Mercury Enrichment Characteristics and Rhizosphere Bacterial Community of Ramie (Boehmeria Nivea L. Gaud.) in Mercury-Contaminated Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design
2.3. Sample Collection
2.4. Chemical Analyses
2.5. Extraction, Sequencing, and Processing of Soil Bacterial DNA
2.6. Statistical Analysis
3. Results and Discussion
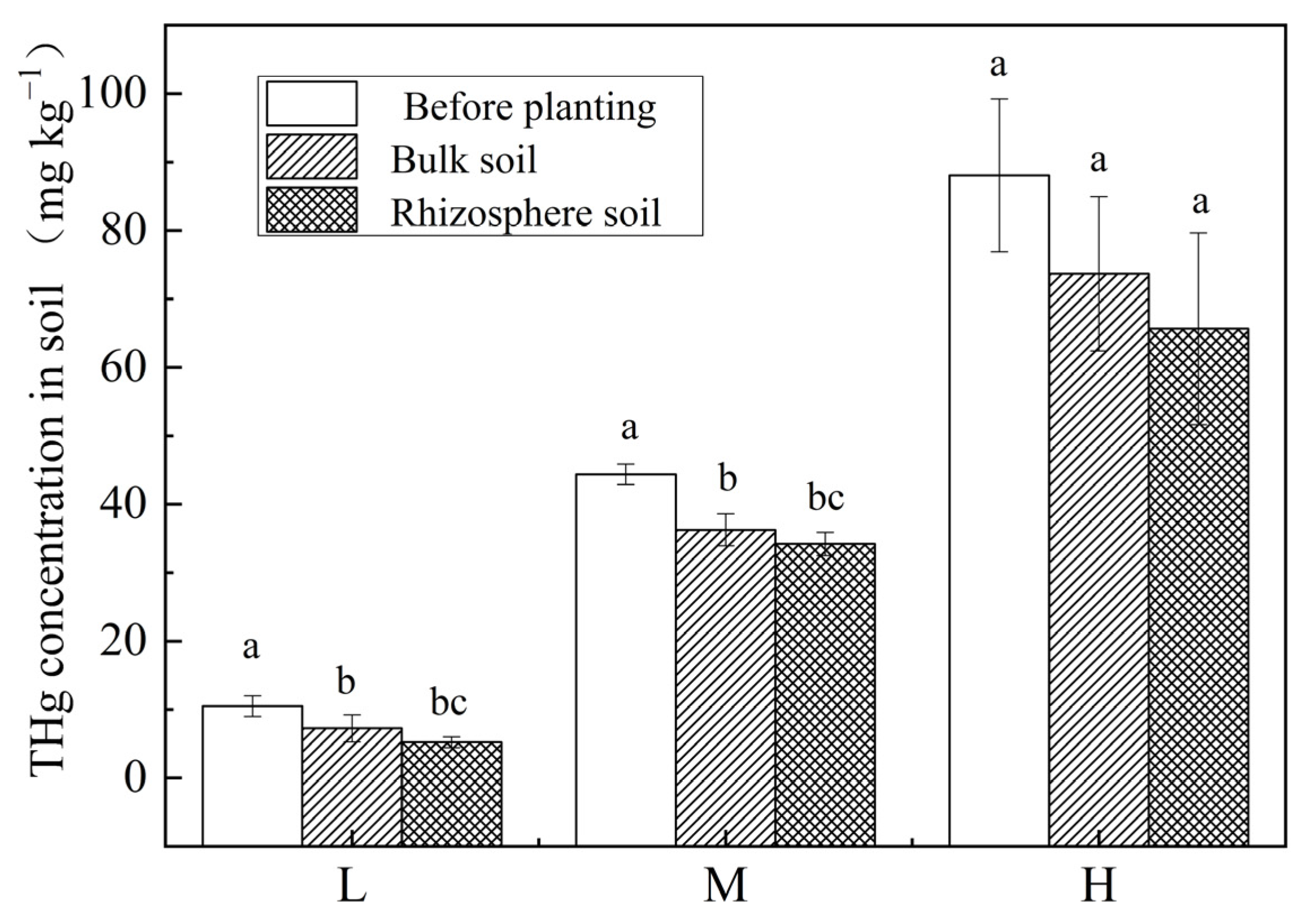
3.1. Total Mercury Concentration in Soil and Plants
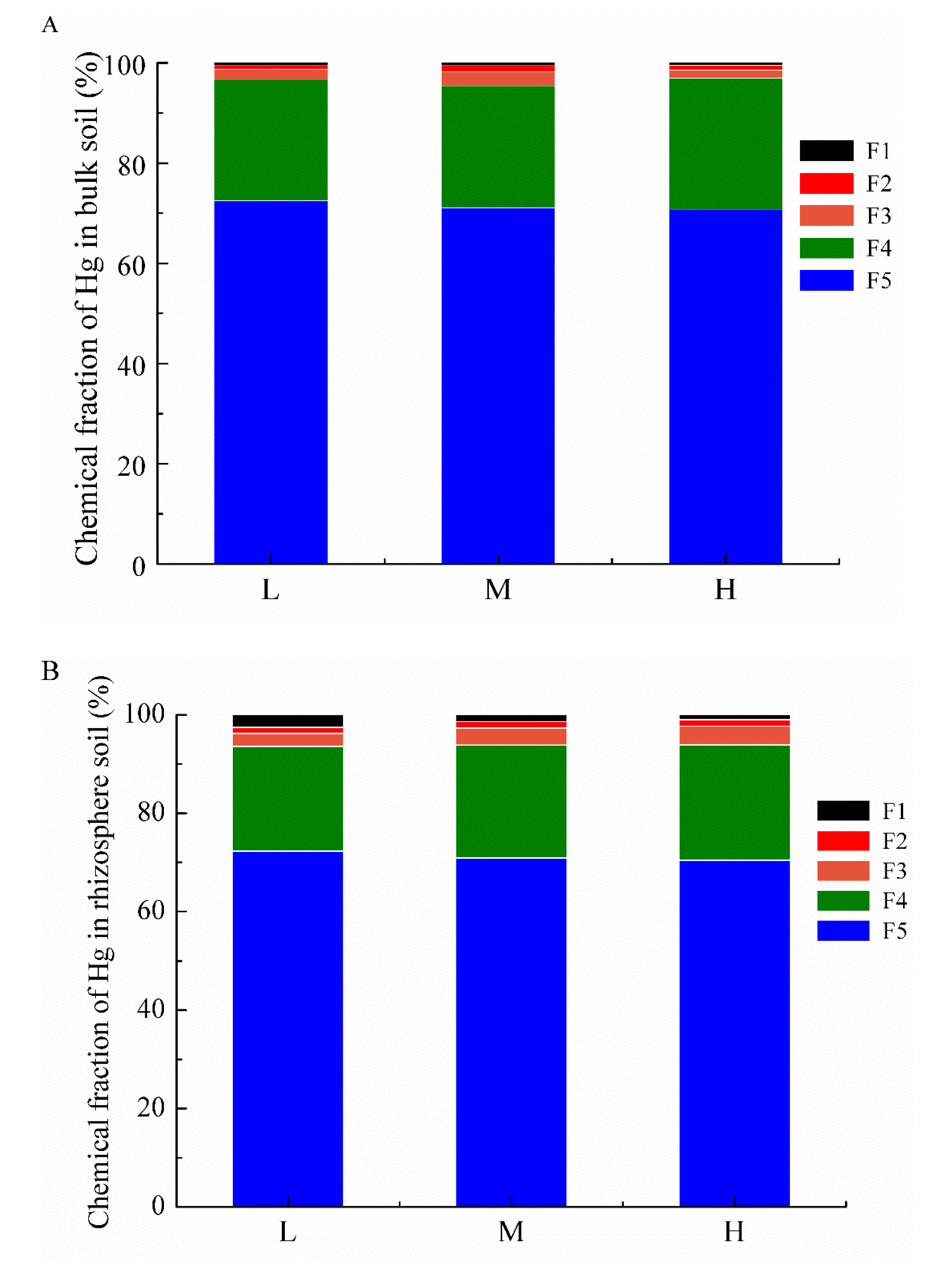
3.2. Changes of Hg Chemical Fractions between Bulk Soil and Rhizosphere Soil
3.3. Diversity and Structure of the Soil Bacterial Community in Hg-Contaminated Soils
3.3.1. Bacterial α-Diversity
3.3.2. Bacterial β-Diversity
3.3.3. Abundances of Bacterial Phyla
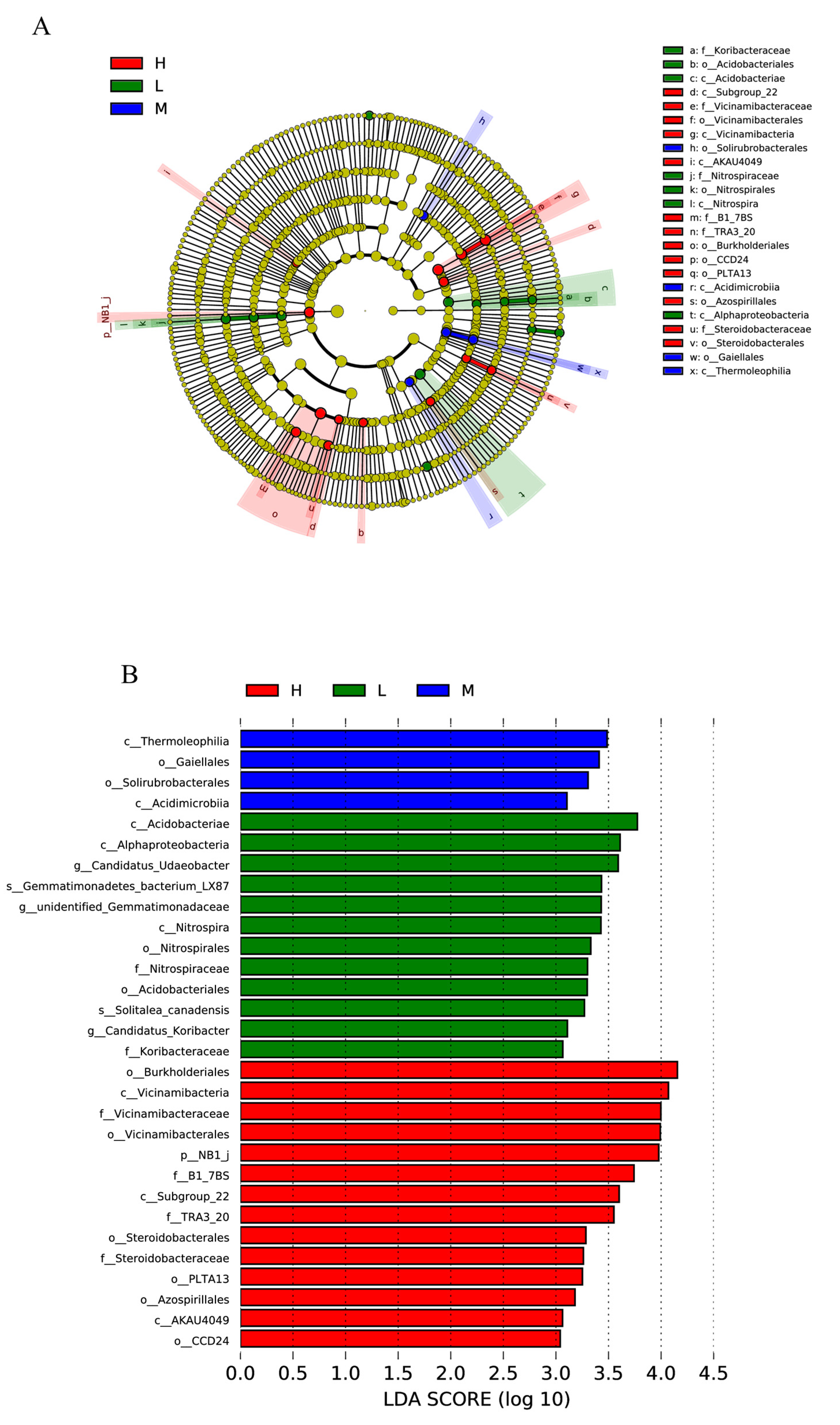
3.4. Differentially Abundant Biomarkers in Plots with Different Levels of Hg Contamination
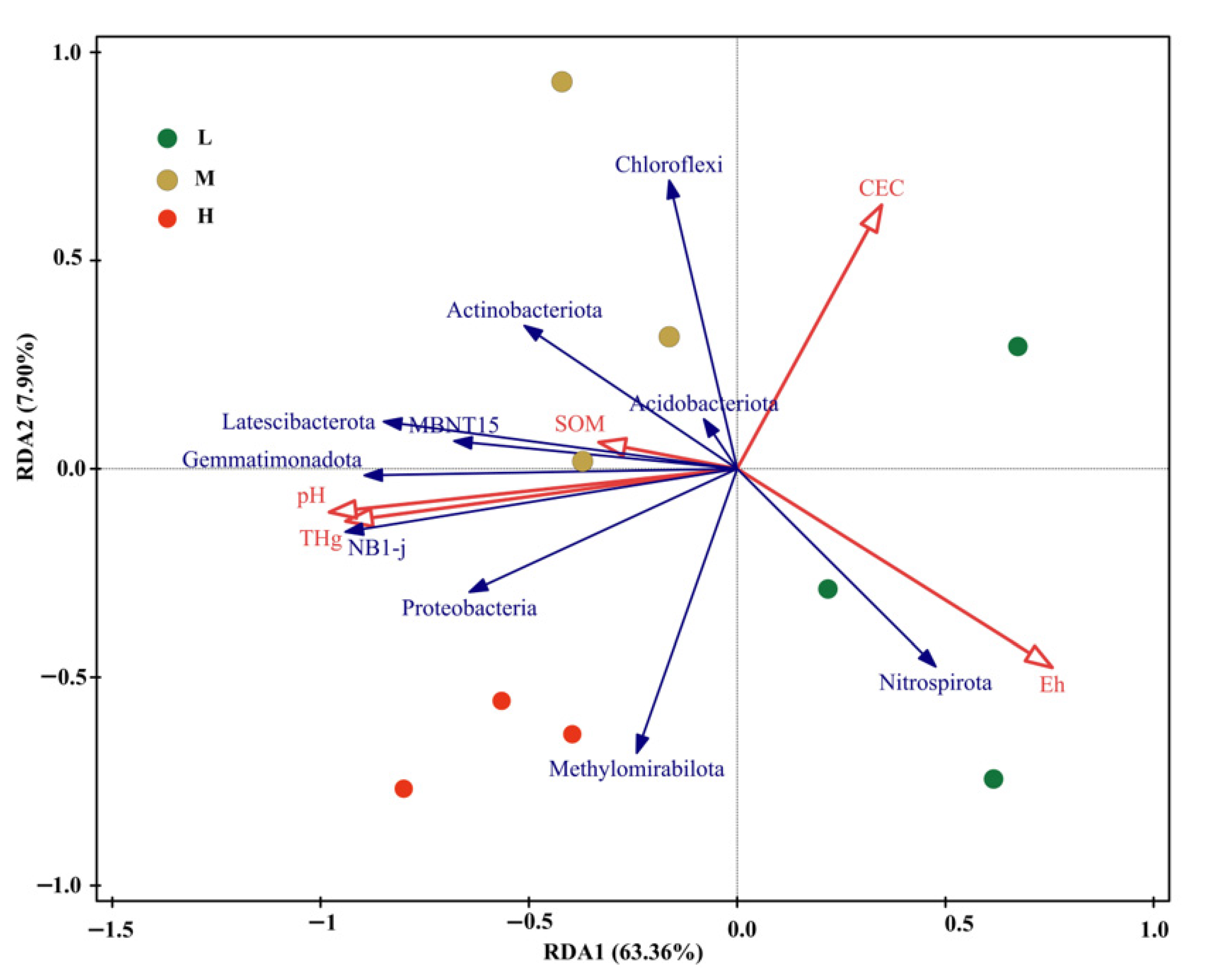
3.5. Effects of Environmental Variables on the Bacterial Community
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; Shaheen, S.M.; Jing, M.; Anderson, C.W.N.; Swertz, A.-C.; Wang, S.-L.; Feng, X.; Rinklebe, J. Mobilization, Methylation, and Demethylation of Mercury in a Paddy Soil Under Systematic Redox Changes. Environ. Sci. Technol. 2021, 55, 10133–10141. [Google Scholar] [CrossRef]
- Singh, A.D.; Khanna, K.; Kour, J.; Dhiman, S.; Bhardwaj, T.; Devi, K.; Sharma, N.; Kumar, P.; Kapoor, N.; Sharma, P.; et al. Critical review on biogeochemical dynamics of mercury (Hg) and its abatement strategies. Chemosphere 2023, 319, 137917. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, X.; Larssen, T.; Shang, L.; Vogt, R.D.; Rothenberg, S.E.; Li, P.; Zhang, H.; Lin, Y. Fractionation, distribution and transport of mercury in rivers and tributaries around Wanshan Hg mining district, Guizhou province, southwestern China: Part 1-Total mercury. Appl. Geochem. 2010, 25, 633–641. [Google Scholar] [CrossRef]
- Saldarriaga, J.F.; Lopez, J.E.; Diaz-Garcia, L.; Montoya-Ruiz, C. Changes in Lolium perenne L. rhizosphere microbiome during phytoremediation of Cd- and Hg-contaminated soils. Sci. Poll. Res. Int. 2023. [Google Scholar] [CrossRef]
- Yang, B.; Zhou, M.; Shu, W.S.; Lan, C.Y.; Ye, Z.H.; Qiu, R.L.; Jie, Y.C.; Cui, G.X.; Wong, M.H. Constitutional tolerance to heavy metals of a fiber crop, ramie (Boehmeria nivea), and its potential usage. Environ. Pollut. 2010, 158, 551–558. [Google Scholar] [CrossRef]
- Pandey, V.C.; Bajpai, O.; Singh, N. Energy crops in sustainable phytoremediation. Renew. Sust. Energy Rev. 2016, 54, 58–73. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Huang, D.Y.; Liu, S.L.; Luo, Z.C.; Rao, Z.X.; Cao, X.L.; Ren, X.F. Accumulation and subcellular distribution of cadmium in ramie (Boehmeria nivea L. Gaud.) planted on elevated soil cadmium contents. Plant Soil Environ. 2013, 59, 57–61. [Google Scholar] [CrossRef]
- Zhu, S.J.; Shi, W.J.; Zhang, J. Effect of different ramie (Boehmeria nivea L. Gaud) cultivars on the adsorption of heavy metal ions cadmium and lead in the remediation of contaminated farmland soils. Open Chem. 2022, 20, 444–454. [Google Scholar] [CrossRef]
- Gong, X.M.; Huang, D.L.; Liu, Y.G.; Zeng, G.M.; Wang, R.Z.; Wan, J.; Zhang, C.; Cheng, M.; Qin, X.; Xue, W.J. Stabilized Nanoscale Zerovalent Iron Mediated Cadmium Accumulation and Oxidative Damage of Boehmeria nivea (L.) Gaudich Cultivated in Cadmium Contaminated Sediments. Environ. Sci. Technol. 2017, 51, 11308–11316. [Google Scholar] [CrossRef]
- Xu, J.; Xing, Y.; Wang, J.; Yang, Y.; Ye, C.; Sun, R. Effect of poly-gamma-glutamic acid on the phytoremediation of ramie (Boehmeria nivea L.) in the Hg-contaminated soil. Chemosphere 2023, 312, 137280. [Google Scholar] [CrossRef]
- Wu, B.H.; Luo, S.H.; Luo, H.Y.; Huang, H.Y.; Xu, F.; Feng, S.; Xu, H. Improved phytoremediation of heavy metal contaminated soils by Miscanthus floridulus under a varied rhizosphere ecological characteristic. Sci. Total Environ. 2022, 808, 151995. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.L.; Li, Z.H.; Xi, B.D.; Xu, Q.G.; Tan, W.B. Responses of bacterial taxonomic attributes to mercury species in rhizosphere paddy soil under natural sulphur-rich biochar amendment. Ecotoxicol. Environ. Saf. 2022, 229, 151995. [Google Scholar] [CrossRef] [PubMed]
- Frossard, A.; Hartmann, M.; Frey, B. Tolerance of the forest soil microbiome to increasing mercury concentrations. Soil Biol. Biochem. 2017, 105, 162–176. [Google Scholar] [CrossRef]
- Zhao, S.W.; Qin, L.Y.; Wang, L.F.; Sun, X.Y.; Yu, L.; Wang, M.; Chen, S.B. Soil bacterial community responses to cadmium and lead stabilization during ecological restoration of an abandoned mine. Soil Use Manag. 2022, 38, 1459–1469. [Google Scholar] [CrossRef]
- Bach, E.M.; Williams, R.J.; Hargreaves, S.K.; Yang, F.; Hofmockel, K.S. Greatest soil microbial diversity found in micro-habitats. Soil Biol. Biochem. 2018, 118, 217–226. [Google Scholar] [CrossRef]
- Liu, Y.R.; Delgado-Baquerizo, M.; Bi, L.; Zhu, J.; He, J.Z. Consistent responses of soil microbial taxonomic and functional attributes to mercury pollution across China. Microbiome 2018, 6, 183. [Google Scholar] [CrossRef]
- Pignataro, A.; Moscatelli, M.C.; Mocali, S.; Grego, S.; Benedetti, A. Assessment of soil microbial functional diversity in a coppiced forest system. Appl. Soil Ecol. 2012, 62, 115–123. [Google Scholar] [CrossRef]
- Luo, J.P.; Tao, Q.; Jupa, R.; Liu, Y.K.; Wu, K.R.; Song, Y.C.; Li, J.X.; Huang, Y.; Zou, L.Y.; Liang, Y.C.; et al. Role of Vertical Transmission of Shoot Endophytes in Root-Associated Microbiome Assembly and Heavy Metal Hyperaccumulation in Sedum alfredii. Environ. Sci. Technol. 2019, 53, 6954–6963. [Google Scholar] [CrossRef]
- Frossard, A.; Donhauser, J.; Mestrot, A.; Gygax, S.; Baath, E.; Frey, B. Long- and short-term effects of mercury pollution on the soil microbiome. Soil Biol. Biochem. 2018, 120, 191–199. [Google Scholar] [CrossRef]
- Hou, J.Y.; Liu, W.X.; Wu, L.H.; Ge, Y.Y.; Hu, P.J.; Li, Z.; Christie, P. Rhodococcus sp. NSX2 modulates the phytoremediation efficiency of a trace metal-contaminated soil by reshaping the rhizosphere microbiome. Appl. Soil Ecol. 2019, 133, 62–69. [Google Scholar] [CrossRef]
- Li, X.H.; Zhao, L.; Teng, Y.; Luo, Y.M.; Huang, B.; Liu, B.L.; Zhao, Q.G. Characteristics, spatial distribution and risk assessment of combined mercury and cadmium pollution in farmland soils surrounding mercury mining areas in Guizhou. Ecol. Environ. Sci. 2022, 31, 1629–1636. (In Chinese) [Google Scholar]
- Ata-Ul-Karim, S.T.; Cang, L.; Wang, Y.J.; Zhou, D.M. Interactions between nitrogen application and soil properties and their impacts on the transfer of cadmium from soil to wheat (Triticum aestivum L.) grain. Geoderma 2020, 357, 113923. [Google Scholar] [CrossRef]
- Zhu, H.; Teng, Y.; Wang, X.; Zhao, L.; Ren, W.; Luo, Y.; Christie, P. Changes in clover rhizosphere microbial community and diazotrophs in mercury-contaminated soils. Sci. Total Environ. 2021, 767, 145473. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Gong, P.; Ding, W.; Wu, S.C.; Yu, X.W.; Liang, P. Mercury uptake by Paspalum distichum L. in relation to the mercury distribution pattern in rhizosphere soil. Environ. Sci. Poll. Res. 2021, 28, 66990–66997. [Google Scholar] [CrossRef]
- Hou, J.Y.; Liu, W.X.; Wang, B.B.; Wang, Q.L.; Luo, Y.M.; Franks, A.E. PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere 2015, 138, 592–598. [Google Scholar] [CrossRef]
- Bussan, D.D.; Douvris, C.; Cizdziel, J.V. Mercury methylation potentials in sediments of an ancient cypress wetland using species-specific isotope dilution GC-ICP-MS. Molecules 2022, 27, 4911. [Google Scholar] [CrossRef]
- Qiao, D.M.; Lu, H.F.; Zhang, X.X. Change in phytoextraction of Cd by rapeseed (Brassica napus L.) with application rate of organic acids and the impact of Cd migration from bulk soil to the rhizosphere. Environ. Pollut. 2020, 267, 115452. [Google Scholar] [CrossRef]
- Sas-Nowosielska, A.; Galimska-Stypa, R.; Kucharski, R.; Zielonka, U.; Malkowski, E.; Gray, L. Remediation aspect of microbial changes of plant rhizosphere in mercury contaminated soil. Environ. Monit. Assess. 2008, 137, 101–109. [Google Scholar] [CrossRef]
- Hao, X.D.; Bai, L.Y.; Liu, X.D.; Zhu, P.; Liu, H.W.; Xiao, Y.H.; Geng, J.B.; Liu, Q.J.; Huang, L.H.; Jiang, H.D. Cadmium Speciation Distribution Responses to Soil Properties and Soil Microbes of Plow Layer and Plow Pan Soils in Cadmium-Contaminated Paddy Fields. Front. Microbiol. 2021, 12, 774301. [Google Scholar] [CrossRef]
- Moreno, F.N.; Anderson, C.W.N.; Stewart, R.B.; Robinson, B.H. Phytofiltration of mercury-contaminated water: Volatilisation and plant-accumulation aspects. Environ. Exp. Bot. 2008, 62, 78–85. [Google Scholar] [CrossRef]
- Liu, Y.R.; Wang, J.J.; Zheng, Y.M.; Zhang, L.M.; He, J.Z. Patterns of Bacterial Diversity Along a Long-Term Mercury-Contaminated Gradient in the Paddy Soils. Microb. Ecol. 2014, 68, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.S.; Gu, C.H.; Feng, X.B.; Hurley, J.P.; Krabbenhoft, D.P.; Lepak, R.F.; Zhu, W.; Zheng, L.R.; Hu, T.D. Distribution and geochemical speciation of soil mercury in Wanshan Hg mine: Effects of cultivation. Geoderma 2016, 272, 32–38. [Google Scholar] [CrossRef]
- Wang, L.; Wang, L.A.; Zhan, X.Y.; Huang, Y.K.; Wang, J.; Wang, X. Response mechanism of microbial community to the environmental stress caused by the different mercury concentration in soils. Ecotoxicol. Environ. Saf. 2020, 188, 109906. [Google Scholar] [CrossRef]
- Li, P.; Feng, X.; Shang, L.; Qiu, G.; Meng, B.; Liang, P.; Zhang, H. Mercury pollution from artisanal mercury mining in Tongren, Guizhou, China. Appl. Geochem. 2008, 23, 2055–2064. [Google Scholar]
- Pu, Q.; Zhang, K.; Poulain, A.J.; Liu, J.; Zhang, R.; Abdelhafiz, M.A.; Meng, B.; Feng, X.B. Mercury drives microbial community assembly and ecosystem multifunctionality across a Hg contamination gradient in rice paddies. J. Hazard. Mater. 2022, 435, 129055. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.; Benimeli, C.S.; Saez, J.M.; Fuentes, M.S.; Cuozzo, S.A.; Polti, M.A.; Amoroso, M.J. Bacterial Bio-Resources for Remediation of Hexachlorocyclohexane. Int. J. Mol. Sci. 2012, 13, 15086–15106. [Google Scholar] [CrossRef]
- Rane, N.R.; Tapase, S.; Kanojia, A.; Watharkar, A.; Salama, E.-S.; Jang, M.; Yadav, K.K.; Amin, M.A.; Cabral-Pinto, M.M.S.; Jadhav, J.P.; et al. Molecular insights into plant-microbe interactions for sustainable remediation of contaminated environment. Bioresour. Technol. 2022, 344, 126246. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, X.; Zhao, L.; Wang, Z.; Teng, Y.; Luo, Y. Mercury Enrichment Characteristics and Rhizosphere Bacterial Community of Ramie (Boehmeria Nivea L. Gaud.) in Mercury-Contaminated Soil. Sustainability 2023, 15, 6009. https://doi.org/10.3390/su15076009
Li X, Wang X, Zhao L, Wang Z, Teng Y, Luo Y. Mercury Enrichment Characteristics and Rhizosphere Bacterial Community of Ramie (Boehmeria Nivea L. Gaud.) in Mercury-Contaminated Soil. Sustainability. 2023; 15(7):6009. https://doi.org/10.3390/su15076009
Chicago/Turabian StyleLi, Xiuhua, Xiaomi Wang, Ling Zhao, Zuopeng Wang, Ying Teng, and Yongming Luo. 2023. "Mercury Enrichment Characteristics and Rhizosphere Bacterial Community of Ramie (Boehmeria Nivea L. Gaud.) in Mercury-Contaminated Soil" Sustainability 15, no. 7: 6009. https://doi.org/10.3390/su15076009
APA StyleLi, X., Wang, X., Zhao, L., Wang, Z., Teng, Y., & Luo, Y. (2023). Mercury Enrichment Characteristics and Rhizosphere Bacterial Community of Ramie (Boehmeria Nivea L. Gaud.) in Mercury-Contaminated Soil. Sustainability, 15(7), 6009. https://doi.org/10.3390/su15076009




