Abstract
The cucumber (Cucumis sativa L.) is often subjected to several fungal diseases. Rhizoctonia solani-induced cucumber damping-off and root rot are the most common diseases reported from the commercial greenhouses of the eastern area of Saudi Arabia. The objective of the current study is to explore the antagonistic activity of four Trichoderma species against R. solani in vitro and in vivo. Ten R. solani isolates (eight belonging to AG-4 and two belonging to AG-A and AG2-1) were studied. AG4 isolates were pathogenic to cucumber plants, while AG-A and AG2-1 isolates were non-pathogenic. Seven isolates of Trichoderma spp., named T. hamatum KSATR8, T. harzianum (KSATR9 and KSATR10), T. asperellum (KSATR11, KSATC, and KSAT1E), and T. longibrachiatum KSATS were isolated, and the identities of both R. solani and Trichoderma isolates were confirmed based on the phylogenetic analysis of the DNA sequence of the ITS region. The dual culture findings indicated that T. asperellum KSATC and KSAT1E exhibited the most significant inhibitory activities against R. solani, with values of 79.33 and 70.89%, respectively. Scanning electron microscope (SEM) images showed a considerable degradation in the cell wall and collapsing of R. solani hyphae by all Trichoderma species. Under greenhouse conditions, the application of T. asperellum KSATC and KSAT1E at concentrations of 2 × 108 conidia/mL revealed a reduction in root rot and damping-off incidence percentages with values that did not reveal a significant (p < 0.05) difference from those of Rizolex-T fungicide. Nevertheless, the efficacy of the fungicide attained 86.67%, being higher than that of T. asperellum KSATC, which reached 80%. Trichoderma asperellum KSATC and KSAT1E were the greatest in increasing peroxidase, catalase, and chitinase enzymes activities in cucumber plants. Conversely, a significant (p < 0.05) elevation in polyphenol oxidase enzyme (0.762 and 0.97 U/g FW) and total phenol content (0.55 and 0.62 mg/g FW) was recorded in cucumber plants treated with T. harzianum KSATR9 and KSATR10, respectively. The statistical analysis results displayed no considerable variations among cucumber plants regarding total chlorophyll content as a response to treatments with Trichoderma species and fungicides. Therefore, we endorse using T. asperellum KSATC and KSAT1E as an alternative to fungicides to manage root rot and damping-off in cucumbers.
1. Introduction
Cucumber (Cucumis sativa L.) is amongst the most popular and economic vegetable crops used for fresh consumption [1]. Cucumber is vulnerable to various fungal diseases, including root rot and damping-off, which are among the main destructive diseases which infect cucumber worldwide [2]. Numerous fungal genera are responsible for cucumber damping-off and root rot diseases, such as Macrophomina phaseolina, Fusarium oxysporum, F. solani, Phytopathora sp., Rhizoctonia solani, and Sclerortium rolfsi [3,4,5,6,7,8]. R. solani Kühn is a root rot causal agent in many plants worldwide. Although the symptoms brought on by R. solani vary within different species, it mainly infects underground tissues. Damping-off is the most prevalent symptom of Rhizoctonia disease and is distinguished by its inhibiting infected seeds’ germination, and infected seedlings could be destroyed prior to or after emerging from the soil [9]. Rhizoctonia solani is responsible for high losses in the yield of horticultural and agricultural crops [6,7].
Depending upon the nuclei number of the hyphal cells, the basidiomycetous fungal Rhizoctonia genus is categorized as either uninucleate, binucleate, and multinucleate. Binucleate Rhizoctonia comprises 19 anastomosis groups (from AG-A to AG-U) [10], whereas multinucleate Rhizoctonia includes 13 anastomosis groups (from AG-1 to AG-13) [11] depending upon its hyphal anastomosis reactions. The multinucleate R. solani is extensively widespread, causing serious damage to over 200 plant species [12]. An increasing number of uninucleate and binucleate isolates have been recognized which also cause serious damage in various plants [13]. Rhizoctonia solani is a species complex, and is mostly classified according to its hyphal anastomosis reactions and host range. However, identification based on these interactions requires experience and may be affected by many factors, which can be time-consuming [14]. Different molecular markers were utilized to differentiate R. solani AGs [15,16,17]. However, the rDNA ITS1-5.8 S-ITS2 region sequence analysis has been verified as an increasingly effective proxy for genetic diversity and phylogenetic studies of R. solani AG and AG-subgroups [18,19].
Controlling R. solani has been erratic, challenging, and costly because of its consistent, long-living sclerotial structures in soil [20]. Several management strategies, such as cultural, chemical, and physical control techniques, have been adopted to manage root rot diseases [21,22]. Nevertheless, up until now, such approaches have been only partly successful [9]. Root rot pathogens are commonly host-specific, but some exhibit different hosts. Consequently, crop rotation cannot be entirely successful as a control method [23]. Fungicide application poses risks to human health, increases environmental pollution, and induces pathogen resistance [24]. Biological control using numerous microbes, especially fungal or bacterial strains, has been commonly utilized against airborne and soil-borne diseases in the last two decades. Since the 1920s, Trichoderma species are also well-recognized for their advantageous potential in managing many plant pathogens [25]. Besides their direct antagonistic potentialities in plant pathogens, numerous investigations have provided a robust indication that Trichoderma species, incorporated into crop production, may improve general plant growth, health, yield, and resistance to disease [21]. Trichoderma has been widely used to control the pathogenic R. solani on cucumber [26,27] and other hosts [28,29,30]. Mechanisms that have been implemented for their Trichoderma antagonistic activity have been discussed by many authors, such as mycoparasitism [31,32] and the induction of plant resistance [33,34]. Furthermore, Trichoderma can penetrate the root system without causing damage and elevate the plant’s defense ability through increasing its peroxidase and chitinase enzyme activity. Therefore, plants treated with Trichoderma are more resistant to R. solani [35]. Moreover, Trichoderma grows faster than fungal plant pathogens and can antagonize many soil-borne plant pathogens via the secreting of many compounds that are antifungal pathogens. Therefore, their application in biological control will easily induce a defense system and increase the growth rate of greenhouse plants and other agricultural crops [36,37].
Hence, the principal goal of the present research is to inspect the antagonistic activity of four indigenous species of Trichoderma against R. solani through laboratory and greenhouse trials. Additional goals of this study are to explore the genetic identity of R. solani AG accompanied with cucumber, depending upon the phylogenetic analysis of the rDNA ITS1-5.8S-ITS2 dataset; exploring the genetic identity of natively isolated Trichoderma species based on the rDNA ITS1-5.8S-ITS2 sequence data; exploring the indirect effects of Trichoderma species on the antioxidant enzymes and phenolic as well as chlorophyll content in healthy and infected cucumber plants.
2. Materials and Methods
2.1. Sampling and Isolation of R. solani and Trichoderma
Cucumber plants exhibiting root rot symptoms were collected from 17 greenhouses in Al-Ahsa, Saudi Arabia during two seasons, 2020/2021. Samples were washed with tap water, followed by surface disinfection with NaOCl (1%) solution for 2 min and washing via sterile distilled water, and were cut into small pieces. Root pieces were then air-dried, placed into Petri dishes supplemented with potato dextrose agar (PDA) and streptomycin sulphate (0.3 g L−1), and left for about 4 days incubation at 25 ± 2 °C. The incubated plates were regularly inspected daily to visualize the emerged fungal growth. The developed fungi colonies were purified utilizing the hyphal tip method [38] and preserved in the same medium (PDA) for additional work. Obtained isolates were first identified as Rhizoctonia by relying upon mycelial features following Sneh et al. [12]. Trichoderma fungi were recovered via their isolation from soil and root samples collected from different Saudi Arabian geographical areas, i.e., Al Karj, Wadi Aldawaser, and the Research Station of King Faisal University. The isolates were kept in the dark on a PDA medium at 25 °C and were initially characterized based on their colony and conidia morphology using the taxonomic criteria of Barnett and Hunter [39]. The Trichoderma species and R. solani isolates have been permanently placed in the Pests and Plant Diseases Unit (PPDU) culture collection at the College of Agricultural and Food Sciences, King Faisal University, Al-Ahsa, Saudi Arabia.
2.2. Molecular Characterization of R. solani and Trichoderma Species
Extraction of total DNA from R. solani and Trichoderma isolates was performed following the Dellaporta protocol for genomic DNA isolation [40]. PCR sequencing and amplification of the rRNA’s internal transcribed spacer region (ITS), involving the 3’ end of 18S (small subunit), the complete 5.8S rRNA gene, the first internal transcribed spacer (ITS1), and the second internal transcribed spacer (ITS2) region, were done with primer pairs ITS4 and ITS5 [41]. The PCR reaction was accomplished in 25 µL reaction volume, containing 10 µL PCR Master Mix (amaR OnePCR, GeneDirex, Inc.), 11 µL of ddH2O, 1.5 µL of each primer, and 1 µL of template DNA. PCR amplification was done in a 2720 Thermal Cycler (Applied Biosystems, Foster City, California) with cycling conditions consisting of 95 °C for 15 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 1 min, extension at 72 °C for 1 min, and a final extension for 5 min at 72 °C [42]. The obtained PCR products were cleaned and sequenced in two directions via the Macrogene Inc. Sequencing Service (Seoul, Korea). MEGA 11 was used to edit and curate the sequences as required [43]. The sequence’s homology was analyzed with BLAST® against the NCBI sequence database (National Center for Biotechnology Information, GenBank) (www.ncbi.nlm.nih.gov/genbank accessed on 20 December 2022.). The possible phylogenetic relations among AGs of R. solani and Trichoderma species were determined via the maximum-likelihood (ML) technique for the ITS sequence data using MEGA 11 [43]. The entire deleting was adjusted as gap/missing data treatment, and the initial ML tree was automatically adjusted. The obtained ML trees were assessed by bootstrap analysis utilizing 1000 replications. The acquired trees were established utilizing MEGA.
2.3. Pathogenicity Test
2.3.1. Preparing of Inoculum
The R. solani inoculum was equipped in well-sterilized 500-mL glass flasks. Each one contained a sorghum grain and sand (2:1 v/v) mixture. The constituents had been admixed thoroughly and moistened via tap water. Afterwards, the mixture was placed in the flasks and autoclaved at 121 °C for 30 min. A 5 mm mycelial plug of 5 day-old R. solani culture was inoculated in each flask and incubated for 15 days at 25 ± 2 °C [44].
2.3.2. Preparation of Soil and Pots
A sand and peat moss (1:2 w/w) soil mixture was formulated, treated via a 5% formalin solution for sterilization, and enclosed via a polyethylene sheet. The sheet was released after two weeks, and the mixture was aerated for 10 days to ensure formalin evaporation. Then, 30 cm diameter plastic pots were immersed for 15 min in 5% formalin solution for sterilization and then dried in air for an entire day. The sterilized soil mixture was placed in these pots, with the pots each containing 3 kg.
2.3.3. Inoculation Procedure
The plastic pots were inoculated with 10 g of sorghum colonized with R. solani /kg soil and irrigated for 4 days prior to planting [45]. The autoclaved sorghum–sand mixture inoculated within the control pots was not supplemented with R. solani. Three replicates (five pots per replicate) were utilized in each isolate and control. The pots were retained in an air-conditioned greenhouse under controlled circumstances at 25 ± 2 °C temperature and 60–75% relative humidity. Cucumber seeds cv. Diala (Nickerson-Zwaan, Netherlands) were submerged in 1% sodium hypochlorite solution for 2 min for surface sterilization, followed by washing with sterile water. Seeds were planted within the pots (5 seeds/pot) and sown in non-infested soil to act as a control. The pots were allocated in the greenhouse randomly, where they were irrigated and fertilized when required. The experiment was carried out two times in an entirely random block design.
2.3.4. Disease Assessment
Disease assessment was done according to Hassan et al. [44], as follows:
- (a)
- Pre-emergence damping-off percentage was estimated after 15 days:Pre-emergence (%) = Number of non-germinated seeds/Number of sown seeds × 100
- (b)
- Post-emergence damping-off percentage was estimated after 30 days:Post-emergence (%) = Number of dead seedlings/Number of survival plants × 100
- (c)
- Survival seedlings (%) = Number of survived plants/ Number of sown seeds × 100
- (d)
- Disease incidence (DI) percentage was estimated after 45 days of seeding:DI (%) = Number of infected plants/ Total number of examined plants × 100.
2.4. Evaluation of Trichoderma Species against R. solani In Vitro
The dual culture procedure [28,45] was adopted to evaluate the antagonistic impacts of T. asperellum, T. harzianum, T. hamatum, and T. longibrachiatum against R. solani on PDA medium under laboratory conditions. In this experiment, a 5 mm mycelial plug was taken from fresh R. solani culture (5-days old) and placed at a 2 cm distance from the edges of the 90-mm Petri dishes. Then, 5 mm of the fresh mycelial plug of Trichoderma was placed on the opposite side of the plate. This was followed by incubating the inoculated plates for 5 days at 25 ± 2 °C. This experiment was performed with three replicates (five plates per each) for each treatment, as well as for the control. When the R. solani mycelial growing reached the petri dish edge, the diameters of the developed R. solani colonies were measured and assessed using the suggested formula according to Hassan et al. [44]:
- I = [(C − T)/C] × 100;
- I = inhibition of radial mycelia growth;
- C = the pathogen radial growth measurement in control;
- T = the pathogen radial growth in the existence of Trichoderma isolates.
2.5. Scanning Electron Microscopy (SEM) Examination
Morphological variations in R. solani hyphae due to treatment with T. harzianum, T. hamatum, T. asperellum, and T. longibrachiatum were observed using SEM [46]. Three mycelial discs approximately 5 mm from the interaction region were placed for 3 h at 28 °C in 20 g glutaradehyde L−1 buffered with 0.1 M cacodylate buffer (pH 7.4), 10 g sucrose L−1, and 1 mM CaCl. Specimens were rinsed with the same buffer and post-fixed in this buffer for 2 h with 10 g osmium tetroxide L−1 at 28 °C. A graded ethanol series was used to acquire sample dehydration. Entirely dehydrated samples were transferred from pure ethanol through an ethanol and propylene oxide (1:1) mixture into pure propylene oxide. Discs were then attached onto stubs utilizing double-phase sticky tape and subjected to gold-palladium coating. SEM images were attained through JEOL GM 5200, Japan.
2.6. Effect of Trichoderma Species on Root Rot and Damping off Diseases under Greenhouse Conditions
Spore suspensions of the tested T. harzianum, T. hamatum, T. asperellum, and T. longibrachiatum isolates were prepared via culturing 5 mm mycelial discs from 4-days old culture on 250 mL potato dextrose broth (PDB) medium and incubation for 10 days at 25 ± 2 °C. Using a haemocytometer, spore suspensions were accustomed to 2 × 108 spores /mL [47]. Preparation of soil mixture, inoculum, and inoculation was done as described in the pathogenicity trial. The pre-inoculated pots with R. solani were treated by soil drench with 100 mL fungal spore suspension (2 × 108 spore/mL) [48]. The fungicide Rizolex-T 50% WP (Sumitomo Chemical Co., Tokyo, Japan) was applied at the endorsed dosage (2 g L−1). Cucumber seeds cv. Diala (Nickerson-Zwaan, the Netherlands) were then planted in the pots (5 seeds/pot). The experiment was repeated two times and established in three replicates (five pots per replicate) for each treatment as well as for the control. The pots were randomly distributed in an air-conditioned greenhouse at 25 ± 2 °C and 60–75% relative humidity. Irrigation and fertilization were done when needed. Based on the abovementioned equations, the pre- and post-emergence damping-off and DI proportions were assessed after 15, 30, and 45 days, respectively. The efficacy of each treatment compared to the control was assessed based on the equation proposed by Ismail [49]: Efficacy (%) = Control − Treatment/Control × 100.
2.7. Assessment of Defense-Related Enzymes
The defense-related enzymes in cucumber plants were assessed after 30 days of sowing by blending 1 g leaves in 100 mL phosphate buffer solution (7.1 pH) and filtrated through five layers of cheesecloth. The filtrate was then placed in a centrifuge which was operated at 3000× g for 15 min. Clear supernatants were utilized as crude enzymes to determine the enzyme’s activity. Measurements were taken for each enzyme at a specified wavelength every 10 s for 1 min using APEL Co., Ltd. Japan PD-303. Spectrophotometer. measurements were taken in triplicate, and the mean for the three measurements was expressed as the change in absorbance per min. per gram fresh weight (g FW) [50].
2.7.1. Polyphenol-Oxidase Activity
The polyphenol-oxidase (PPO) enzyme activity was estimated following Matta and Dimond [51]. The mixture used for the reaction comprised 0.1 mL enzyme extract + 1 mL sodium phosphate buffer solution (0.1 M at pH 7.0) + 1 mL catechol (0.03 M), and was completed with distilled water to be 6.0 mL and incubated at 30 °C for 3 min. The PPO activity was measured as a change in the absorbance at 495 nm.
2.7.2. Peroxidase Activity
The peroxidase (PO) activity was appraised following the technique described by Allam and Hollis [52]. The mixture used for the reaction comprised 0.3 mL enzyme extract + 0.5 mL phosphate buffer (0.1 M at pH 7.0) + 0.3 mL pyrogallol (0.1 M) + 1 mL 0.1% H2O2. The distilled water was added until the mixture was completed up to 3.0 mL. The change in the absorbance at 422 nm denotes the PO activity.
2.7.3. Catalase Activity
The catalase (CAT) enzyme was appraised utilizing the technique described by Aebi [53]. First, 0.1 mL crude extract was admixed with 0.5 mL sodium phosphate buffer (0.2 M at pH 7.6) and 0.3 mL 0.5% H2O2 in the sample cuvette. Afterwards, distilled water was added until the mixture reached an ultimate volume of 3.0 mL. The H2O2 breakdown was then carried out with the absorbance estimation at 240 nm.
2.7.4. Chitinase Activity
Chitinase (CHT) activity assessment was achieved following Wirth and Wolf [54] and utilizing chitin as a substrate. In this method, 1 mL of 1% (w/v) colloidal chitin was added to sodium phosphate buffer (0.05 M at pH 6.6), and 1 mL of enzyme extract was then mixed by shaking for 1 hour at 500 rpm and 37 °C. The tubes were incubated for an hour at 37 °C. The reaction was stopped in boiling water, then the mixture cooled and centrifuged for 5 min at 10,000× g. The supernatant was collected to appraise CHT activity as the change in the absorbance at 540 nm.
2.8. Estimation of Total Phenol Content
Total phenolic compounds (TPC) were calorimetrically estimated after 30 days of sowing using the phosphotungstic-phosphomolybdic acid reagent Folin-Cioalteu as described by Snell and Snell [55]. The optical density was calorimetrically measured at wavelength 520 nm. The measurements have been presented as mg of gallic acid per gram of fresh weight (mg/g FW) [49].
2.9. Estimation of Total Chlorophyll Content
The total chlorophyll contents (TCC) of Chl. a and Chl. b were measured in cucumber leaves after 30 days of sowing utilizing the technique described by Nagata and Yamashita [56] at wavelengths of 645 and 663 nm, respectively. The total chlorophyll content (TCC) was expressed in mg/g F.W and determined using the equation of Arnon [57]:
- Chl. a = 12.72A663 − 2.59A645
- Chl. b = 22.9A663 − 4.67A645
- Chl. t = 20.31A645 + 8.05A663
2.10. Statistical Analysis
All attained data were analyzed utilizing analysis of variance (ANOVA) among treatments following Snedecor and Cochran [58]. Means were compared via least significant differences (LSD) utilizing SPSS software v. 8.0. (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. R. solani Isolation and Molecular Characterization
Ten R. solani isolates were recovered from cucumber plants with infected roots (Figure 1a,b) which were collected from various commercial greenhouses in the eastern area of Saudi Arabia between September and April 2021. Ten isolates of R. solani were recovered from the roots of cucumber plants with a frequency which reached 95–100%. Microscopic examination revealed that nine isolates were multinucleate while one isolate was bi-nucleate. Eight isolates belonged to the AG-4 anastomosis group, while one isolate was AG2-1. The only binucleate isolate was AG-A (Figure 1c–g). Blast analysis of the obtained sequences against known sequences of R. solani AGs from NCBI GenBank confirmed that eight R. solani isolates belonged to the multinucleated AG-4 group, which showed 99.07% similarity to R. solani (Accession No. OQ357845) (Figure 1d). Further, the isolate PPDURS10 (Figure 1e) showed 99.8% homology with R. solani (AG2-1) (Accession No. MF474260), and PPDURS9 was a binucleated cell (Figure 1c) and showed 100% homology with R. solani (AG-A) (Accession No. OQ410437). The ML tree derived from the ITS sequences revealed a distinct differentiation between anastomosis groups of R. solani (Figure 2). The isolates attained in the current study and those retrieved from GenBank (Table S1) were categorized into three major groups with strong bootstrap values (99–100%). Based on the ML analysis (Figure 2), eight R. solani isolates (Figure 1d,f) belonged to the multinucleated AG-4 group. In contrast, the isolates PPDURS9 (Figure 1c,g) and PPDURS10 (Figure 1e,g) of R. solani belonged to the binucleated AG-A and AG2-1 groups, respectively. Only two representative R. solani (AG-4) isolates (PPDURS1 and PPDURS4) and the PPDURS10 and PPDURS9 isolates were deposited in the NCBI database (Table 1 and Table S1).
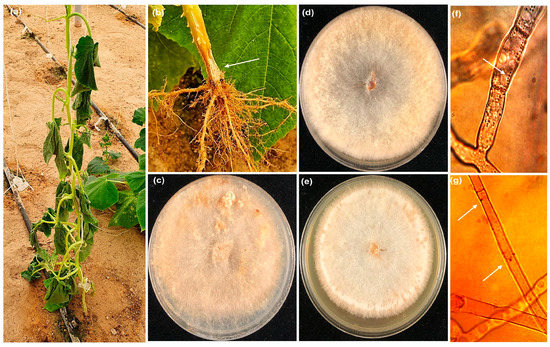
Figure 1.
Root rot symptoms (arrow) observed on cucumber plants in a commercial greenhouse (a,b), cultures of R. solani (AG-A) (c), AG-4 (d), AG2-1 (e), multinucleate (arrow) (f) and binucleate (arrows), and (g) hyphae of R. solani.
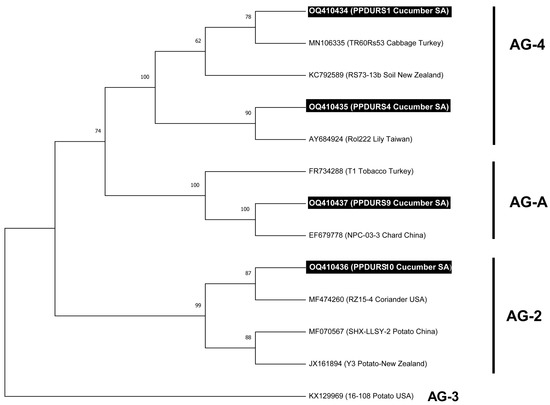
Figure 2.
Maximum likelihood (ML) tree of 13 R. solani isolates obtained from the heuristic search by applying the Maximum Parsimony technique through MEGA 11. The accession numbers in the tree are followed by the name of the host and the origin of R. solani isolates. The numbers above branches are the bootstrap values of 1000 replications. The tree was rooted with the outgroup taxa R. solani 16-108 (AG-3) (GenBank Accession No. KX129969). The isolates obtained in this study are emphasized with a black background.

Table 1.
Results of pathogenicity assessment of R. solani isolates on cucumber plants under greenhouse conditions.
3.2. Trichoderma Species Isolation and Molecular Characterization
Seven Trichoderma isolates, representing four species, were recovered from soil rhizospheres and the roots of cucumber plants (Table S1). The blast search results showed that one isolate, KSATR8, was 100% similar to T. hamatum (Accession No. KY951914). In contrast, the two isolates KSATR9 and KSATR10 showed 100% similarity with T. harzianum (Accession No. MT529404). Three isolates, KSATR11, KSATRTC, and KSA1E, showed 100% similarity with T. asperellum (Accession No. MK928418). The isolate KSATS exhibited 100% homology with T. longibrachitum (Accession No. MT520646). Trichoderma isolates were deposited in the NCBI database under accession numbers OQ423222 (T. hamatum), OQ423223, and OQ423224 for T. harzianum KSATR9 and KSATR10, respectively. The evolutionary history specified from the ML technique revealed that the Trichoderma isolates were accommodated in four discrete clades (Figure 3). The three isolates KSATR11, KSATC, and KSA1E resided in three subclades within T. asperellum in a clade which was moderately maintained with a bootstrap (BS) value of 80%. The single isolate KSATR8 grouped with reference sequences of T. hamatum in a highly supported clade with BS values reaching 99%, whereas the isolate KSATS sub-clustered with T. longibrachitum in a clade which was strongly maintained by BS of 100%. In contrast, the isolates KSATR9 and KSATR10 grouped with T. harzianum in a clade which was strongly maintained by BS 100%. The Trichoderma isolates obtained in the present investigation were deposited in the NCBI database (Table S1).
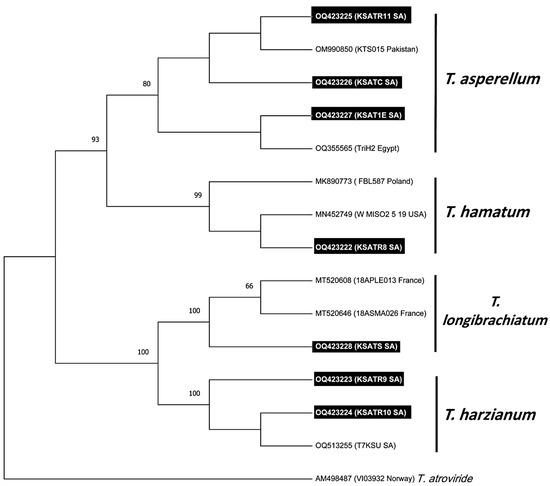
Figure 3.
Maximum likelihood (ML) tree of 15 isolates of Trichoderma species obtained from the heuristic search by applying the Maximum Parsimony method using MEGA 11. The numbers above branches are the bootstrap values of 1000 replications. The tree was rooted with the outgroup taxa T. atroviride VI03932 (GenBank Accession No. AM498487). The isolates obtained in this study are emphasized with black background.
3.3. Pathogenicity Test
The recovered R. solani isolates were tested for their pathogenic potentiality on cucumbers under greenhouse conditions (Table 1). In this respect, R. solani PPDURS10 of the AG2-1 group was non-pathogenic and did not induce pre- nor post-emergence root rot or damping-off of cucumber seedlings. R. solani PPDURS9 exhibited non-significant pre- and post-damping-off values relative to healthy control and PPDURS10, suggesting that this isolate is non-pathogenic to cucumber plants. Nevertheless, all isolates of AG-4 were pathogenic to cucumber with significant variations (Table 1). All AG-4 isolates caused different degrees of pre-damping-off, post-root rot, and post-damping-off symptoms. Nevertheless, the isolates PPDURS1 and PPDURS2 of the AG-4 group were the most severe among those tested, and significantly decreased the survival of cucumber seedlings either pre- or post-emergence. Therefore, the isolate PPDURS2 was selected for additional experiments due to its aggressiveness on cucumber seedlings, which caused 76.67% pre- and 23.33% post-damping-off for all sown seeds, with no surviving seedlings. There was no DI recorded for this isolate because no seedlings survived.
3.4. Antagonistic Activity of Trichoderma Species on the R. solani Mycelial Growth
The in vitro findings of the dual culture procedure specified that the tested Trichoderma species considerably repressed the R. solani mycelial growth (Figure 4a,b). In this respect, the highest inhibitory spectrum was T. asperellum KSATC and T. asperellum KSA1E, which revealed the highest inhibition percentages of 79.33% and 70.89%, respectively. In contrast, T. asperellum KSATR11 exhibited the lowest inhibitory effect against R. solani (Figure 4a,b).
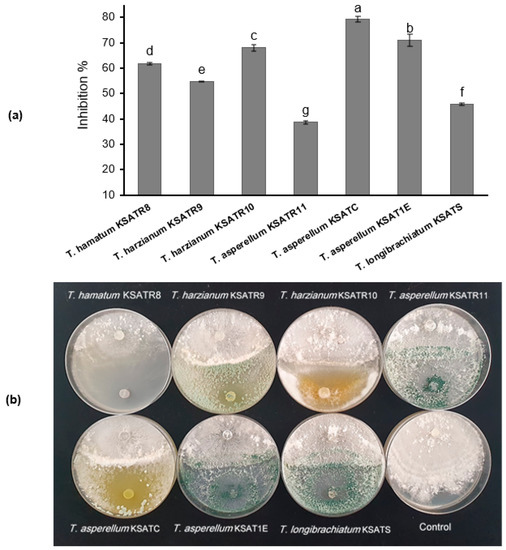
Figure 4.
R. solani inhibition rate in dual culture with Trichoderma (a). In vitro antagonistic activity of four species of Trichoderma against R. solani mycelial growth (PPDURS2) (b). Expressed values represent the mean of three replicates (five plates per each) ± stanard deviation. Bars designated by different letters significantly differ (p < 0.05) depending upon the least significant difference (LSD) test.
3.5. Scanning Electron Microscopy (SEM) Examination
A scanning electron microscope observed mycelial discs in the interaction regions between R. solani and Trichoderma species (Figure 5). The diameter of R. solani hyphae (Figure 5a,b) is larger than the hyphae of Trichoderma species; thus, they are easily distinguishable. Following the interaction, different patterns of mycoparasitism were observed, such as overgrowth, degradation in the cell wall, and the collapsing (arrows) of R. solani hyphae by T. harzianum KSATR10, T. hamatum KSATR8, and T. longibrachiatum KSATS (Figure 5c–e). In comparison, T. asperellum KSATC formed coiled hyphae and wrapping (arrows) around R. solani and revealed a possible digestion of the penetrated mycelium and hyphal lysis (Figure 5e).
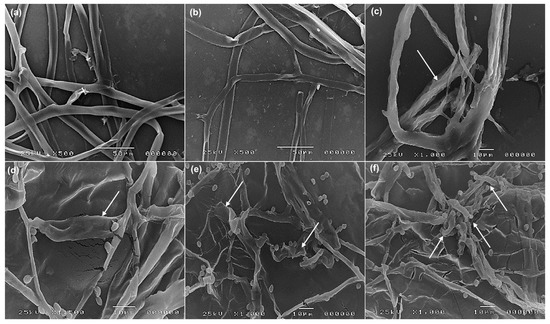
Figure 5.
SEM images demonstrating the parasitic process of the tested Trichoderma species. Untreated R. solani cultures in control (a,b), dual culture interaction showing mycoparasitism process of T. harzianum KSATR10 (c), T. hamatum KSATR8 (d), T. longibrachiatum KSATS (e) causing turgidity, shrinkage, and degradation (arrows) of the hyphal cell wall of R. solani. Mycoparasitism process showing coiling of T. asperellum KSATC (arrows) over the hyphae of R. solani (f).
3.6. Effect of Trichoderma Species on Damping-Off and Root Rot Disease under Greenhouse Conditions
The results outlined in Table 2 exhibit that the examined Trichoderma species demonstrated a significant (p < 0.05) decrease in pre- and post-root rot and damping-off relative to control and untreated plants. In this respect, the highest significant decrease in total percentages of pre- (10%) and post-damping-off (13.33%) were recorded for T. asperellum KSATC, with efficacy reaching 80%. It is clear from the obtained data that no significant (p < 0.05) differences occurred between T. asperellum KSATC and fungicide treatment regarding pre- and post-damping-off as well as DI values. In addition, the treatments with T. asperellum KSATC showed a survival percentage of up to 76.67%, with a low incidence of root rot disease that reached 4.76%. However, the efficacy of the fungicide treatment was recorded as 86.67%, which is higher than for the treatment with T. asperellum KSATC, which touched 80%. Furthermore, a comparable trend of results was detected with the treatment of T. asperellum KSAT1E, which exhibited non-significant (p < 0.05) values of pre- (13.33%) and post-damping-off (23.33%) when compared to Rizolex-T fungicide treatment. In the infected control, R. solani caused pre-damping-off for 86.66% of sown seeds and post-damping-off of 13.33% of the remaining germinated seeds. This explains why the post-damping-off value of the infected control is lower than any treatments except Rizolex-T 50% and healthy control.

Table 2.
Trichoderma species antagonistic activity against R. solani-induced damping-off and root rot under greenhouse conditions.
3.7. Assessment of Defense-Related Enzymes
The results demonstrated in Figure 6 indicate that the activities of the PO, PPO, CAT, and CHT enzymes were significantly (p < 0.05) improved in cucumber plants after treatment with Trichoderma species compared to either healthy or infected as well as fungicide-treated plants. Among the tested Trichoderma species, T. asperellum KSATC and T. asperellum KSAT1E exhibited a significant (p < 0.05) increment in the activity of PO (Figure 6a), CAT (Figure 6c), and CHT (Figure 6d) in cucumber plants, followed by T. harzianum KSATR10. Cucumber plants treated with T. asperellum KSATR11 showed the lowest PO, CAT, and CHT activity in comparison with other Trichoderma treatments, but were still significantly higher than either healthy or infected as well as fungicide-treated plants (Figure 6a,c,d). Meanwhile, T. harzianum KSATR10-treated cucumber plants revealed the greatest elevation in PPO activity, followed by those treated with T. harzianum KSATR9, T. asperellum KSATC, and T. asperellum KSAT1E (Figure 6b).
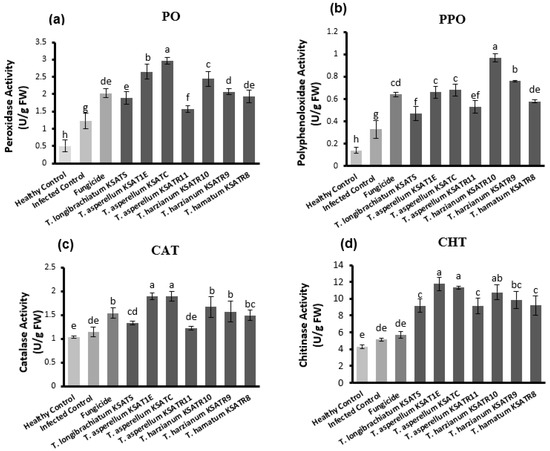
Figure 6.
Enzymatic activity levels of PO (a), PPO (b), CAT (c), and CHT (d) estimated in diseased and healthy cucumber plants in response to treatments with Trichoderma species and Rizolex-T fungicide compared to control. The expressed values (U/g FW) are the mean of three replicates ± standard deviation. Bars with various letters significantly (p < 0.05) differ depending upon the least significant difference (LSD) test.
3.8. Estimation of TPC and TCC
The obtained data in Figure 7a revealed that cucumber plants exhibited significant (p < 0.05) variances within the TPC compounds in response to treatment with Trichoderma species. The greatest increase in TPC was recognized in cucumber plants treated with T. harzianum KSATR10, followed by those treated with T. harzianum KSATR9 and T. asperellum KSATC (Figure 7a). Inversely, the statistical analysis showed no great variations among cucumber plants in terms of TCC in response to treatments with Trichoderma species (Figure 7b).
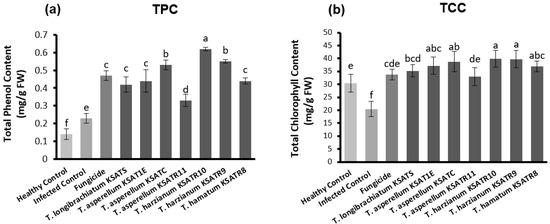
Figure 7.
Levels of TPC (a) and TCC (b) in diseased and healthy cucumber plants in response to treatments with Trichoderma species and Rizolex-T fungicide compared to control. Values are presented as the mean of three replicates ± standard deviation. Bars with various letters significantly (p < 0.05) differ depending upon the least significant difference (LSD) test.
4. Discussion
Root rot diseases are a global problem in the production of agricultural crops. Rhizoctonia solani is a significant root rot pathogen which is capable of causing severe damage to cucumber [26,27,28,34]. In the current investigation, 10 R. solani isolates were recovered from cucumber root rot samples. Most isolates were multinucleate and belonged to AG4, while one isolate belonged to AG2-1, and the only detected binucleated isolate was AG-A [59,60]; their identities were confirmed based on their phylogenetic relationship inferred by MEGA 11 software. Most (80%) isolates belonged to the multinucleate AG-4 group. According to the literature, AG-4 R. solani frequently causes root rot on cucumber [44,61,62,63]. Other investigators have reported similar results on cucumber. For example, R. solani AG-4 was frequently (61.9%) isolated from the surveyed cucumber greenhouses [62]. Moreover, R. solani AG-4 is the increasingly predominant (97.9%) anastomosis group isolated from cucumber plants [59]. The results of the current work revealed two isolates belonging to the binucleate AG-A and AG2-1 groups, these being less frequently isolated anastomosis groups. As support for our finding, other studies have reported the association of AG-A with cucumber with a very low frequency of 1.9% [62].
The tested R. solani AG-4 isolates exhibited variability in causing pre- and post-damping-off and root rot in comparison with the control treatments. This variability in virulence can be attributed to the variances in the growth speed and their parasitic nature [64]. The pathogenicity test findings revealed that the variances in the virulence of multinucleate and binucleate R. solani isolates were statistically significant (p < 0.05). Our findings are congruent with those previously reported [60,62], in which the binucleate Rhizoctonia AG2-1 was weakly or not pathogenic to cucumber and tomato. In this regard, Yıldırım and Erper [62] discovered, through a pathogenicity trial, that the binucleate Rhizoctonia AG2-1 was moderately virulent on cucumber. Furthermore, Eken and Tuncer [60] demonstrated that the binucleate Rhizoctonia isolates of AG-A and AG-K were weakly aggressive on tomato.
The current investigation explored the antagonistic activity of four Trichoderma species against damping–off and root rot disease in cucumber, attempting to find an effective and secure technique for controlling such diseases. The Trichoderma isolates exhibit an increasingly variable antagonistic ability [65]. This was clear in this study, in which T. asperellum KSATC and T. asperellum KSA1E revealed the highest inhibition percentage, 79.33% and 70.89%, respectively, against R. solani when compared to other tested species. In a related study, T. asperellum T-5A revealed a great inhibitory activity against R. solani under laboratory and greenhouse conditions [66]. Moreover, the antagonistic activity of T. asperellum TA1 revealed the highest (84.44%) inhibitory effect against R. solani, the principal cause for cucumber root rot and damping-off, relative to other species [67]. Furthermore, it was confirmed that T. aggressivum f. europaeum had great antagonistic activity against R. solani and different phytopathogens, greater than 80% [28]. Trichoderma longibrachiatum caused 82% and 75% in vitro mycelial growth inhibition of R. solani and B. cinerea, respectively [68]. Generally, the outcomes from the comparison differ among Trichoderma species and isolates against different phytopathogens [28]. This coincides with our study case, in which the strain of T. asperellum KSATR11 displayed the lowest activity against R. solani in laboratory and greenhouse trials. Additionally, these findings are also influenced by the temperature, growth medium, and other parameters.
The greenhouse trials also revealed promising results, in which T. asperellum KSATC and T. asperellum KSA1E were the greatest treatments for reducing root rot and damping-off in cucumber. The efficacy of fungicide treatment is still the highest, but the values of pre- and post-damping-off and DI are statistically insignificant (p < 0.05) compared to those of T. asperellum KSATC. In vivo trials against R. solani and other soil-borne pathogens have reported comparable results to those described in this study. For example, Rini and Sulochana [29] found that applying T. harzianum or T. pseudokoningii resulted in a 25% reduction in Rhizoctonia root rot of chili. Correspondingly, the T. asperellum strain (T-34) decreased the R. solani disease incidence when maintained at 103 conidia ml−1 [69]. Also, the findings of Huang et al. [70] point out that the T. harzianum strain SQR-T37 was an effective antagonist against R. solani in a mycoparasitic manner and reduced damping-off disease in cucumber seedlings. Thus, our findings are of great interest for considering T. asperellum KSATC as a secure and effective alternate for fungicides in managing such diseases.
The obtained SEM images revealed the dense coiling and penetration of T. asperellum KSATC inside the R. solani hyphae, causing digestion and lysis of its cell wall. Inbar et al. [71] reported similar observations for the T. harzianum and Sclerotinia sclerotiorum interaction using a light microscope. Using a SEM, it was demonstrated that T. harzianum Th-9 overgrew and coiled around R. solani, causing hyphal damage. Consistently, using the same method [72], T. longibrachiatum caused lysis and overgrowth of R. solani and F. oxysporum f.sp. capsici in a dual cultures trial [67].
Mechanisms through which Trichoderma maintains resistance to plant pathogens have been clarified by numerous researchers [31,32,68,73]. Trichoderma can combat phytopathogenic fungi by mycoparasitism, which implicates growth suppression or destroying the pathogen and its structures at the same site [32]. Other mechanisms were reported, involving the triggering of defense-related genes within T. harzianum and T. asperellum-treated plants [30], producing volatile and nonvolatile enzymes and antibiotics [74,75], or inducing molecular and biochemical variations within the plants [33]. In our study, increased concentrations of defense-related enzymes PPO, PO, CAT, and CHT were considerably observed in plants treated with Trichoderma relative to the untreated and fungicide-treated plants under infection with R. solani. Similar to our findings, Hassan et al. [67] stated that T. asperellum and other bacterial bio-agents conferred resistance to cucumber plants against R. solani via elevating the PO, PPO, and CAT enzymes. Furthermore, Konappa et al. [76] specified that T. asperellum treatment conferred protection against R. solanacearum through initiating defense-related enzymes PPO, PO, and PAL-1,3-glucanase. This finding could support our results that the reduction in root rot and damping-off due to T. asperellum KSATC and T. asperellum KSAT1E were correlated with increased defense-related enzymes, thereby conferring resistance to cucumber plants. Moreover, the indirect effects of Trichoderma in controlling diseases through inducing plant-systemic resistance have been previously detailed in many studies [33,77]. The indirect effects include the induction of systemic plant resistance through products (elicitors) released from the cell walls of the plant host, improving plant growth, enhancing stress tolerance, and improving the uptake of nutrients.
In the present study, treatment with Trichoderma revealed an increased accumulation of TPC in cucumber plants due to R. solani infection. The highest elevation in TPC and PPO enzyme was attributed to the treatment with T. harzianum KSATR10 and KSATR9. The role of polyphenol oxidase activity in resistance to plant disease likely originates from its capability to oxidize phenolic compounds to quinines, which often reveal higher toxicity toward microorganisms relative to the original phenols [78]. This could explain our results that increases in TPC by T. harzianum KSATR10 and KSATR9 are associated with increased PPO enzyme. Also, the accumulation of phenolic compounds in Trichoderma-treated plants has been correlated with oxidative biochemical defense against pathogenic fungi [77,79]. Additionally, phenolic compounds can serve as electron and hydrogen donors, preventing root tissue from oxidative damage throughout pathogen attacks [80]. Therefore, we suggest that the accumulation of TPC may significantly contribute to cucumber protection against R. solani, as previously documented [81]. Additionally, all Trichoderma-treated plants, especially those treated with T. harzianum KSATR10 and KSATR9, experienced an elevated TCC level relative to healthy and infected control plants. These results coincide with previous studies on T. harzianum application on maize plants. When applied directly to the seeds or the soil, the fungus induced increased levels of chlorophyll and growth parameters [82].
Trichoderma species also have a diverse set of effectors and elicitors which are recognized by plant receptors to activate signaling and gene regulation, which serves as the foundation for Trichoderma spp. to develop R. solani defense responses [83]. For example, Doni et al. [84] stated that the inoculation of rice seedlings with T. asperellum coincides with the upregulation of some genes in rice that were recognized to be included for photosynthesis and chlorophyll biosynthesizing. Moreover, a recent study [79] indicated that 15 days after inoculation, promising increases in the relative expression levels of three defense-related genes (PAL, CHS, and HQT) were observed in all T. pubescens-treated tomato plants. Very recent findings also demonstrated that seeds bioprimed with T. asperellum, T. harzianum, and their combination induced the strengthening of the cell wall by the lignification and expression of six defense-related genes: CaPDF1.2, SOD, APx, GPx, PR-2, and PR-5 in pepper against anthracnose diseases caused by Colletotrichum truncatum [79].
5. Conclusions
Our study documented the association of three AGs of R. solani with the root rot of cucumber. It is interesting to note that, through pathogenicity tests, the non-pathogenic AG2-1 did not induce pre- or post-damping-off nor root rot in cucumber. Surprisingly, these non-pathogenic groups improved the growth of cucumber plants. In the current study, the treatment with T. asperellum KSATC and KSAT1E revealed a reduction in root rot and damping-off incidence percentages with values not significantly (p < 0.05) different from Rizolex-T fungicide. They also showed a marked increase in the concentrations of three defense-related enzymes (PO, CAT, and CHT) in treated cucumber plants. PPO enzyme and TPC significantly (p < 0.05) increased in cucumber plants treated with T. harzianum KSATR9 and KSATR10. The output of this study can be considered a step toward developing eco-friendly control measures using a combination between T. asperellum (KSATC and KSAT1E) and T. harzianum (KSATR9 and KSATR10) to control R. solani, enhance cucumber growth, and induce systemic resistance. Following this pilot study, future research can be focused on studying the combinatory effects of both T. asperellum and T. harzianum for the commercial production of bio-fertilizers using indigenous resources to promote sustainable agriculture in Saudi Arabia.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/su15097250/s1, Table S1: Rhizoctonia solani and Trichoderma species of the current study and those retrieved from GenBank used in phylogenetic analysis.
Author Contributions
Conceptualization, A.M.I. and S.M.E.-G.; methodology, S.M.E.-G., A.M.I. and M.I.A.; software, A.M.I. and. S.M.E.-G.; writing—original draft preparation, A.M.I. and M.I.A.; writing—review and editing, S.M.E.-G., A.M.I. and M.I.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia, project number [INST124].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data related to the current study are represented in the manuscript.
Acknowledgments
The authors thank the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia, for funding this research work (Project number INST124).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Crane, M.; Wehner, T.C.; Naegele, R.P. Cucumber Cultivars for Container Gardening and the Value of Field Trials for Predicting Cucumber Performance in Containers. HortScience 2018, 53, 16–22. [Google Scholar] [CrossRef]
- Mahmoud, A.F.; Abdalla, O.A. Biological Control of Fungi Associated with Damping-off and Root Rot Disease of Cucumber (Cucumis sativus L.). Arch. Phytopathol. Plant Prot. 2021, 54, 870–885. [Google Scholar] [CrossRef]
- El-Komy, M.H.; Al-Qahtani, R.M.; Widyawan, A.; Molan, Y.; Almasrahi, A. First Report of Fusarium Root and Stem Rot Caused by Fusarium oxysporum f. sp. radicis-cucumerinum on Greenhouse Cucumbers in Saudi Arabia. Plant Dis. 2021, 105, 3758. [Google Scholar] [CrossRef] [PubMed]
- Morsy, S.M.; Drgham, E.A.; Mohamed, G.M. Effect of Garlic and Onion Extracts or Their Intercropping on Suppressing Damping-off and Powdery Mildew Diseases and Growth Characteristics of Cucumber. Egypt. J. Phytopathol. 2009, 37, 35–46. [Google Scholar]
- Ristaino, J.B.; Johnston, S.A. Ecologically Based Approaches to Management of Phytophthora Blight on Bell Pepper. Plant Dis. 1999, 83, 1080–1089. [Google Scholar] [CrossRef]
- Simsek Ersahin, Y.; Haktanir, K.; Yanar, Y. Vermicompost suppresses Rhizoctonia solani Kühn in cucumber seedlings. J. Plant Dis. Prot. 2009, 116, 182–188. [Google Scholar] [CrossRef]
- Heflish, A.A.; Abdelkhalek, A.; Al-Askar, A.A.; Behiry, S.I. Protective and Curative Effects of Trichoderma asperelloides Ta41 on Tomato Root Rot Caused by Rhizoctonia solani Rs33. Agronomy 2021, 11, 1162. [Google Scholar] [CrossRef]
- Wallon, T.; Sauvageau, A.; Van der Heyden, H. Detection and Quantification of Rhizoctonia solani and Rhizoctonia solani AG1-IB Causing the Bottom Rot of Lettuce in Tissues and Soils by Multiplex QPCR. Plants 2021, 10, 57. [Google Scholar] [CrossRef]
- Williamson-Benavides, B.A.; Dhingra, A. Understanding Root Rot Disease in Agricultural Crops. Horticulturae 2021, 7, 33. [Google Scholar] [CrossRef]
- Erper, I.; Ozer, G.; Kalendar, R.; Avci, S.; Yildirim, E.; Alkan, M.; Turkkan, M. Genetic Diversity and Pathogenicity of Rhizoctonia spp. Isolates Associated with Red Cabbage in Samsun (Turkey). J. Fungi 2021, 7, 234. [Google Scholar] [CrossRef]
- Yang, G.; Li, C. General Description of Rhizoctonia Species Complex. In Plant pathology; Citeseer: Princeton, NJ, USA, 2012; ISBN 9535104896. [Google Scholar]
- Sneh, B.; Burpee, L.; Ogoshi, A. Identification of Rhizoctonia Species; APS Press: College Park, MD, USA, 1991; ISBN 089054123X. [Google Scholar]
- Yang, Y.; Zhao, C.; Guo, Z.; Wu, X. Anastomosis Groups and Pathogenicity of Binucleate Rhizoctonia Isolates Associated with Stem Canker of Potato in China. Eur. J. Plant Pathol. 2014, 139, 535–544. [Google Scholar] [CrossRef]
- Stodart, B.J.; Harvey, P.R.; Neate, S.M.; Melanson, D.L.; Scott, E.S. Genetic Variation and Pathogenicity of Anastomosis Group 2 Isolates of Rhizoctonia solani in Australia. Mycol. Res. 2007, 111, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Guleria, S.; Aggarwal, R.; Thind, T.S.; Sharma, T.R. Morphological and Pathological Variability in Rice Isolates of Rhizoctonia solani and Molecular Analysis of Their Genetic Variability. J. Phytopathol. 2007, 155, 654–661. [Google Scholar] [CrossRef]
- Gonzalez-Vera, A.D.; Bernardes-de-Assis, J.; Zala, M.; McDonald, B.A.; Correa-Victoria, F.; Graterol-Matute, E.J.; Ceresini, P.C. Divergence between Sympatric Rice-and Maize-Infecting Populations of Rhizoctonia solani AG-1 IA from Latin America. Phytopathology 2010, 100, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Ciampi, M.B.; Gale, L.R.; Lemos, E.G.; Ceresini, P.C. Distinctively Variable Sequence-Based Nuclear DNA Markers for Multilocus Phylogeography of the Soybean-and Rice-Infecting Fungal Pathogen Rhizoctonia solani AG-1 IA. Genet. Mol. Biol. 2009, 32, 840–846. [Google Scholar] [CrossRef]
- Carling, D.E.; Kuninaga, S.; Brainard, K.A. Hyphal Anastomosis Reactions, RDNA-Internal Transcribed Spacer Sequences, and Virulence Levels among Subsets of Rhizoctonia solani Anastomosis Group-2 (AG-2) and AG-BI. Phytopathology 2002, 92, 43–50. [Google Scholar] [CrossRef]
- Fenille, R.C.; Ciampi, M.B.; Kuramae, E.E.; Souza, N.L. Identification of Rhizoctonia solani Associated with Soybean in Brazil by RDNA-ITS Sequences. Fitopatol. Bras. 2003, 28, 413–419. [Google Scholar] [CrossRef]
- Zachow, C.; Grosch, R.; Berg, G. Impact of Biotic and A-Biotic Parameters on Structure and Function of Microbial Communities Living on Sclerotia of the Soil-Borne Pathogenic Fungus Rhizoctonia solani. Appl. Soil Ecol. 2011, 48, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, N.S.; Doni, F.; Mispan, M.S.; Saiman, M.Z.; Yusuf, Y.M.; Oke, M.A.; Suhaimi, N.S.M. Harnessing Trichoderma in Agriculture for Productivity and Sustainability. Agronomy 2021, 11, 2559. [Google Scholar] [CrossRef]
- Kumari, N.; Katoch, S. Wilt and Root Rot Complex of Important Pulse Crops: Their Detection and Integrated Management. In Management of Fungal Pathogens in Pulses: Current Status and Future Challenges; Springer: Cham, Switzerland, 2020; pp. 93–119. [Google Scholar]
- Gaulin, E.; Jacquet, C.; Bottin, A.; Dumas, B. Root Rot Disease of Legumes Caused by Aphanomyces euteiches. Mol. Plant Pathol. 2007, 8, 539–548. [Google Scholar] [CrossRef]
- Myresiotis, C.K.; Karaoglanidis, G.S.; Tzavella-Klonari, K. Resistance of Botrytis cinerea Isolates from Vegetable Crops to Anilinopyrimidine, Phenylpyrrole, Hydroxyanilide, Benzimidazole, and Dicarboximide Fungicides. Plant Dis. 2007, 91, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Saba, H.; Vibhash, D.; Manisha, M.; Prashant, K.S.; Farhan, H.; Tauseef, A. Trichoderma—A Promising Plant Growth Stimulator and Biocontrol Agent. Mycosphere 2012, 3, 524–531. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhuang, W.-Y. Trichoderma brevicrassum Strain TC967 with Capacities of Diminishing Cucumber Disease Caused by Rhizoctonia solani and Promoting Plant Growth. Biol. Control 2020, 142, 104151. [Google Scholar] [CrossRef]
- Singh, S.; Balodi, R. Bio-Management of Soil Borne Pathogens Infesting Cucumber (Cucumis Sativus L.) under Protected Cultivation System. Biol. Control 2021, 157, 104569. [Google Scholar] [CrossRef]
- Sánchez-Montesinos, B.; Santos, M.; Moreno-Gavíra, A.; Marín-Rodulfo, T.; Gea, F.J.; Diánez, F. Biological Control of Fungal Diseases by Trichoderma aggressivum f. europaeum and Its Compatibility with Fungicides. J. Fungi 2021, 7, 598. [Google Scholar] [CrossRef]
- Rini, C.R.; Sulochana, K.K. Management of Seedling Rot of Chilli (Capsicum annuum L.) Using Trichoderma spp. and Fluorescent pseudomonads (Pseudomonas fluorescens). J. Trop. Agric. 2006, 44, 79–82. [Google Scholar]
- Gallou, A.; Cranenbrouck, S.; Declerck, S. Trichoderma harzianum Elicits Defence Response Genes in Roots of Potato Plantlets Challenged by Rhizoctonia solani. Eur. J. Plant Pathol. 2009, 124, 219–230. [Google Scholar] [CrossRef]
- Fernandes, T.; Lopes, F.A.C.; Steindorff, A.S.; Brandao, R.S.; Jesuino, R.S.A.; Ulhoa, C.J. Mycoparasitism Studies of Trichoderma Species against Three Phytopathogenic Fungi: Evaluation of Antagonism and Hydrolytic Enzyme Production. Biotechnol. Lett. 2013, 35, 1461–1468. [Google Scholar]
- Howell, C.R. Understanding the Mechanisms Employed by Trichoderma virens to Effect Biological Control of Cotton Diseases. Phytopathology 2006, 96, 178–180. [Google Scholar] [CrossRef]
- Nawrocka, J.; Małolepsza, U. Diversity in Plant Systemic Resistance Induced by Trichoderma. Biol. Control 2013, 67, 149–156. [Google Scholar] [CrossRef]
- Nawrocka, J.; Gromek, A.; Małolepsza, U. Nitric Oxide as a Beneficial Signaling Molecule in Trichoderma atroviride TRS25-Induced Systemic Defense Responses of Cucumber Plants against Rhizoctonia solani. Front. Plant Sci. 2019, 10, 421. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, N.; Yong, X.; Yang, X.; Shen, Q. Biocontrol of Rhizoctonia solani Damping-off Disease in Cucumber with Bacillus pumilus SQR-N43. Microbiol. Res. 2012, 167, 135–143. [Google Scholar] [CrossRef]
- Verma, M.; Brar, S.K.; Tyagi, R.D.; Surampalli, R.Y.; Valero, J.R. Antagonistic Fungi, Trichoderma spp.: Panoply of Biological Control. Biochem. Eng. J. 2007, 37, 1–20. [Google Scholar] [CrossRef]
- Lorito, M.; Woo, S.L.; Harman, G.E.; Monte, E. Translational Research on Trichoderma: From’omics to the Field. Annu. Rev. Phytopathol. 2010, 48, 395–417. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, O.D.; Sinclair, J.B. Basic Plant Pathology Methods, 2nd ed.; Lewis Publishers: Boca Raton, FL, USA, 1995. [Google Scholar]
- Barnett, H.L.; Hunter, B.B. Illustrated Genera of Imperfect Fungi; Burgess Publishing Company: Minneapolis, MN, USA, 1972. [Google Scholar]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A Plant DNA Minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J.; Nnis, M.A.; Gelfand, D.H.; Sninsky, J.J. PCR Protocols-A Guide to Methods and Applications; The University of Michigan: Ann Arbor, MI, USA; Academic Press: Cambridge, UK, 1990. [Google Scholar]
- Misawa, T.; Kurose, D.; Shishido, K.; Toda, T.; Kuninaga, S. Characterization of a New Subgroup of Rhizoctonia solani Anastomosis Group 3 (AG-3 TM) Associated with Tomato Leaf Blight. J. Gen. Plant Pathol. 2020, 86, 457–467. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.; Elsharkawy, M.M.; Villajuan-Abgona, R.; Hyakumachi, M. A Nonpathogenic Species of Binucleate Rhizoctonia Inhibits the Formation of Infection Structures Caused by Rhizoctonia solani on Cucumber. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2015, 65, 208–214. [Google Scholar]
- Montealegre, J.; Valderrama, L.; Sánchez, S.; Herrera, R.; Besoain, X.; Pérez, L.M. Biological Control of Rhizoctonia solani in Tomatoes with Trichoderma harzianum Mutants. Electron. J. Biotechnol. 2010, 13, 1–2. [Google Scholar] [CrossRef]
- Ghosh, P.; Roy, A.; Hess, D.; Ghosh, A.; Das, S. Deciphering the Mode of Action of a Mutant Allium sativum Leaf Agglutinin (MASAL), a Potent Antifungal Protein on Rhizoctonia solani. BMC Microbiol. 2015, 15, 1–16. [Google Scholar] [CrossRef]
- Elad, Y.; Chet, I.; Katan, J. Trichoderma harzianum: A Biocontrol Agent Effective against Sclerotium rolfsii and Rhizoctonia solani. Phytopathology 1980, 70, 119–121. [Google Scholar] [CrossRef]
- Ketta, H.A.; Elkhateeb, N.M.; Saleh, M.M.; Kamel, S.M. Efficiency Assessment of Combinations between Rhizobium leguminosarum and Trichoderma spp. for Controlling of Pea (Pisum sativum L.) Damping-off Disease. Egypt. J. Phytopathol. 2021, 49, 1–14. [Google Scholar] [CrossRef]
- Ismail, A.M. Efficacy of Copper Oxide and Magnesium Oxide Nanoparticles on Controlling Black Scurf Disease on Potato. Egypt. J. Phytopathol. 2021, 49, 116–130. [Google Scholar] [CrossRef]
- Ismail, A.M.; El-Gawad, A.; Mona, E. Antifungal Activity of MgO and ZnO Nanoparticles against Powdery Mildew of Pepper under Greenhouse Conditions. Egypt. J. Agric. Res. 2021, 99, 421–434. [Google Scholar]
- Matta, A.; Dimond, A.E. Symptoms of Fusarium Wilt in Relation to Quantity of Fungus and Enzyme Activity in Tomato Stems. Phytopathology 1963, 53, 574. [Google Scholar]
- Allam, A.I.; Hollis, J.P. Sulfide Inhibition of Oxidases in Rice Roots. Phytopathology 1972, 62, 634–639. [Google Scholar] [CrossRef]
- Aebi, H.E. Catalase. In Methods of Enzymatic Analysis; Academic Press: Cambridge, MA, USA, 1983; pp. 273–286. [Google Scholar]
- Wirth, S.J.; Wolf, G.A. Dye-Labelled Substrates for the Assay and Detection of Chitinase and Lysozyme Activity. J. Microbiol. Methods 1990, 12, 197–205. [Google Scholar] [CrossRef]
- Snell, F.D.; Snell, C.T. Colorimetric Methods; D. Van Nostrand Co. Inc.: Toronto, ON, Canada; New York, NY, USA; London, UK, 1953; Volume III, 606p. [Google Scholar]
- Nagata, M.; Yamashita, I. Simple Method for Simultaneous Determination of Chlorophyll and Carotenoids in Tomato Fruit. Nippon. Shokuhin Kogyo Gakkaishi 1992, 39, 925–928. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1. [Google Scholar] [CrossRef]
- Snedecor, G.W.; Cochran, W.G. Statistical Methods, 7th ed.; Iowa State University Press: Ames, IA, USA, 1980. [Google Scholar]
- Erper, I.; Hatat Karaca, G.; Özkoc, I. Characterization of Rhizoctonia species Causing Root-Rot of Cucumber Plants Grown in Greenhouses in Samsun, Turkey. In II Balkan Symposium on Vegetables and Potatoes; ISHS: Thessaloniki, Greece, 2000; pp. 531–534. [Google Scholar]
- Eken, C.; Tuncer, S. Rhizoctonia Species and Anastomosis Groups Isolated from Tomato and Cucumber in Erzincan, Turkey. Int. J. Res. Agric. For. 2019, 6, 26–31. [Google Scholar]
- Ersahin, Y.S.; Haktanir, K.; Yanar, Y. Vermicompost Suppresses Rhizoctonia solani Kühn in Cucumber Seedlings/Wurmkompost Unterdrückt Die Entwicklung von Rhizoctonia solani Kühn an Gurkenkeimlingen. J. Plant Dis. Prot. 2009, 116, 182–188. [Google Scholar] [CrossRef]
- Yıldırım, E.; Erper, I. Characterization and Pathogenicity of Rhizoctonia spp. Isolated from Vegetable Crops Grown in Greenhouses in Samsun Province, Turkey. Biosci. J. 2017, 33, 257–267. [Google Scholar]
- Mirmajlessi, S.M.; Safaie, N.; Mostafavi, H.A.; Mansouripour, S.M.; Mahmoudy, S.B. Genetic Diversity among Crown and Root Rot Isolates of Rhizoctonia solani Isolated from Cucurbits Using PCR Based Techniques. Afr. J. Agric. Res. 2012, 7, 583–590. [Google Scholar]
- Desvani, S.D.; Lestari, I.B.; Wibowo, H.R.; Supyani; Poromarto, S.H. Hadiwiyono Morphological Characteristics and Virulence of Rhizoctonia solani Isolates Collected from Some Rice Production Areas in Some Districts of Central Java. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2018; Volume 2014, p. 20068. [Google Scholar]
- Mukhopadhyay, A. Biological control of soil borne plant pathogens by Trichoderma spp. Indian J. Mycol. Plant Pathol. 1987, 17, 1–10. [Google Scholar]
- Ryu, J.-Y.; Jin, R.-D.; Kim, Y.-W.; Lee, H.-B.; Kim, K.-Y. Biocontrol of Damping-off (Rhizoctonia solani) in Cucumber by Trichoderma asperellum T-5. Korean J. Soil Sci. Fertil. 2006, 39, 185–194. [Google Scholar]
- Hassan, M.; Ahmed, H.; Kamel, S.; El-Hamed, A.; Yousef, H. Biological Control of Damping-off and Root Rot Disease Caused by Rhizoctonia solani on Cucumber Plants. Fayoum J. Agric. Res. Dev. 2021, 35, 525–541. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, B.; Zhang, J.; Gan, Y. Identification of the Antifungal Activity of Trichoderma longibrachiatum T6 and Assessment of Bioactive Substances in Controlling Phytopathgens. Pestic. Biochem. Physiol. 2018, 147, 59–66. [Google Scholar] [CrossRef]
- Trillas, M.I.; Casanova, E.; Cotxarrera, L.; Ordovás, J.; Borrero, C.; Avilés, M. Composts from Agricultural Waste and the Trichoderma asperellum Strain T-34 Suppress Rhizoctonia solani in Cucumber Seedlings. Biol. Control 2006, 39, 32–38. [Google Scholar] [CrossRef]
- Huang, X.; Chen, L.; Ran, W.; Shen, Q.; Yang, X. Trichoderma harzianum Strain SQR-T37 and Its Bio-Organic Fertilizer Could Control Rhizoctonia solani Damping-off Disease in Cucumber Seedlings Mainly by the Mycoparasitism. Appl. Microbiol. Biotechnol. 2011, 91, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Inbar, J.; Menendez, A.N.A.; Chet, I. Hyphal Interaction between Trichoderma harzianum and Sclerotinia sclerotiorum and Its Role in Biological Control. Soil Biol. Biochem. 1996, 28, 757–763. [Google Scholar] [CrossRef]
- De Melo, I.S.; Faull, J.L. Parasitism of Rhizoctonia solani by Strains of Trichoderma spp. Sci. Agric. 2000, 57, 55–59. [Google Scholar] [CrossRef]
- El-Kazzaz, M.K.; Ghoneim, K.E.; Agha, M.K.M.; Helmy, A.; Behiry, S.I.; Abdelkhalek, A.; Saleem, M.H.; Al-Askar, A.A.; Arishi, A.A.; Elsharkawy, M.M. Suppression of Pepper Root Rot and Wilt Diseases Caused by Rhizoctonia solani and Fusarium oxysporum. Life 2022, 12, 587. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Ashraf, S. Role of Trichoderma spp. as a Biocontrol Agent of Fungal Plant Pathogens. In Probiotics and Plant Health; Kumar, V., Kumar, M., Sharma, S., Prasad, R., Eds.; Springer: Singapore, 2017; pp. 497–506. ISBN 978-981-10-3473-2. [Google Scholar]
- Nawrocka, J.; Małolepsza, U.; Szymczak, K.; Szczech, M. Involvement of Metabolic Components, Volatile Compounds, PR Proteins, and Mechanical Strengthening in Multilayer Protection of Cucumber Plants against Rhizoctonia solani Activated by Trichoderma atroviride TRS25. Protoplasma 2018, 255, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Konappa, N.; Krishnamurthy, S.; Siddaiah, C.N.; Ramachandrappa, N.S.; Chowdappa, S. Evaluation of Biological Efficacy of Trichoderma asperellum against Tomato Bacterial Wilt Caused by Ralstonia solanacearum. Egypt. J. Biol. Pest Control. 2018, 28, 63. [Google Scholar] [CrossRef]
- Yedidia, I.; Shoresh, M.; Kerem, Z.; Benhamou, N.; Kapulnik, Y.; Chet, I. Concomitant Induction of Systemic Resistance to Pseudomonas syringae Pv. lachrymans in Cucumber by Trichoderma asperellum (T-203) and Accumulation of Phytoalexins. Appl. Environ. Microbiol. 2003, 69, 7343–7353. [Google Scholar] [CrossRef]
- Taranto, F.; Pasqualone, A.; Mangini, G.; Tripodi, P.; Miazzi, M.M.; Pavan, S.; Montemurro, C. Polyphenol Oxidases in Crops: Biochemical, Physiological and Genetic Aspects. Int. J. Mol. Sci. 2017, 18, 377. [Google Scholar] [CrossRef]
- Behiry, S.; Soliman, S.A.; Massoud, M.A.; Abdelbary, M.; Kordy, A.M.; Abdelkhalek, A.; Heflish, A. Trichoderma pubescens Elicit Induced Systemic Resistance in Tomato Challenged by Rhizoctonia solani. J. Fungi 2023, 9, 167. [Google Scholar] [CrossRef]
- Singh, B.N.; Singh, A.; Singh, S.P.; Singh, H.B. Trichoderma harzianum-Mediated Reprogramming of Oxidative Stress Response in Root Apoplast of Sunflower Enhances Defence against Rhizoctonia solani. Eur. J. Plant Pathol. 2011, 131, 121–134. [Google Scholar] [CrossRef]
- Han, H.S.; Lee, K.D. Plant Growth Promoting Rhizobacteria Effect on Antioxidant Status, Photosynthesis, Mineral Uptake and Growth of Lettuce under Soil Salinity. Res. J. Agric. Biol. Sci. 2005, 1, 210–215. [Google Scholar]
- Akladious, S.A.; Abbas, S.M. Application of Trichoderma harziunum T22 as a Biofertilizer Supporting Maize Growth. Afr. J. Biotechnol. 2012, 11, 8672–8683. [Google Scholar]
- Abbas, A.; Mubeen, M.; Zheng, H.; Sohail, M.A.; Shakeel, Q.; Solanki, M.K.; Iftikhar, Y.; Sharma, S.; Kashyap, B.K.; Hussain, S. Trichoderma spp. Genes Involved in the Biocontrol Activity against Rhizoctonia solani. Front. Microbiol. 2022, 13, 884469. [Google Scholar] [CrossRef] [PubMed]
- Doni, F.; Fathurrahman, F.; Mispan, M.S.; Suhaimi, N.S.M.; Yusoff, W.M.W.; Uphoff, N. Transcriptomic Profiling of Rice Seedlings Inoculated with the Symbiotic Fungus Trichoderma asperellum SL2. J. Plant Growth Regul. 2019, 38, 1507–1515. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).