, 198, 7–14. [Google Scholar] [CrossRef]
- Zhan, L.; Douglas, E.; Strahler, A.; Schaaf, C.; Xiaoyuan, Y.; Zhuosen, W.; Tian, Y.; Feng, Z.; Saenz, E.J.; Paynter, I.; et al. Separating leaves from trunks and branches with dual-wavelength terrestrial lidar scanning. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS 2013), Melbourne, Australia, 21–26 July 2013; pp. 3383–3386. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, A.; Orts-Escolano, S.; Oprea, S.; Villena-Martinez, V.; Garcia-Rodriguez, J. A Review on Deep Learning Techniques Applied to Semantic Segmentation. 2017. Available online: https://arxiv.org/abs/1704.06857 (accessed on 7 June 2018).
- Boulch, A.; Saux, B.L.; Audebert, N. Unstructured Point Cloud Semantic Labeling Using Deep Segmentation Networks. 2017. Available online: http://www.boulch.eu/files/2017_3dor-point.pdf (accessed on 7 June 2018).
- Lawin, F.J.; Danelljan, M.; Tosteberg, P.; Bhat, G.; Khan, F.S.; Felsberg, M. Deep Projective 3D Semantic Segmentation. 2017. Available online: https://link.springer.com/chapter/10.1007/978-3-319-64689-3_8 (accessed on 7 June 2018).
- Yang, Z.; Jiang, W.; Xu, B.; Zhu, Q.; Jiang, S.; Huang, W. A convolutional neural network-based 3D semantic labeling method for ALS point clouds. Remote Sens. 2017, 9, 936. [Google Scholar] [CrossRef]
- Huang, J.; You, S. Point Cloud Labeling Using 3D Convolutional Neural Network. 2016. Available online: http://graphics.usc.edu/cgit/publications/papers/point_cloud_3dcnn.pdf (accessed on 7 June 2018).
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Graham, B. Sparse 3D Convolutional Neural Networks. 2015. Available online: https://arxiv.org/abs/1505.02890 (accessed on 7 June 2018).
- Riegler, G.; Ulusoys, A.O.; Geiger, A. OctNet: Learning Deep 3D Representations at High Resolutions. 2016. Available online: http://openaccess.thecvf.com/content_cvpr_2017/papers/Riegler_OctNet_Learning_Deep_CVPR_2017_paper.pdf (accessed on 7 June 2018).
- Hochreiter, S.; Bengio, Y.; Frasconi, P.; Schmidhuber, J. Gradient Flow In Recurrent Nets: The Difficulty Of Learning Long-Term Dependencies. 2001. Available online: https://pdfs.semanticscholar.org/2e5f/2b57f4c476dd69dc22ccdf547e48f40a994c.pdf (accessed on 7 June 2018).
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. Pointnet: Deep Learning on Point Sets for 3D Classification and Segmentation. 2016. Available online: http://openaccess.thecvf.com/content_cvpr_2017/papers/Qi_PointNet_Deep_Learning_CVPR_2017_paper.pdf (accessed on 7 June 2018).
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. Pointnet++: Deep Hierarchical Feature Learning on Point Sets in a Metric Space. 2017. Available online: http://papers.nips.cc/paper/7095-pointnet-deep-hierarchical-feature-learning-on-point-sets-in-a-metric-space (accessed on 7 June 2018).
- Xi, Z.; Hopkinson, C.; Chasmer, L. Automating plot-level stem analysis from terrestrial laser scanning. Forests 2016, 7, 252. [Google Scholar] [CrossRef]
- Blomley, R.; Weinmann, M.; Leitloff, J.; Jutzi, B. Shape distribution features for point cloud analysis—A geometric histogram approach on multiple scales. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2014. [Google Scholar] [CrossRef]
- Sorkine, O. Laplacian Mesh Processing. 2005. Available online: https://pdfs.semanticscholar.org/3ae1/e9b3e39cc8c6e51ef9a36954051845e18d3c.pdf (accessed on 7 June 2018).
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. 2015. Available online: https://www.cv-foundation.org/openaccess/content_cvpr_2015/html/Long_Fully_Convolutional_Networks_2015_CVPR_paper.html (accessed on 7 June 2018).
- Bridle, J.S. Probabilistic Interpretation of Feedforward Classification Network Outputs, with Relationships to Statistical Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 1990; pp. 227–236. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. 2015. Available online: https://arxiv.org/pdf/1412.6980.pdf (accessed on 7 June 2018).
- Gander, W.; Golub, G.H.; Strebel, R. Least-squares fitting of circles and ellipses. BIT Numer. Math. 1994, 34, 558–578. [Google Scholar] [CrossRef]
- Zanne, A.E.; Lopez-Gonzalez, G.; Coomes, D.A.; Ilic, J.; Jansen, S.; Lewis, S.L.; Miller, R.B.; Swenson, N.G.; Wiemann, M.C.; Chave, J. Data from: Towards a worldwide wood economics spectrum. Dryad Data Repos. 2009. [Google Scholar] [CrossRef]
- Lambert, M.; Ung, C.; Raulier, F. Canadian national tree aboveground biomass equations. Can. J. For. Res. 2005, 35, 1996–2018. [Google Scholar] [CrossRef]
- Ung, C.H.; Lambert, M.C.; Raulier, F.; Guo, J.; Bernier, P.Y. Biomass of Trees Sampled Across Canada as Part of the Energy from the Forest Biomass (ENFOR) Program. 2017. Available online: https://open.canada.ca/data/en/dataset/fbad665e-8ac9-4635-9f84-e4fd53a6253c (accessed on 10 January 2018).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 8–10 June 2015; pp. 1–9. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI-17) AAAI, San Francisco, CA, USA, 4–9 February 2017; pp. 4278–4284. [Google Scholar]
- Zheng, S.; Jayasumana, S.; Romera-Paredes, B.; Vineet, V.; Su, Z.; Du, D.; Huang, C.; Torr, P.H. Conditional random fields as recurrent neural networks. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 1529–1537. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; pp. 91–99. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).