Tree Species Classification of the UNESCO Man and the Biosphere Karkonoski National Park (Poland) Using Artificial Neural Networks and APEX Hyperspectral Images
Abstract
1. Introduction
- Are hyperspectral data and artificial neural networks useful for mapping tree species?
- What are the differences between forest inventory and Airborne Prism Experiment (APEX) derived tree species compositions of forest growing in Karkonoski National Park (KNP)?
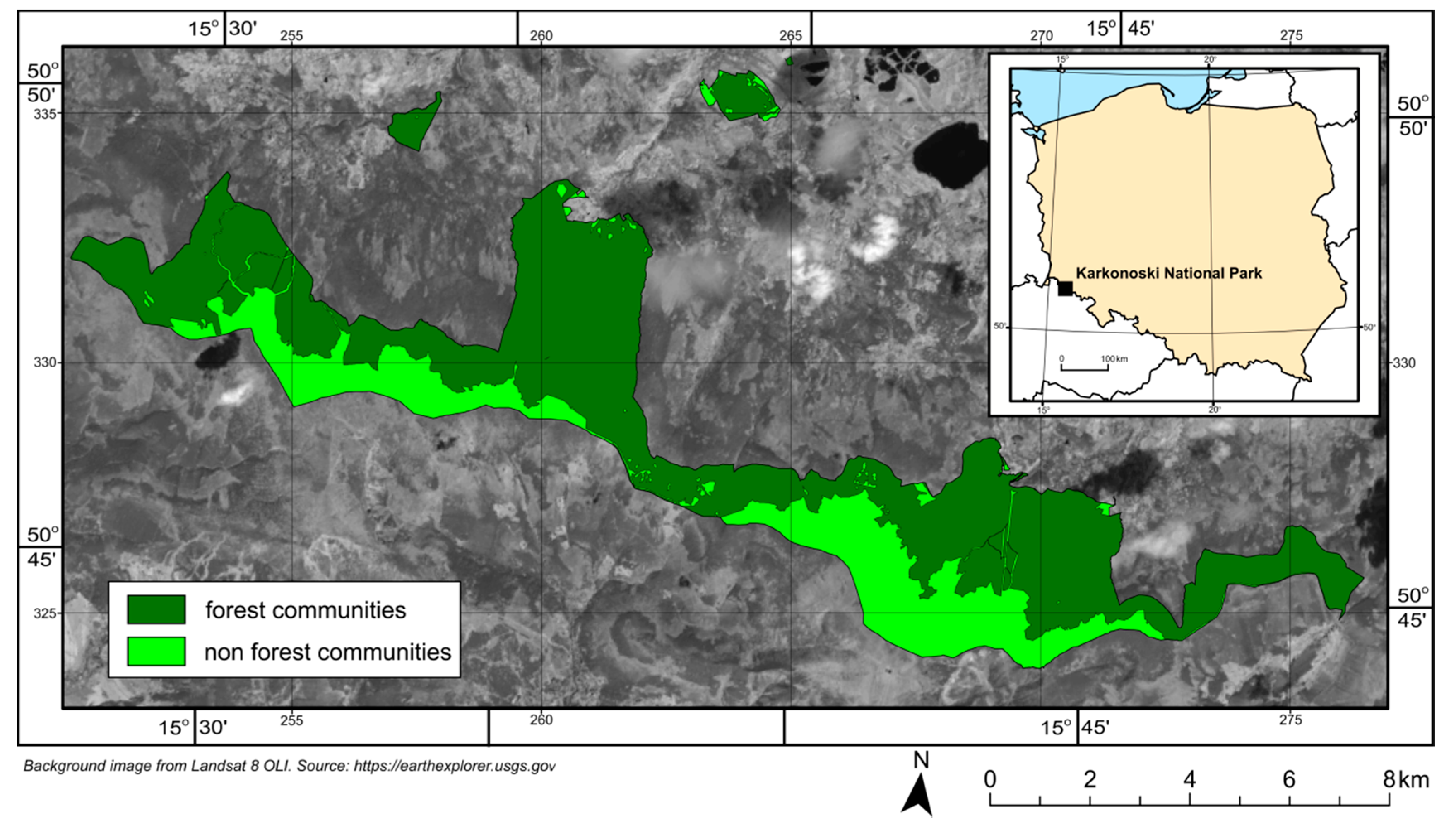
2. Research Area
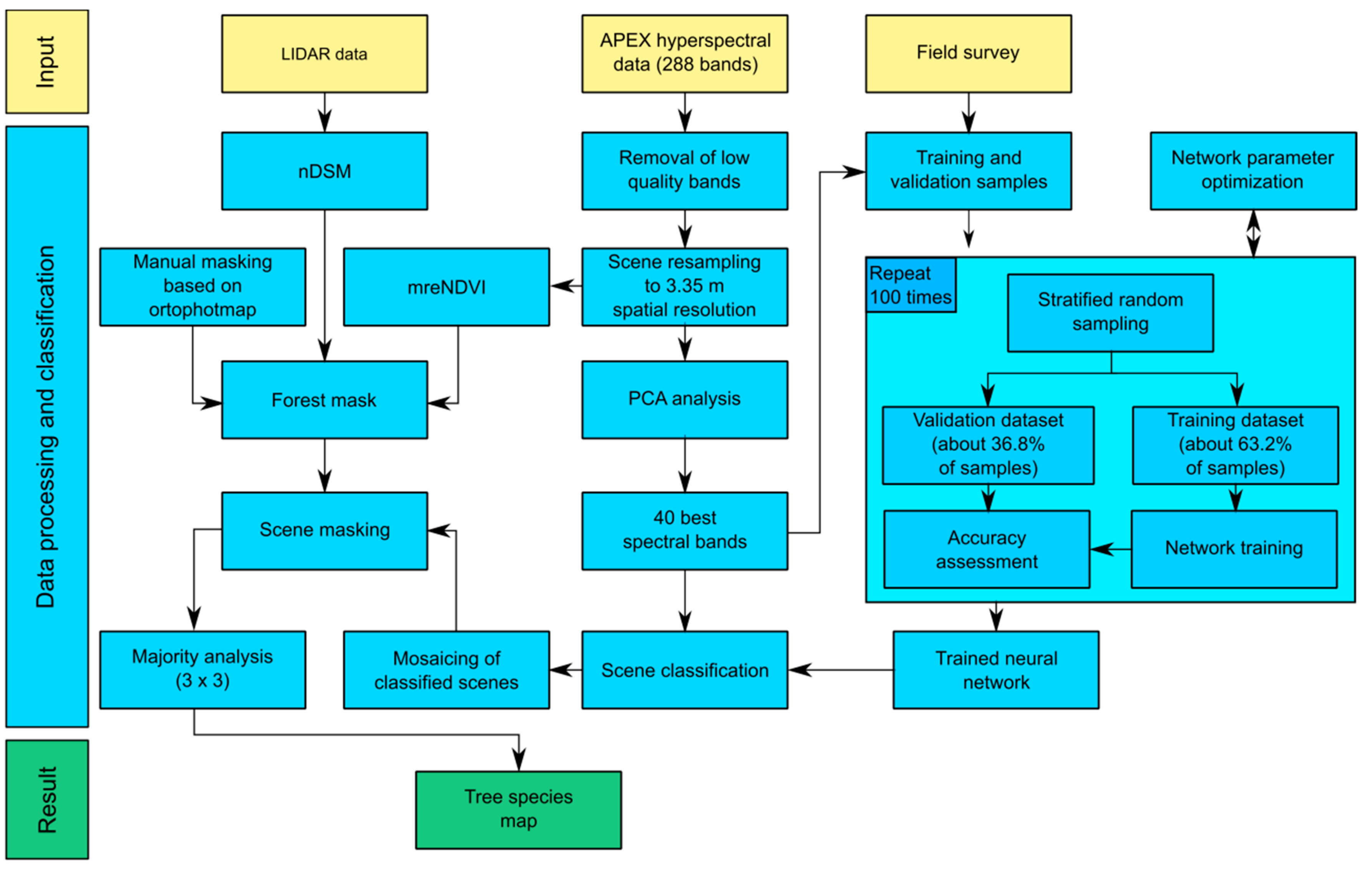
3. Materials and Methods
- Acquisition of APEX hyperspectral data and LiDAR data.
- Creation of normalized digital surface model (nDSM) using LiDAR data.
- Acquisition of a large and representative sample of geographic locations of field-identified tree species during field surveys. Collected data were used to extract classified tree species spectral characteristics from images.
- Band selection using principal component analysis (PCA).
- Creation of forest mask used to limit classification scope only to forested areas.
- Optimization of ANN learning parameters.
- Assessment of iterative accuracy.
- Classification of APEX scenes using trained and validated ANNs.
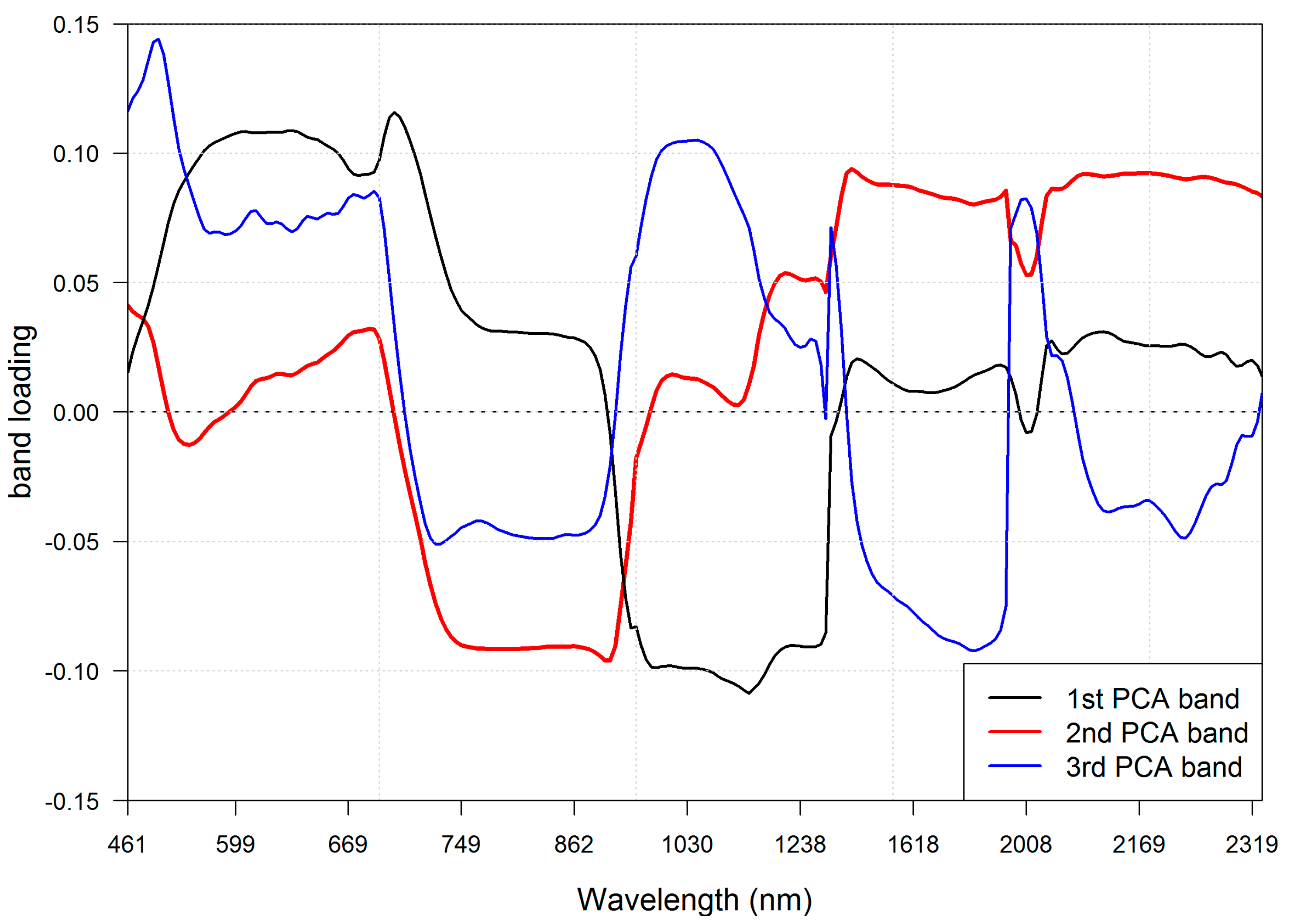
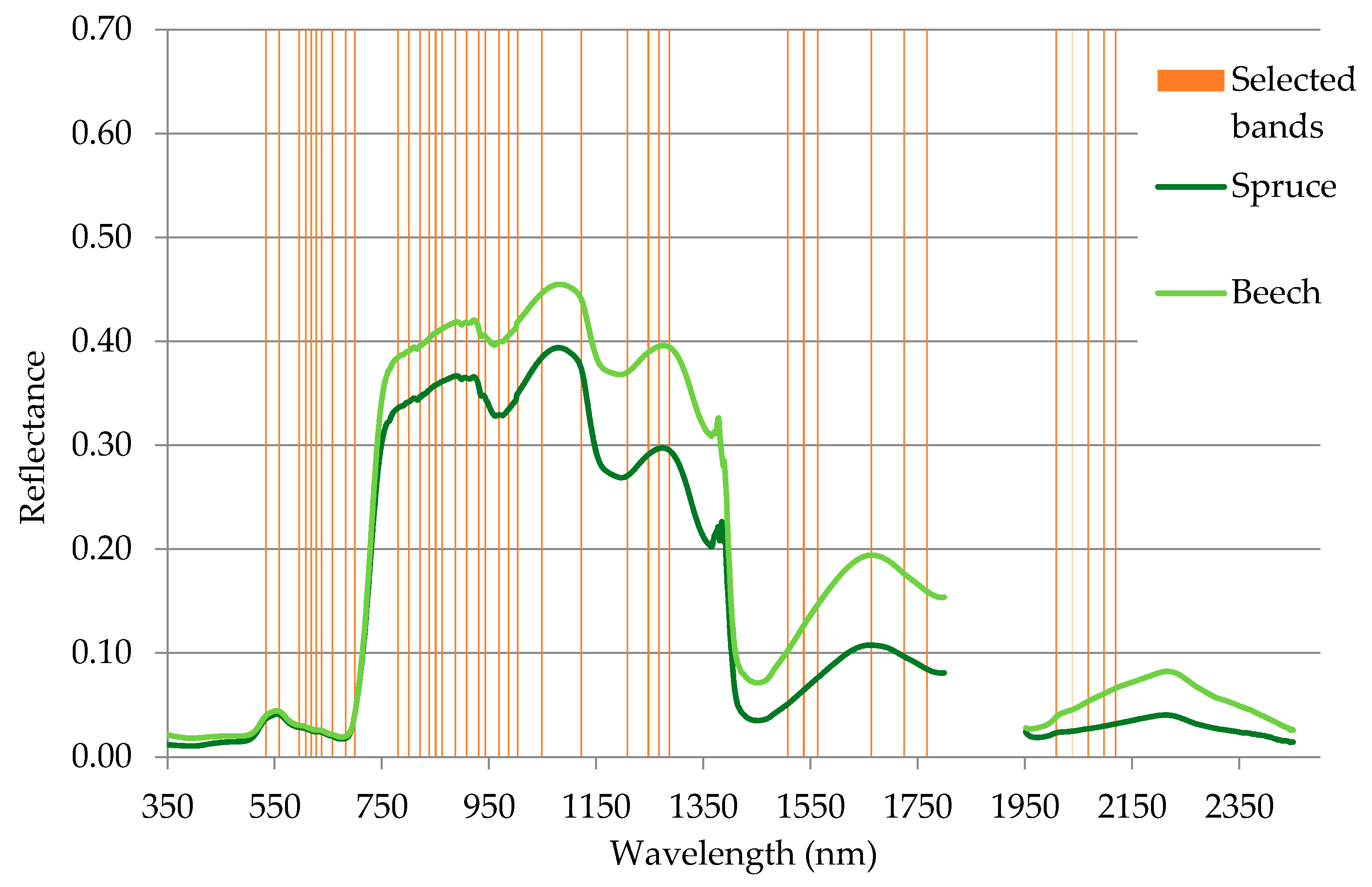
3.1. Band Selection
3.2. Field Data Collection
3.3. Artificial Neural Network Parameter Tuning
3.4. Iterative Accuracy Assessment
3.5. Forest Mask Creation
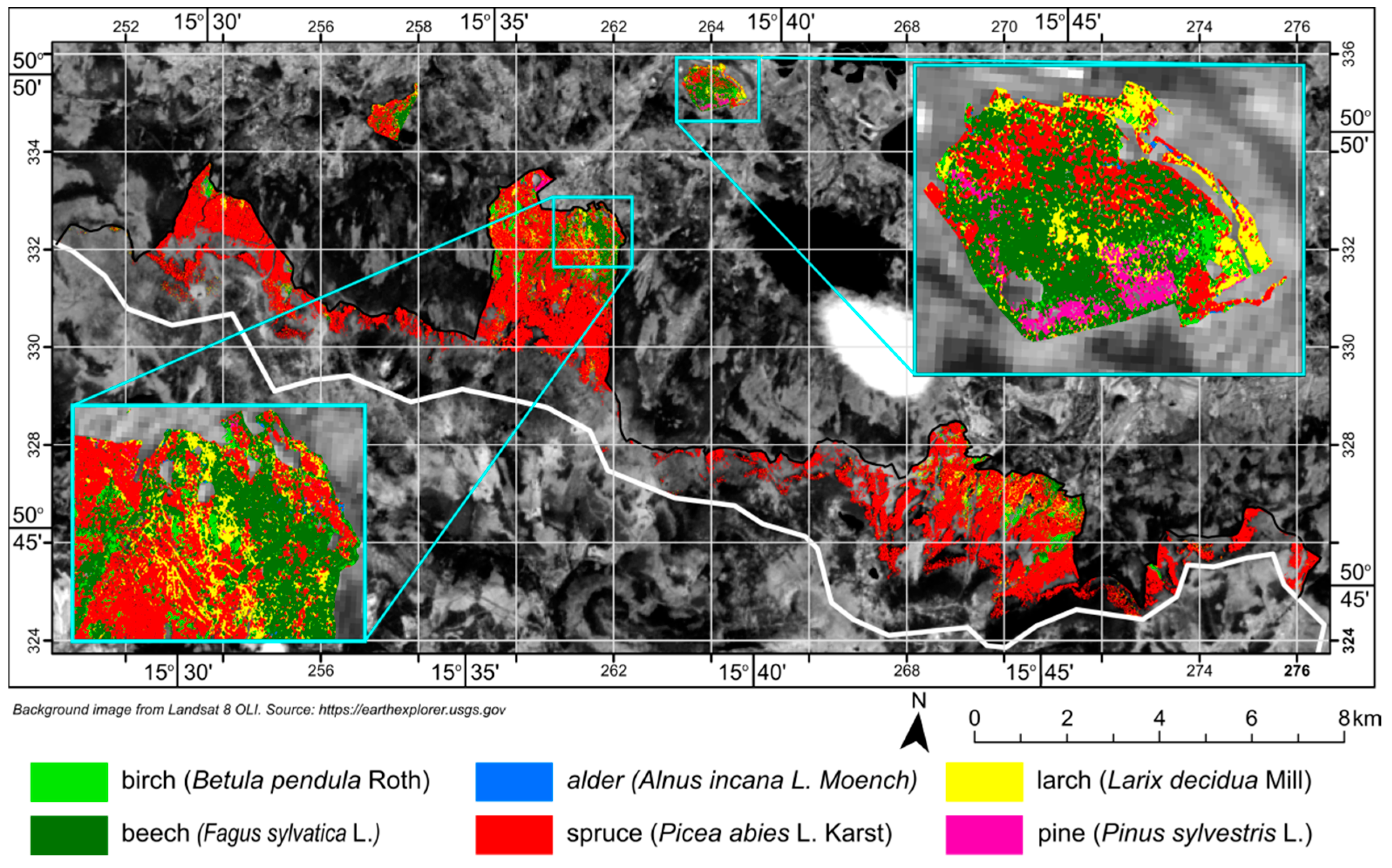
3.6. Scene Classification
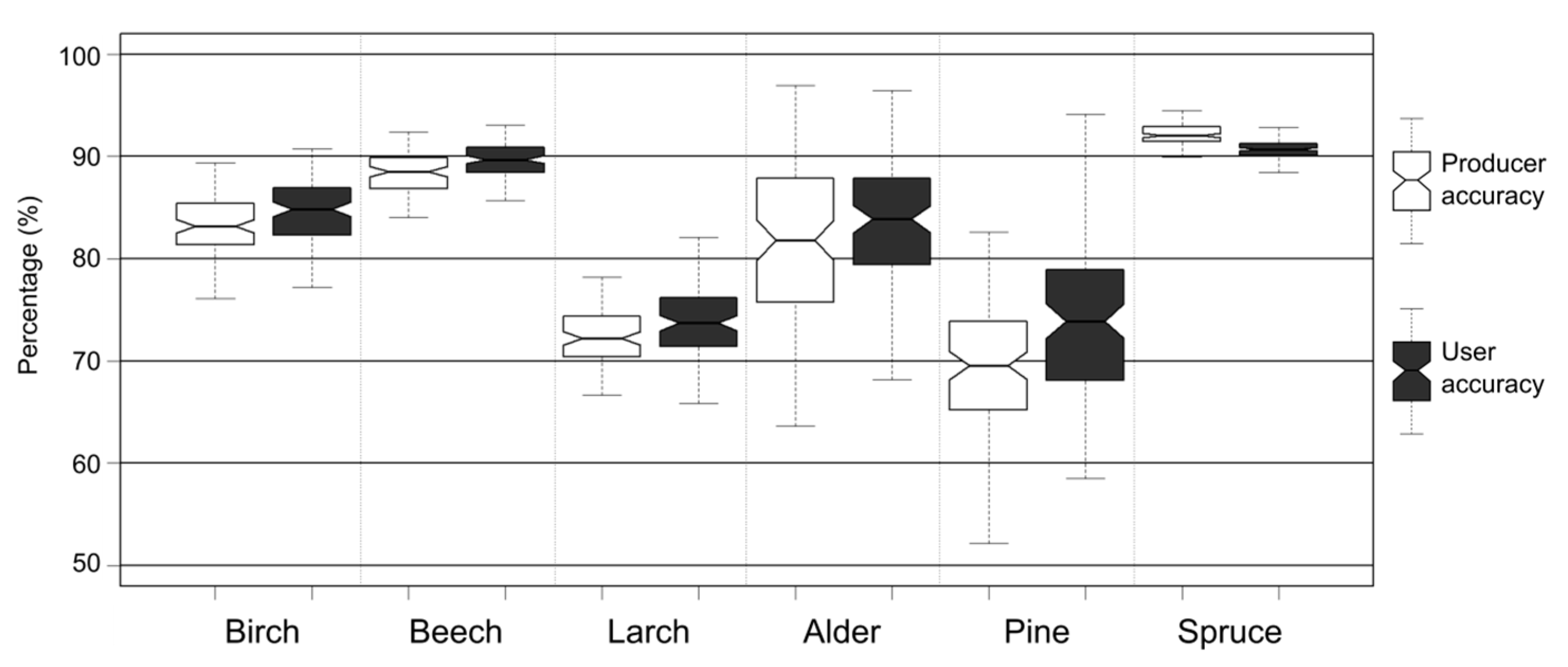
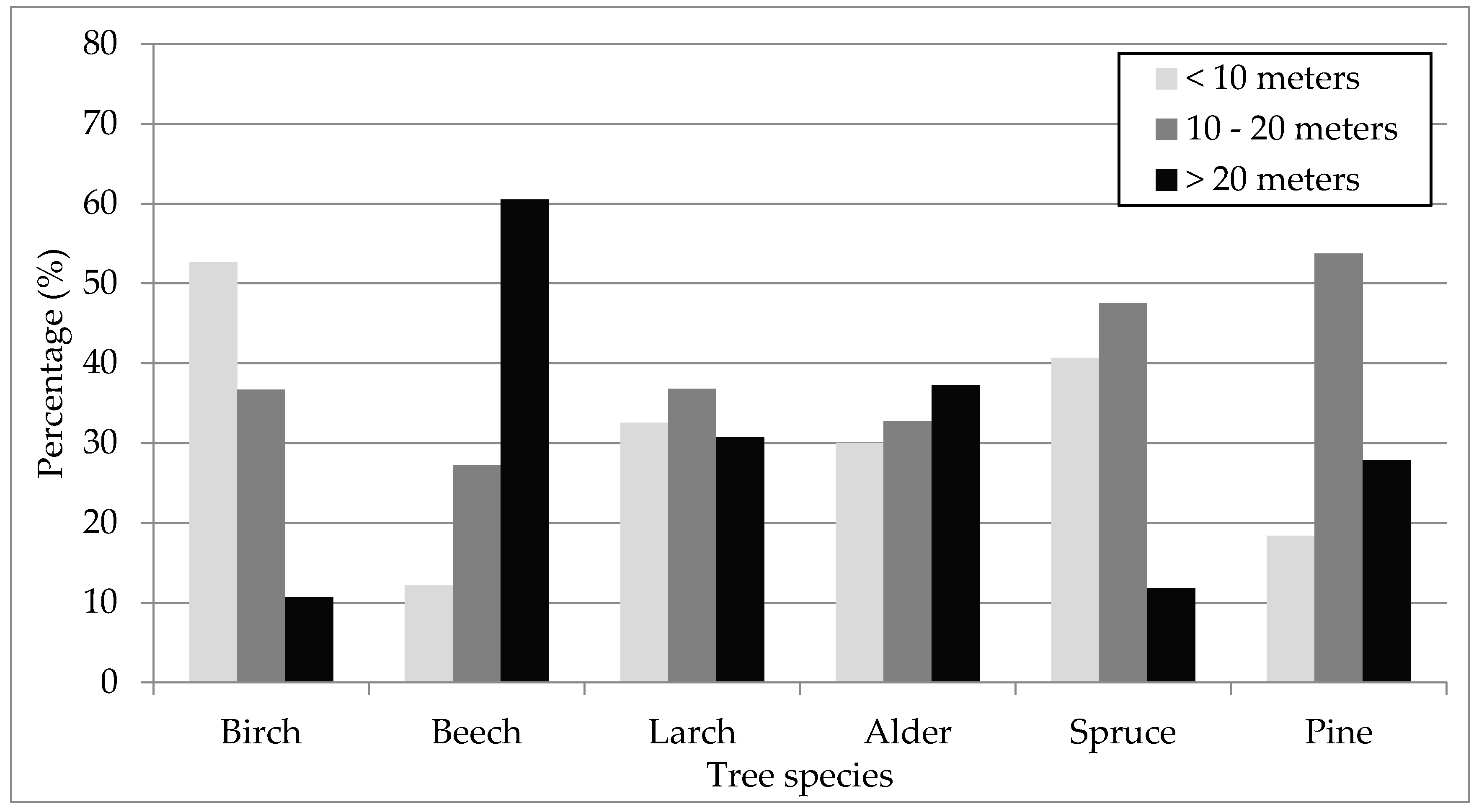
4. Results
5. Discussion
6. Conclusions
- Despite some difficulty in classifying protected areas (due to the less orderly occurrence of species as compared to commercial forests), the classification achieved an overall accuracy of 87%. This allows the usefulness of hyperspectral data to be confirmed for studies of protected and difficult-to-access areas.
- Hyperspectral data are undoubtedly useful in classifying tree species in protected areas. A large number of very narrow spectral channels allows tree species to be successfully distinguished, although the wealth of data requires an answer to the question of which data to use.
- The results allowed the dominant tree species in the research area (spruce, beech) to be classified effectively, as well as species that occur there far less frequently (larch, pine, birch, alder).
- The algorithm used to select hyperspectral channels allowed us to select the 40 most informative bands. Using the entire dataset and ANNs would have resulted in very long training times, which would have seriously hindered the optimization of learning parameters and the assessment of accuracy. Proper band selection is particularly important when analyzing large areas using advanced data-processing techniques.
- Instead of relying on a one-time training and validation process, it was decided to randomly select multiple sets of samples for training and validation of the results. This method allows unintended influence by the researcher on the results to be reduced and frees the researcher from the tedious process of selecting samples for the training and validation set. Moreover, the accuracy assessment results can be better understood. Its use not only provides information on measures of accuracy, but also allows changes in accuracy and their impact on training and validation sets to be observed. One of the method’s downsides is the requirement to repeat the training and validation procedure a number of times, which translates to longer processing times. There remain open questions about the spatial autocorrelation of randomly-selected pixels, which can bias results.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shen, G.; Sakai, K.; Hoshino, Y. High Spatial Resolution Hyperspectral Mapping for Forest Ecosystem at Tree Species Level. Agric. Inf. Res. 2010, 19, 71–78. [Google Scholar] [CrossRef]
- Peerbhay, K.; Mutanga, O.; Ismail, R. Commercial tree species discrimination using airborne AISA Eagle hyperspectral imagery and partial least squares discriminant analysis (PLS-DA) in KwaZulu–Natal, South Africa. ISPRS J. Photogramm. Remote Sens. 2013, 79, 19–28. [Google Scholar] [CrossRef]
- Martinez del Castillo, E.; García-Martin, A.; Longares Aladrén, L.; de Luis, M. Evaluation of forest cover change using remote sensing techniques and landscape metrics in Moncayo Natural Park (Spain). Appl. Geogr. 2015, 62, 247–255. [Google Scholar] [CrossRef]
- Martin, M.; Newman, S.; Aber, J.; Congalton, R. Determining forest species composition using high spectral resolution remote sensing data. Remote Sens. Environ. 1998, 65, 249–254. [Google Scholar] [CrossRef]
- Bergseng, E.; Ørka, H.; Næsset, E.; Gobakken, T. Assessing forest inventory information obtained from different inventory approaches and remote sensing data sources. Ann. For. Sci. 2015, 72, 33–45. [Google Scholar] [CrossRef]
- Jarocińska, A.M.; Kacprzyk, M.; Marcinkowska-Ochtyra, A.; Ochtyra, A.; Zagajewski, B.; Meuleman, K. The application of APEX images in the assessment of the state of non-forest vegetation in the Karkonosze Mountains. Misc. Geogr. 2016, 20, 21–27. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.; Carranza, L.; Martinez, P.; Houcheime, M.; Sinca, F.; Weiss, P. Spectroscopy of canopy chemicals in humid tropical forests. Remote Sens. Environ. 2011, 115, 3587–3598. [Google Scholar] [CrossRef]
- Zagajewski, B.; Tømmervik, H.; Bjerke, J.W.; Raczko, E.; Bochenek, Z.; Kłos, A.; Jarocińska, A.; Lavender, S.; Ziółkowski, D. Intraspecific Differences in Spectral Reflectance Curves as Indicators of Reduced Vitality in High-Arctic Plants. Remote Sens. 2017, 9, 1289. [Google Scholar] [CrossRef]
- Kycko, M.; Zagajewski, B.; Zwijacz-Kozica, M.; Cierniewski, J.; Romanowska, E.; Orłowska, K.; Ochtyra, A.; Jarocińska, A. Assessment of Hyperspectral Remote Sensing for Analyzing the Impact of Human Trampling on Alpine Swards. Mt. Res. Dev. 2017, 37, 66–74. [Google Scholar] [CrossRef]
- Kycko, M.; Zagajewski, B.; Lavender, S.; Romanowska, E.; Zwijacz-Kozica, M. The Impact of Tourist Traffic on the Condition and Cell Structures of Alpine Swards. Remote Sens. 2018, 10, 220. [Google Scholar] [CrossRef]
- Guidici, D.; Clark, M.L. One-Dimensional Convolutional Neural Network Land-Cover Classification of Multi-Seasonal Hyperspectral Imagery in the San Francisco Bay Area, California. Remote Sens. 2017, 9, 629. [Google Scholar] [CrossRef]
- Kupková, L.; Červená, L.; Suchá, R.; Jakešová, L.; Zagajewski, B.; Březina, S.; Albrechtová, J. Classification of Tundra Vegetation in the Krkonoše Mts. National Park Using APEX, AISA Dual and Sentinel-2A Data. Eur. J. Remote Sens. 2017, 50, 29–46. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Raczko, E.; Ochtyra, A.; Jarocińska, A. Classification of High-Mountain Vegetation Communities within a Diverse Giant Mountains Ecosystem Using Airborne APEX Hyperspectral Imagery. Remote Sens. 2018, 10, 570. [Google Scholar] [CrossRef]
- Price, J.C. How unique are spectral signatures? Remote Sens. Environ. 1994, 49, 181–186. [Google Scholar] [CrossRef]
- Masaitis, G.; Mozgeris, G.; Augustaitis, A. Spectral reflectance properties of healthy and stressed coniferous trees. iForest 2013, 6, 30–36. [Google Scholar] [CrossRef]
- Cochrane, M.A. Using vegetation reflectance variability for species level classification of hyperspectral data. Int. J. Remote Sens. 2000, 10, 2075–2087. [Google Scholar] [CrossRef]
- Sommer, C.; Holzwarth, S.; Heiden, U.; Heurich, M.; Mueller, J.; Mauser, W. Feature-based tree species classification using airborne hyperspectral and LiDAR data in the Bavarian Forest National Park. EARSeL eProc. 2015, 14, 49–70. [Google Scholar]
- Tagliabue, G.; Panigada, C.; Colombo, R.; Fava, F.; Cilia, C.; Baret, F.; Vreys, K.; Meuleman, K.; Rossini, M. Forest species mapping using airborne hyperspectral APEX data. Misc. Geogr. 2016, 20, 28–33. [Google Scholar] [CrossRef]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Graves, S.J.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Colgan, M.S.; Kalantari, L.; Bohlman, S.A. Tree species abundance predictions in a tropical agricultural landscape with a supervised classification model and imbalanced data. Remote Sens. 2016, 8, 161. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Gobakken, T.; Gianelle, D.; Naesset, E. Tree species classification in boreal forests with hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2013, 51, 2632–2645. [Google Scholar] [CrossRef]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree Species Classification Using Hyperspectral Imagery: A Comparison of Two Classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef]
- Lee, J.; Cai, X.; Dalponte, M.; Schonlieb, C.; Coomes, D.; Lellmann, J.; Malhi, Y.; Butt, N.; Morecroft, M. Individual Tree Species Classification from Airborne Multisensor Imagery Using Robust PCA. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 2554–2567. [Google Scholar] [CrossRef]
- Mas, J.F.; Flores, J.J. The application of artificial neural networks to the analysis of remotely sensed data. Int. J. Remote Sens. 2008, 29, 617–663. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, X.; Jia, X. Spectral–Spatial Classification of Hyperspectral Data Based on Deep Belief Network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2381–2392. [Google Scholar] [CrossRef]
- Paoletti, M.; Haut, M.; Plaza, J.; Plaza, A. A new deep convolutional neural network for fast hyperspectral image classification. ISPRS J. Photog. Remote Sens. 2017. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of Support Vector Machine, Random Forest and Neural Network Classifiers for Tree Species Classification on Airborne Hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef]
- Beluco, A.; Engel, P.; Alexandre, B. Classification of textures in satellite image with Gabor filters and a multilayer perceptron with back propagation algorithm obtaining high accuracy. Int. J. Energy Environ. 2015, 6, 437–459. [Google Scholar] [CrossRef]
- Jain, A.K.; Duin, R.P.W.; Mao, J. Statistical pattern recognition: A review. IEEE Trans. Pattern Anal. Mach. Intel. 2000, 22, 4–37. [Google Scholar] [CrossRef]
- Miguez, G.; Xavier, A.; Maculan, N. An evaluation of the bihyperbolic function in the optimization of the backpropagation algorithm. Int. Trans. Oper. Res. 2014, 21, 835–854. [Google Scholar] [CrossRef]
- Raj, A. Przemiany Krajobrazu Leśnego Karkonoskiego Parku Narodowego w Okresie Ostatnich Kilkudziesięciu Lat; Karkonoski Park Narodowy: Jelenia Góra, Poland, 2014; p. 100. ISBN 978-83-64528-16-3. [Google Scholar]
- Danielewicz, W.; Raj, A.; Zientarski, J. Ekosystemy Leśne Karkonoskiego Parku Narodowego; Karkonoski Park Narodowy: Jelenia Góra, Poland, 2012; p. 96. ISBN 978-83-926933-5-2. [Google Scholar]
- Itten, K.I.; Dell’Endice, F.; Hueni, A.; Kneubuhler, M.; Schlaepfer, D.; Odermatt, D.; Seidel, F.; Huber, S.; Schopfer, J.; Kellenberger, T.; et al. APEX—The Hyperspectral ESA Airborne Prism Experiment. Sensors 2008, 8, 6235–6259. [Google Scholar] [CrossRef] [PubMed]
- Vreys, K.; Iordache, M.; Biesemans, J.; Meuleman, K. Geometric correction of APEX hyperspectral data. Misc. Geogr. 2016, 20, 11–15. [Google Scholar] [CrossRef]
- Sterckx, S.; Vreys, K.; Biesemans, J.; Iordache, M.; Bertels, L.; Meuleman, K. Atmospheric correction of APEX hyperspectral data. Misc. Geogr. 2016, 20, 16–20. [Google Scholar] [CrossRef]
- Poop, C.; Brunner, D.; Damm, A.; van Roozendael, M.; Fayt, C.; Buchmann, B. High-resolution NO2 remote sensing from the Airborne Prism EXperiment (APEX) imaging spectrometer. Atmos. Meas. Tech. 2012, 5, 2211–2225. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Enclona, E.A.; Ashton, M.S.; van der Meer, B. Accuracy assessments of hyperspectral waveband performance for vegetation analysis applications. Remote Sens. Environ. 2004, 91, 354–376. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. Some issues in the classification of DAIS hyperspectral data. Int. J. Remote Sens. 2006, 27, 2895–2916. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Advances in hyperspectral remote sensing of Vegetation and Agricultural Croplands. In Hyperspectral Remote Sensing of Vegetation; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2012; pp. 3–35. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; p. 462. ISBN 0-387-95457-0. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Braga-Neto, U.M.; Dougherty, E.R. Is cross-validation valid for small sample microarray classification? Bioinformatics 2004, 20, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Marcinkowska-Ochtyra, A.; Dobrowolski, M. Forest species identification of Mount Chojnik (Karkonoski National Park) using airborne hyperspectal APEX data. Sylwan 2015, 159, 593–599. [Google Scholar]
- Kokaly, R.F.; Despain, D.G.; Clark, R.N.; Livo, K.E. Mapping vegetation in Yellowstone National Park using spectral feature analysis of AVIRIS data. Remote Sens. Environ. 2003, 84, 437–456. [Google Scholar] [CrossRef]
- Dalponte, M.; Ørka, H.O.; Ene, L.T.; Gobakken, T.; Næsset, E. Tree crown delineation and tree species classification in boreal forests using hyperspectral and ALS data. Remote Sens. Environ. 2014, 140, 306–317. [Google Scholar] [CrossRef]
- Feret, J.B.; Asner, G.P. Tree species discrimination in tropical forests using Airborne Imaging Spectroscopy. IEEE Trans. Geosci. Remote Sens. 2013, 51, 73–84. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, S.; Zhao, T.; Wang, J. An application of tree species classification using high-resolution remote sensing image based on the rough set theory. Multimed. Tools Appl. 2017, 76, 22999–23015. [Google Scholar] [CrossRef]
- Pu, R.; Landry, S. A comparative analysis of high spatial resolution IKONOS and WorldView-2 imagery for mapping urban tree species. Remote Sens. Environ. 2012, 124, 516–533. [Google Scholar] [CrossRef]
- Deng, S.; Katoh, M.; Hyyppa, J.; Gao, T. Comparison of Tree Species Classifications at the Individual Tree Level by Combining ALS Data and RGB Images Using Different Algorithms. Remote Sens. 2016, 8, 1034. [Google Scholar] [CrossRef]
- Priedītis, G.; Šmits, I.; Daģis, S.; Paura, L.; Krūmiņš, J.; Dubrovskis, D. Assessment Of Hyperspectral Data Analysis Methods To Classify Tree Species. Res. Rural Dev. 2015, 2, 7–13. [Google Scholar]
- Baldeck, C.A.; Asner, G.P.; Martinm, R.E.; Andersonm, C.B.; Knappm, D.E.; Kellner, J.R.; Wright, J.S. Operational Tree Species Mapping in a Diverse Tropical Forest with Airborne Imaging Spectroscopy. PLoS ONE 2015, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Knapik, R.; Raj, A. Monitoring Przyrodniczy w Karkonoskim Parku Narodowym Jako Narzędzie do Śledzenia Zmian w Środowisku; Peckiana, Senckenberg Museum für Naturkunde: Görlitz, Germany, 2014; Volume 9, pp. 3–44. ISSN 1618-1735. [Google Scholar]

| VNIR | SWIR | |
|---|---|---|
| Spectral range | 380.0–971.7 nm | 941.2–2501.5 nm |
| Number of bands | up to 334 | 198 |
| Spectral sampling interval | 0.6–6.3 nm | 6.2–11 nm |
| Field of view (FOV) | 28° | |
| Instantaneous field of view (IFOV) | 0.028° (≈0.5 mrad) | |
| Spatial resolution | 2.5 m at 5000 m above ground level (AGL) | |
| Class | Birch | Beech | Larch | Alder | Pine | Spruce |
|---|---|---|---|---|---|---|
| Number of measurement polygons | 115 | 124 | 193 | 11 | 27 | 806 |
| Number of pixel samples | 615 | 972 | 685 | 90 | 125 | 2677 |
| Class | Birch | Beech | Larch | Alder | Pine | Spruce |
|---|---|---|---|---|---|---|
| Training dataset | 389 | 615 | 433 | 57 | 79 | 1692 |
| Validation dataset | 226 | 357 | 252 | 33 | 46 | 985 |
| Selected spectral bands (nm) | 530, 550, 590, 600, 610, 620, 630, 650, 680, 690,780, 800, 820, 830, 850, 860, 880, 900, 930, 940, 960, 980, 1000, 1040, 1120, 1200, 1240, 1260, 1280, 1500, 1530, 1560, 1660, 1720, 1760, 2000, 2030, 2060, 2090, 2110 |
| Birch | Beech | Larch | Alder | Pine | Spruce | |
|---|---|---|---|---|---|---|
| Classification result | 4.81 | 10.25 | 6,30 | 0.11 | 0.72 | 77.81 |
| Forest inventory | 4.79 | 4.14 | 4,58 | 0.12 | 0.53 | 85.55 |
| Difference (percentage points) | 0.02 | 6.11 | 1.72 | 0.01 | 0.19 | 7.74 |
| Authors | Data | Classification Algorithm | Number of Classified Tree Species | Overall Accuracy (%) | Kappa Coefficient |
|---|---|---|---|---|---|
| Kokaly et al. [46] | Hyperspectral | Expert system | 4 | 74 | 0.62 |
| Peerbhay et al. [2] | Hyperspectral | Partial least squares discriminant analysis (PLS-DA) | 6 | 88 | 0.87 |
| Dalponte et al. [22] | Hyperspectral | Support vector machine (SVM), random forest (RF), Gaussian maximum likelihood | 4 | 90 | <0.8 |
| Feret et al. [48] | Hyperspectral | Support vector machine (radial-kernel) | 17 | 83 | NA |
| Dalponte et al. [47] | Hyperspectral and LiDAR | Support vector machine | 3 | 93 | 0.88 |
| Ghosh et al. [20] | Hyperspectral and LiDAR | Support vector machine, random forest | 5 | 94 | 0.95 |
| Fassnacht et al. [19] | Hyperspectral | Support vector machine, random forest | 5 | 92 | 0.83 |
| Priedītis et al. [52] | Hyperspectral | Linear Discriminant Analysis (LDA)and artificial neural networks (ANN) | 5 | 86 LDA; 71 ANN | NA |
| Sommer et al. [17] | Hyperspectral | Random forest | 13 | 94 | 0.93 |
| Baldeck et al. [53] | Hyperspectral | Support vector machine | 3 | 98 | NA |
| Ballanti et al. [23] | Hyperspectral and LiDAR | Support vector machine | 8 | 95 | NA |
| Graves et al. [21] | Hyperspectral | Support vector machine | 20 | 62 | NA |
| Lee et al. [24] | Hyperspectral and LiDAR | Support vector machine | 6 | 91 | 0.89 |
| Raczko and Zagajewski (present study) | Hyperspectral | Artificial neural networks | 6 | 87 | 0.82 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raczko, E.; Zagajewski, B. Tree Species Classification of the UNESCO Man and the Biosphere Karkonoski National Park (Poland) Using Artificial Neural Networks and APEX Hyperspectral Images. Remote Sens. 2018, 10, 1111. https://doi.org/10.3390/rs10071111
Raczko E, Zagajewski B. Tree Species Classification of the UNESCO Man and the Biosphere Karkonoski National Park (Poland) Using Artificial Neural Networks and APEX Hyperspectral Images. Remote Sensing. 2018; 10(7):1111. https://doi.org/10.3390/rs10071111
Chicago/Turabian StyleRaczko, Edwin, and Bogdan Zagajewski. 2018. "Tree Species Classification of the UNESCO Man and the Biosphere Karkonoski National Park (Poland) Using Artificial Neural Networks and APEX Hyperspectral Images" Remote Sensing 10, no. 7: 1111. https://doi.org/10.3390/rs10071111
APA StyleRaczko, E., & Zagajewski, B. (2018). Tree Species Classification of the UNESCO Man and the Biosphere Karkonoski National Park (Poland) Using Artificial Neural Networks and APEX Hyperspectral Images. Remote Sensing, 10(7), 1111. https://doi.org/10.3390/rs10071111







