COS Attenuates AFB1-Induced Liver Injury in Medaka through Inhibition of Histopathological Damage and Oxidative Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Animals
2.3. Experimental Design
2.4. Hepatic Pathological Examination
2.5. Determination of Biochemical Indicators in Liver
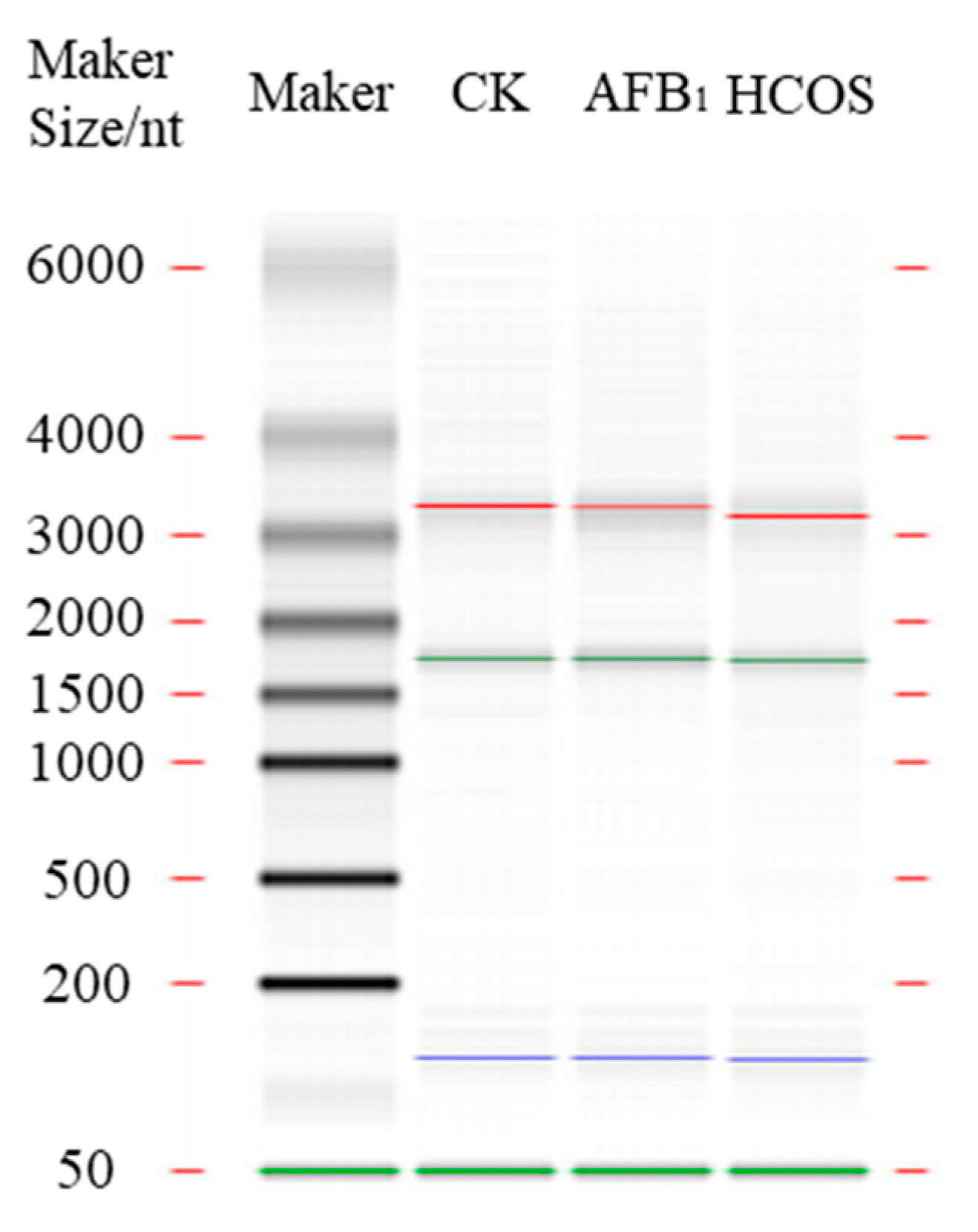
2.6. Illumina RNA Sequencing
2.7. Real-Time Quantitative PCR Analysis
2.8. Statistical Analysis
3. Results
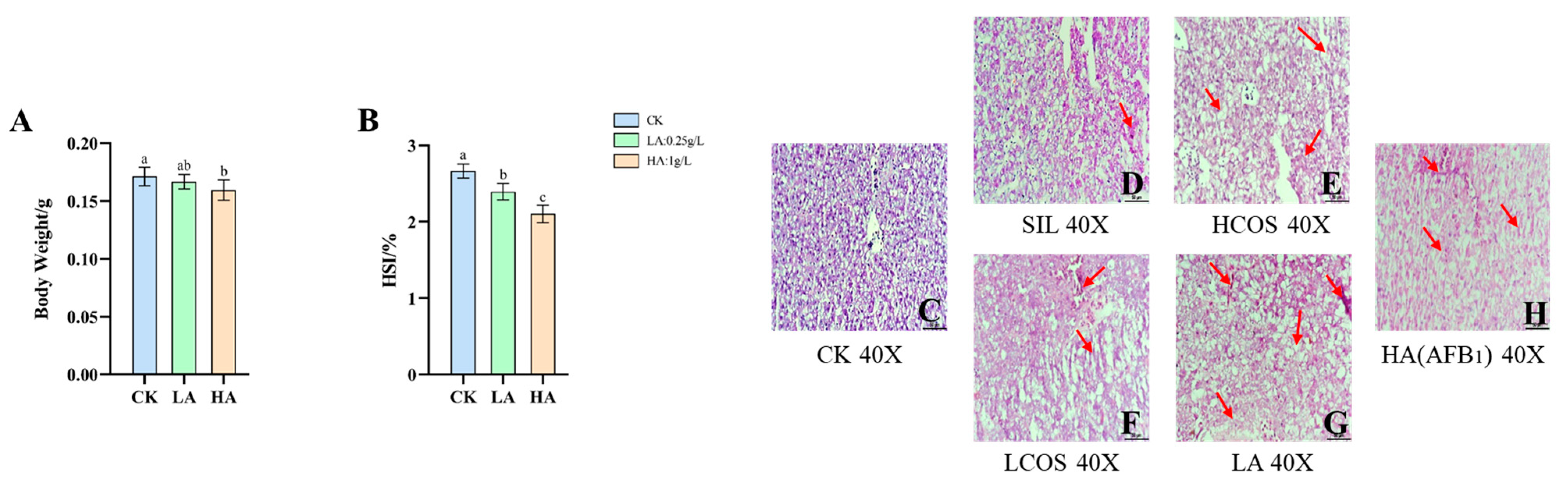
3.1. Body Weight and HSI Changes in Medaka
3.2. Histopathological Changes of Liver Tissue in Medaka
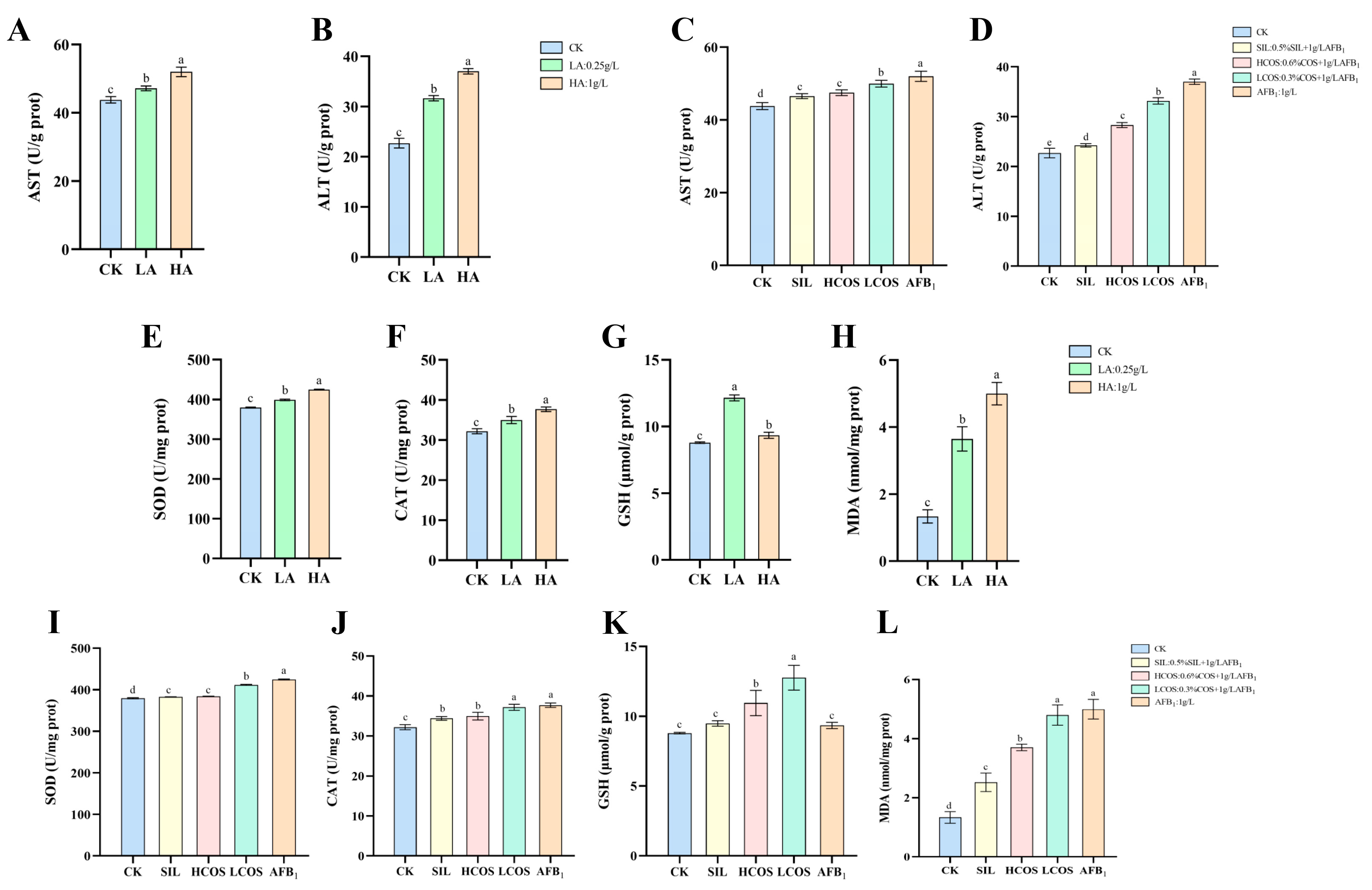
3.3. AFB1-Induced Liver Function Impairment
3.4. AFB1-Induced Oxidative Stress in Liver
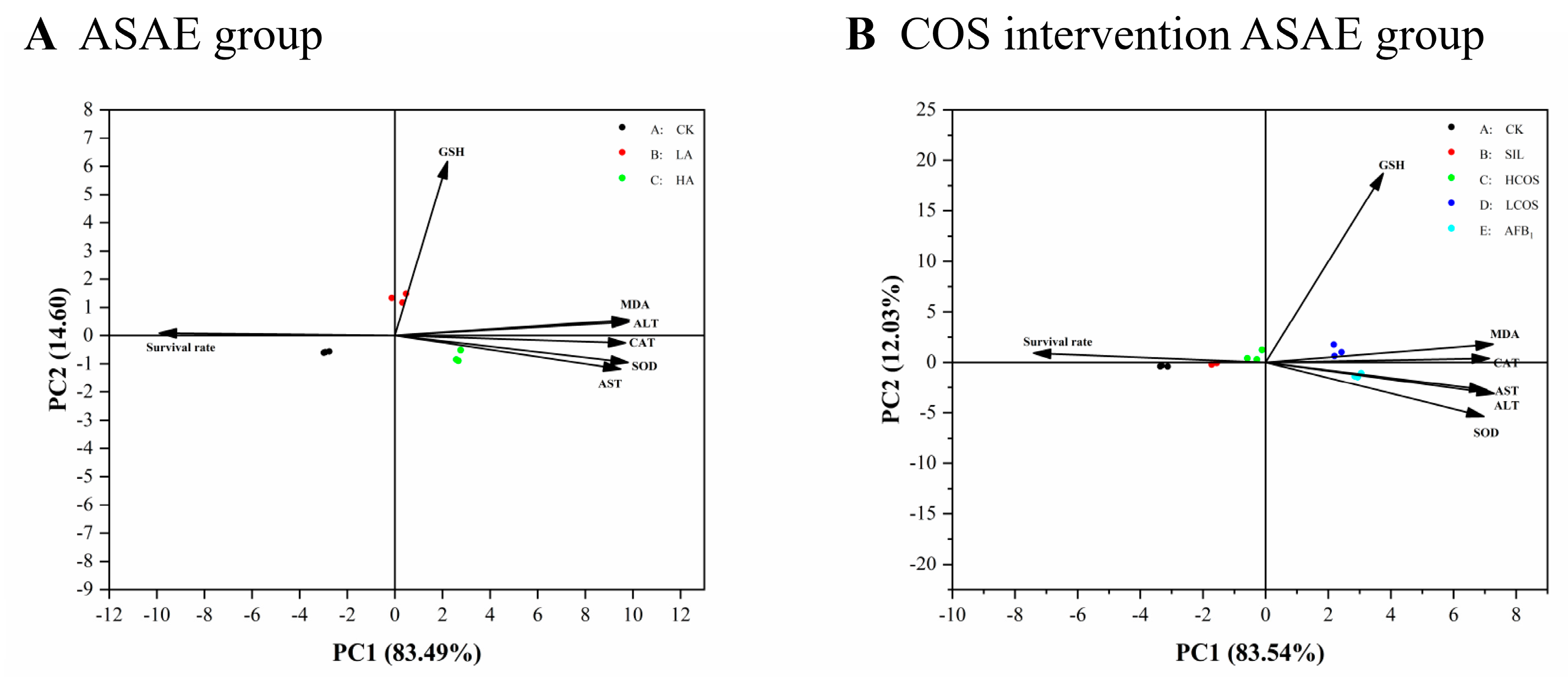
3.5. Principal Component Analysis of Enzyme Activity
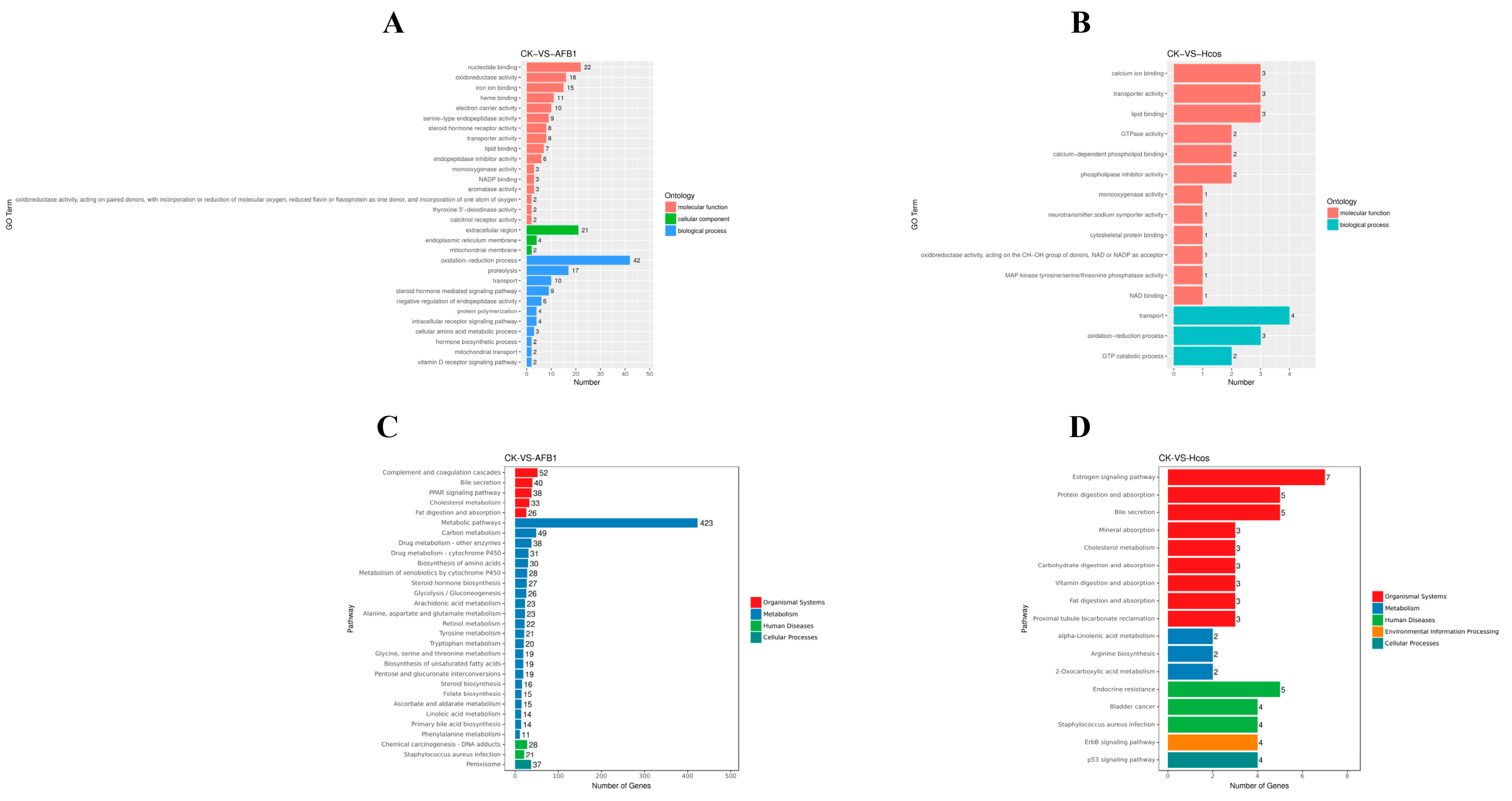
3.6. Expression and Functional Analysis of Differential Genes
3.7. Quantitative Validation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Wu, F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 2010, 118, 818–824. [Google Scholar] [CrossRef] [Green Version]
- Rushing, B.R.; Selim, M.I. Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem. Toxicol. 2019, 124, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Ferencík, M.; Ebringer, L. Modulatory effects of selenium and zinc on the immune system. Folia Microbiol. 2003, 48, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.S.; Brasel, J.M. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology 2001, 167, 101–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Zhan, D.L.; Chen, Y.Y.; Wang, W.H.; He, C.Y.; Lin, Y.; Lin, Y.C.; Lin, Z.N. Aflatoxin B1 enhances pyroptosis of hepatocytes and activation of Kupffer cells to promote liver inflammatory injury via dephosphorylation of cyclooxygenase-2: An in vitro, ex vivo and in vivo study. Arch. Toxicol. 2019, 93, 3305–3320. [Google Scholar] [CrossRef]
- Zhang, N.Y.; Qi, M.; Zhao, L.; Zhu, M.K.; Guo, J.; Liu, J.; Gu, C.Q.; Rajput, S.A.; Krumm, C.S.; Qi, D.S.; et al. Curcumin Prevents Aflatoxin B₁ Hepatoxicity by Inhibition of Cytochrome P450 Isozymes in Chick Liver. Toxins 2016, 8, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neal, G.E.; Eaton, D.L.; Judah, D.J.; Verma, A. Metabolism and toxicity of aflatoxins M1 and B1 in human-derived in vitro systems. Toxicol. Appl. Pharmacol. 1998, 151, 152–158. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Johnson, W.W.; Shimada, T.; Ueng, Y.F.; Langouet, S. Mutagenesis, Activation and detoxication of aflatoxin B1. Mutat. Res. 1998, 402, 121–128. [Google Scholar] [CrossRef]
- Qu, M.; Xu, K.; Li, Y.; Wong, G.; Wang, D. Using acs-22 mutant Caenorhabditis elegans to detect the toxicity of nanopolystyrene particles. Sci. Total Environ. 2018, 643, 119–126. [Google Scholar] [CrossRef]
- Tang, J.; Ni, X.; Zhou, Z.; Wang, L.; Lin, S. Acute microplastic exposure raises stress response and suppresses detoxification and immune capacities in the scleractinian coral Pocillopora damicornis. Environ. Pollut. 2018, 243, 66–74. [Google Scholar] [CrossRef]
- Naveed, M.; Phil, L.; Sohail, M.; Hasnat, M.; Baig, M.; Ihsan, A.U.; Shumzaid, M.; Kakar, M.U.; Mehmood Khan, T.; Akabar, M.D.; et al. Chitosan oligosaccharide (COS): An overview. Int. J. Biol. Macromol. 2019, 129, 827–843. [Google Scholar] [CrossRef]
- Liu, X.; Xia, W.; Jiang, Q.; Yu, P.; Yue, L. Chitosan oligosaccharide-N-chlorokojic acid mannich base polymer as a potential antibacterial material. Carbohydr. Polym. 2018, 182, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Wu, X.; Long, C.; Wang, Q.; Fan, Z.; Li, S.; Yin, Y. Chitosan oligosaccharide affects antioxidant defense capacity and placental amino acids transport of sows. BMC Vet. Res. 2016, 12, 243. [Google Scholar] [CrossRef] [Green Version]
- Saleh, H.; El-Shorbagy, H.M. Chitosan protects liver against ischemia-reperfusion injury via regulating Bcl-2/Bax, TNF-α and TGF-β expression. Int. J. Biol. Macromol. 2020, 164, 1565–1574. [Google Scholar] [CrossRef]
- Karagozlu, M.Z.; Karadeniz, F.; Kim, S.K. Anti-HIV activities of novel synthetic peptide conjugated chitosan oligomers. Int. J. Biol. Macromol. 2014, 66, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Arana, S.; Alves, V.A.; Sabino, M.; Tabata, Y.A.; Nonogaki, S.; Zaidan-Dagli, M.L.; Hernandez-Blazquez, F.J. Immunohistochemical evidence for myofibroblast-like cells associated with liver injury induced by aflatoxin B1 in rainbow trout (Oncorhynchus mykiss). J. Comp. Pathol. 2014, 150, 258–265. [Google Scholar] [CrossRef]
- Federico, A.; Dallio, M.; Loguercio, C. Silymarin/Silybin and Chronic Liver Disease: A Marriage of Many Years. Molecules 2017, 22, 191. [Google Scholar] [CrossRef] [Green Version]
- Tighe, S.P.; Akhtar, D.; Iqbal, U.; Ahmed, A. Chronic Liver Disease and Silymarin: A Biochemical and Clinical Review. J. Clin. Transl. Hepatol. 2020, 8, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Young, A.I.; Benonisdottir, S.; Przeworski, M.; Kong, A. Deconstructing the sources of genotype-phenotype associations in humans. Science 2019, 365, 1396–1400. [Google Scholar] [CrossRef]
- Sun, B.; Gui, L.; Liu, R.; Hong, Y.; Li, M. Medaka oct4 is essential for gastrulation, central nervous system development and angiogenesis. Gene 2020, 733, 144270. [Google Scholar] [CrossRef]
- Troxel, C.M.; Buhler, D.R.; Hendricks, J.D.; Bailey, G.S. CYP1A induction by beta-naphthoflavone, Aroclor 1254, and 2,3,7,8-tetrachlorodibenzo-p-dioxin and its influence on aflatoxin B1 metabolism and DNA adduction in zebrafish. Toxicol. Appl. Pharmacol. 1997, 146, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.W.; Yang, W.Y.; Lin, Y.M.; Jin, S.L.; Yuh, C.H. Hepatitis B virus X antigen and aflatoxin B1 synergistically cause hepatitis, steatosis and liver hyperplasia in transgenic zebrafish. Acta Histochem. 2013, 115, 728–739. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kumar Mishra, S.; Wang, T.; Xu, Z.; Zhao, X.; Wang, Y.; Yin, H.; Fan, X.; Zeng, B.; Yang, M.; et al. AFB1 Induced Transcriptional Regulation Related to Apoptosis and Lipid Metabolism in Liver of Chicken. Toxins 2020, 12, 290. [Google Scholar] [CrossRef]
- Cheng, Y.C.; Wu, T.S.; Huang, Y.T.; Chang, Y.; Yang, J.J.; Yu, F.Y.; Liu, B.H. Aflatoxin B1 interferes with embryonic liver development: Involvement of p53 signaling and apoptosis in zebrafish. Toxicology 2021, 458, 152844. [Google Scholar] [CrossRef]
- Jindal, N.; Mahipal, S.K.; Mahajan, N.K. Toxicity of aflatoxin B1 in broiler chicks and its reduction by activated charcoal. Res. Vet. Sci. 1994, 56, 37–40. [Google Scholar] [CrossRef]
- Li, S.; Muhammad, I.; Yu, H.; Sun, X.; Zhang, X. Detection of Aflatoxin adducts as potential markers and the role of curcumin in alleviating AFB1-induced liver damage in chickens. Ecotoxicol. Environ. Saf. 2019, 176, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chen, L.; Zhang, L.; Zhang, Z.; Zhao, Y.; Wang, Y.; Ou, J. New Insights Into the Persistent Effects of Acute Exposure to AFB(1) on Rat Liver. Front. Microbiol. 2022, 13, 911757. [Google Scholar] [CrossRef] [PubMed]
- Yasmeen, R.; Zahid, B.; Alyas, S.; Akhtar, R.; Zahra, N.; Kouser, S.; Hashmi, A.S.; Athar, M.; Tayyab, M.; Anjum, A.A. Ameliorative effects of Lactobacillus against Aflatoxin B1. Braz. J. Biol. 2021, 84, e250517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Li, M.; Xu, J.; Wang, J.; Li, M.; Wei, L.; Lv, Q.; Chen, X.; Wang, Y.; et al. SeMet attenuates AFB1-induced intestinal injury in rabbits by activating the Nrf2 pathway. Ecotoxicol. Environ. Saf. 2022, 239, 113640. [Google Scholar] [CrossRef]
- Livingstone, D.R. Contaminant-stimulated reactive oxygen species production and oxidative damage in aquatic organisms. Mar. Pollut. Bull. 2001, 42, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Egea, J.; Fabregat, I.; Frapart, Y.M.; Ghezzi, P.; Görlach, A.; Kietzmann, T.; Kubaichuk, K.; Knaus, U.G.; Lopez, M.G.; Olaso-Gonzalez, G.; et al. Corrigendum to “European contribution to the study of ROS: A summary of the findings and prospects for the future from the COST action BM1203 (EU-ROS)” [Redox Biol. 13 (2017) 94-162]. Redox Biol. 2018, 14, 694–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorantes-Aranda, J.J.; Seger, A.; Mardones, J.I.; Nichols, P.D.; Hallegraeff, G.M. Progress in Understanding Algal Bloom-Mediated Fish Kills: The Role of Superoxide Radicals, Phycotoxins and Fatty Acids. PloS ONE 2015, 10, e0133549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Liu, Z.; Li, M.; Jiang, Q.; Wu, D.; Huang, Y.; Jiao, Y.; Zhang, M.; Zhao, Y. Effects of nanoplastics on antioxidant and immune enzyme activities and related gene expression in juvenile Macrobrachium nipponense. J. Hazard. Mater. 2020, 398, 122990. [Google Scholar] [CrossRef]
- Xu, L.; Yu, Y.; Sang, R.; Li, J.; Ge, B.; Zhang, X. Protective Effects of Taraxasterol against Ethanol-Induced Liver Injury by Regulating CYP2E1/Nrf2/HO-1 and NF-κB Signaling Pathways in Mice. Oxidative Med. Cell. Longev. 2018, 2018, 8284107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, W.; Li, K.; Rong, S.; Yao, P.; Hao, L.; Ying, C.; Zhang, X.; Nussler, A.; Liu, L. Curcumin alleviates ethanol-induced hepatocytes oxidative damage involving heme oxygenase-1 induction. J. Ethnopharmacol. 2010, 128, 549–553. [Google Scholar] [CrossRef]
- Li, S.; Liu, R.; Xia, S.; Wei, G.; Ishfaq, M.; Zhang, Y.; Zhang, X. Protective role of curcumin on aflatoxin B1-induced TLR4/RIPK pathway mediated-necroptosis and inflammation in chicken liver. Ecotoxicol. Environ. Saf. 2022, 233, 113319. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Kong, D.; He, K.; Xia, Q. Aurora kinase A regulates liver regeneration through macrophages polarization and Wnt/β-catenin signalling. Liver Int. 2022, 42, 468–478. [Google Scholar] [CrossRef]
- Horiguchi, H.; Yamagata, S.; Rong Qian, Z.; Kagawa, S.; Sakashita, N. Thrombospondin-1 is highly expressed in desmoplastic components of invasive ductal carcinoma of the breast and associated with lymph node metastasis. J. Med. Investig. JMI 2013, 60, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Jayachandran, A.; Anaka, M.; Prithviraj, P.; Hudson, C.; McKeown, S.J.; Lo, P.H.; Vella, L.J.; Goding, C.R.; Cebon, J.; Behren, A. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget 2014, 5, 5782–5797. [Google Scholar] [CrossRef] [Green Version]
- Jeanne, A.; Schneider, C.; Martiny, L.; Dedieu, S. Original insights on thrombospondin-1-related antireceptor strategies in cancer. Front. Pharmacol. 2015, 6, 252. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, L.; Kang, Y.; Huang, S.; Xiao, Y. Circulating THBS1: A Risk Factor for Nonalcoholic Fatty Liver Disease in Obese Children. Ann. Nutr. Metab. 2023, 79, 16–28. [Google Scholar] [CrossRef]
- Liu, R.J.; Xu, Z.P.; Li, S.Y.; Yu, J.J.; Feng, N.H.; Xu, B.; Chen, M. BAP1-Related ceRNA (NEAT1/miR-10a-5p/SERPINE1) Promotes Proliferation and Migration of Kidney Cancer Cells. Front. Oncol. 2022, 12, 852515. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.; Zhang, J.X.; Chen, Y.; Shen, X.D.; Su, C.; Guo, Y.J.; Wang, P.H.; Shi, C.C.; Lei, M.; Cao, Y.O.; et al. LncRNA NKX2-1-AS1 promotes tumor progression and angiogenesis via upregulation of SERPINE1 expression and activation of the VEGFR-2 signaling pathway in gastric cancer. Mol. Oncol. 2021, 15, 1234–1255. [Google Scholar] [CrossRef]
- Miyazaki, H.; Sawada, T.; Kiyohira, M.; Yu, Z.; Nakamura, K.; Yasumoto, Y.; Kagawa, Y.; Ebrahimi, M.; Islam, A.; Sharifi, K.; et al. Fatty acid binding protein 7 regulates phagocytosis and cytokine production in Kupffer cells during liver injury. Am. J. Pathol. 2014, 184, 2505–2515. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.J.; Yang, J.W.; Park, J.H.; Choi, E.S.; Lim, C.S.; Lee, S.; Han, C.Y. Endoplasmic Reticulum Stress Increases DUSP5 Expression via PERK-CHOP Pathway Leading to Hepatocyte Death. Int. J. Mol. Sci. 2019, 20, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene | Primer Sequence (5′ to 3′) | Number of Bases |
|---|---|---|
| β-Actin | Forward: CCAGCCTTCCTTCCTTGGTA Reverse: GTACCTCCAGACAGCACAGT | 20 20 |
| aurka | Forward: GCGATGGAGCCTAGAAGACT Reverse: TGAACTTGGTCTGTCGCTCT | 20 20 |
| thbs1 | Forward: TGTGAGTGCAAGCCAGGATA Reverse: AGAGTTGGGAAGGTTAGGGC | 20 20 |
| serpine1 | Forward: TCCCTTCAACCCCAAACTGA Reverse: TCCCCATAGTTGAAGCGGTT | 20 20 |
| fabp7 | Forward: CACCCAGAGCACCTTCAAAA Reverse: CGTGAACCAACTTGTCTCCG | 20 20 |
| dusp5 | Forward: TTTCCTCTACCTCGGCAGTG Reverse: TTGAGCAAGGCAGTGATGTG | 20 20 |
| Relative Gene Expression Level | Fold Change | ||||
|---|---|---|---|---|---|
| CK | HCOS | AFB1 | |||
| Q-PCR | RNA-Seq | Q-PCR | RNA-Seq | ||
| aurka | 1 | 6.76 ± 0.09 | 43.29 ± 0.76 | 18.00 ± 0.28 | 188.04 ± 0.89 |
| thbs1 | 1 | 5.61 ± 0.21 | 14.34 ± 0.48 | 24.71 ± 0.70 | 101.73 ± 1.08 |
| serpine1 | 1 | 7.44 ± 0.19 | 23.30 ± 0.26 | 61.77 ± 0.67 | 282.11 ± 0.37 |
| fabp1 | 1 | 0.08 ± 0.01 | 0.08 ± 0.01 | 0.01 ± 0.01 | 0.03 ± 0.01 |
| dusp5 | 1 | 5.57 ± 0.25 | 16.34 ± 0.56 | 17.35 ± 0.06 | 86.71 ± 0.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, H.; Chen, L.; Zhang, Z.; Zhao, Y.; Ou, J. COS Attenuates AFB1-Induced Liver Injury in Medaka through Inhibition of Histopathological Damage and Oxidative Stress. Sustainability 2023, 15, 5418. https://doi.org/10.3390/su15065418
Shi H, Chen L, Zhang Z, Zhao Y, Ou J. COS Attenuates AFB1-Induced Liver Injury in Medaka through Inhibition of Histopathological Damage and Oxidative Stress. Sustainability. 2023; 15(6):5418. https://doi.org/10.3390/su15065418
Chicago/Turabian StyleShi, Huijun, Lin Chen, Zhaohuan Zhang, Yong Zhao, and Jie Ou. 2023. "COS Attenuates AFB1-Induced Liver Injury in Medaka through Inhibition of Histopathological Damage and Oxidative Stress" Sustainability 15, no. 6: 5418. https://doi.org/10.3390/su15065418
APA StyleShi, H., Chen, L., Zhang, Z., Zhao, Y., & Ou, J. (2023). COS Attenuates AFB1-Induced Liver Injury in Medaka through Inhibition of Histopathological Damage and Oxidative Stress. Sustainability, 15(6), 5418. https://doi.org/10.3390/su15065418





