The Effect of Plant Diversity and Soil Properties on Soil Microbial Biomass and Activity in a Novel Ecosystem
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
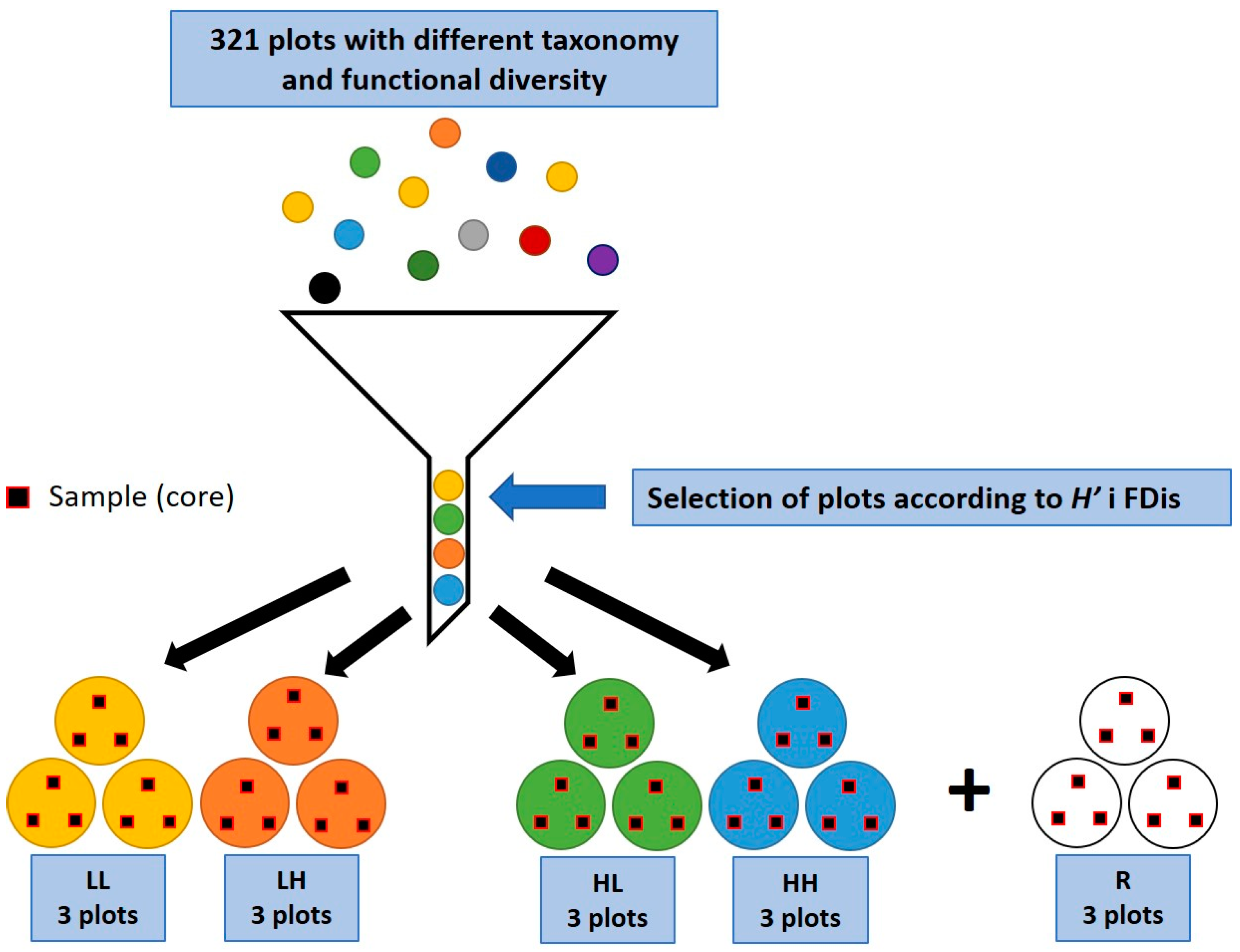
2.2. Criteria for Selection and Sampling of Vegetation Plots
2.3. Substrate Physicochemical Analyses
2.4. Substrate Respiration (Rs) Measurements
2.5. Analysis of Microbial Community Structure Using PLFA
2.6. Statistical Analysis
3. Results
3.1. Substrate Physicochemical Parameters
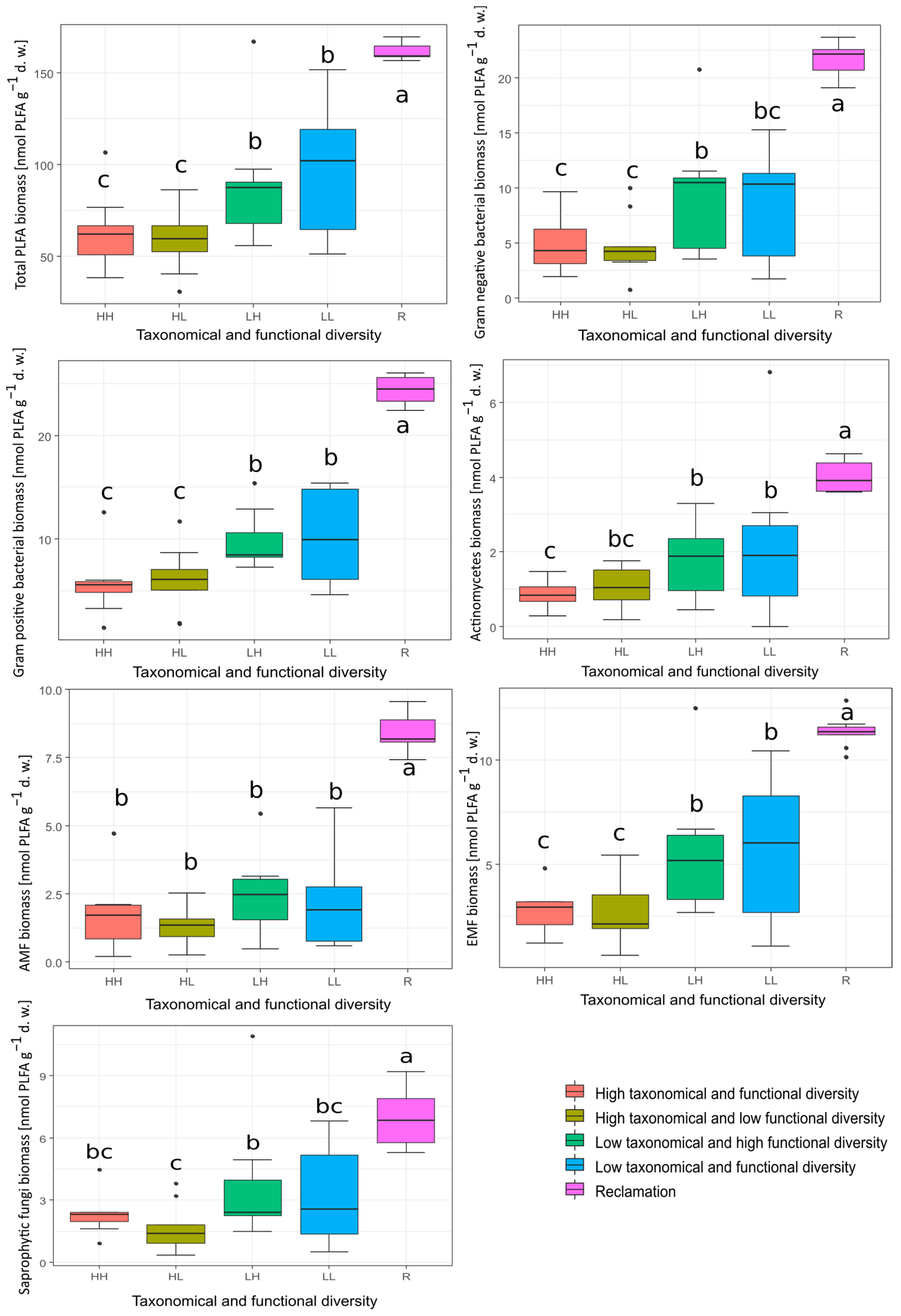
3.2. PLFA Biomass
3.3. Soil Respiration
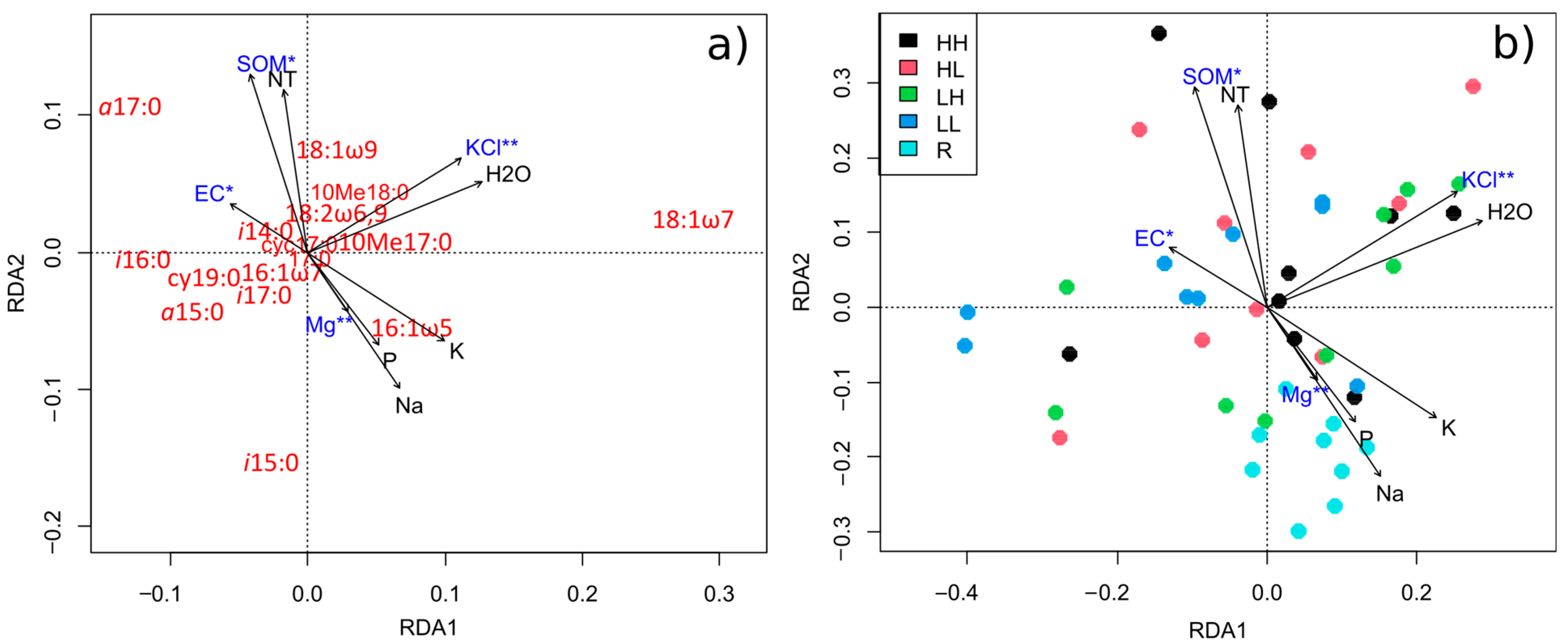
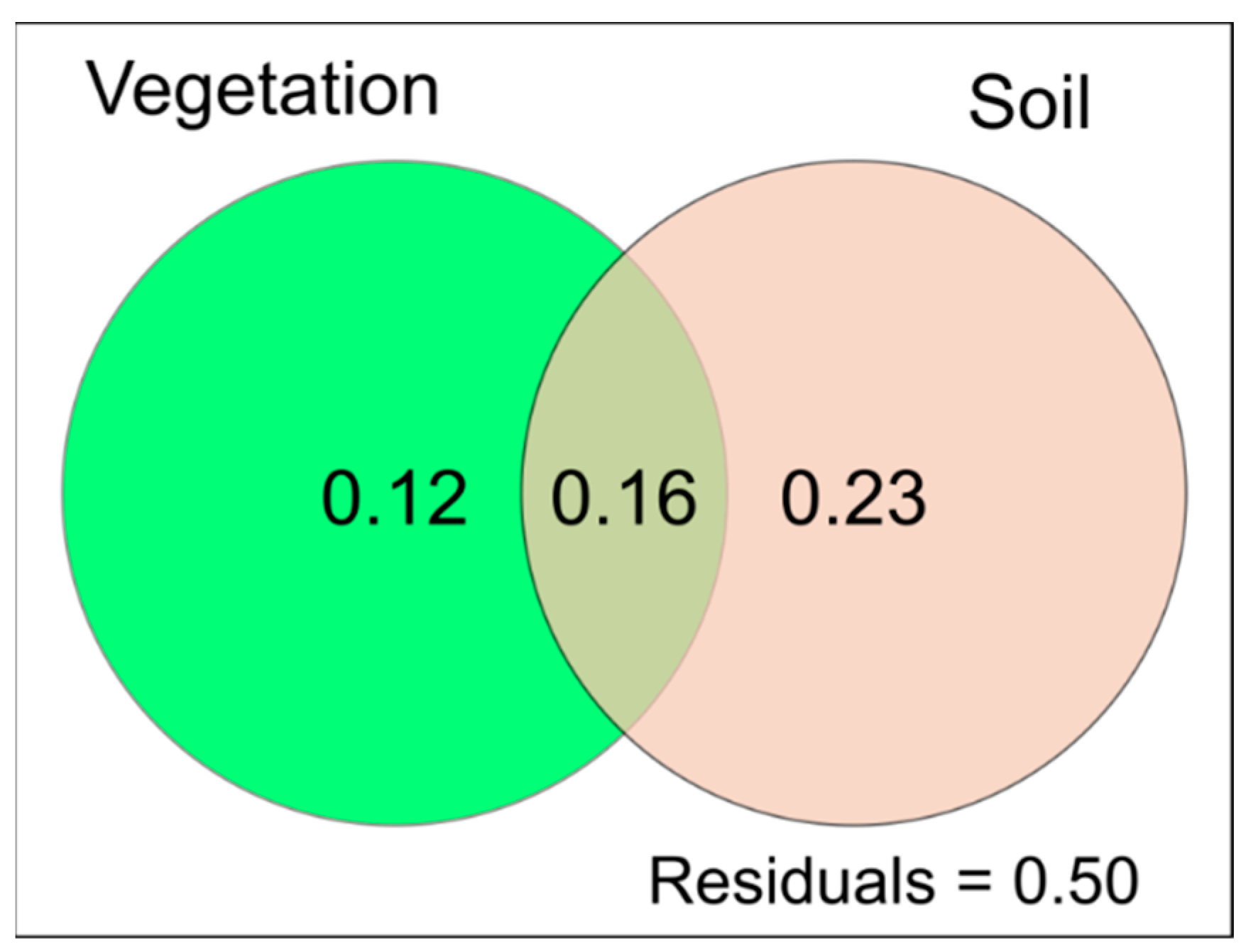
3.4. Relationships between Substrate Physicochemical Parameters, Plant Diversity and PLFA Profiles
4. Discussion
4.1. Explanatory Ability of Functional Traits
4.2. Soil Microorganisms and Physicochemical Substrate Parameters
4.3. Interactions between Plant Taxonomic and Functional Diversity and Soil Microorganisms
4.4. Soil Respiration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bardgett, R.D.; Van Der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Kaiser, C.; Kilburn, M.R.; Clode, P.L.; Fuchslueger, L.; Koranda, M.; Cliff, J.B.; Solaiman, Z.M.; Murphy, D.V. Exploring the transfer of recent plant photosynthates to soil microbes: Mycorrhizal Pathway vs Direct Root Exudation. N. Phytol. 2015, 205, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, K.; Lal, R.; Shipitalo, M.J. Stabilized soil organic carbon pools in subsoils under forest are potential sinks for atmospheric CO2. For. Sci. 2011, 57, 19–25. [Google Scholar]
- Turner, B.L.; Lambers, H.; Condron, L.M.; Cramer, M.D.; Leake, J.R.; Richardson, A.E.; Smith, S.E. Soil microbial biomass and the fate of phosphorus during long-term ecosystem development. Plant Soil 2013, 367, 225–234. [Google Scholar] [CrossRef]
- Terrer, C.; Phillips, R.P.; Hungate, B.A.; Rosende, J.; Pett-Ridge, J.; Craig, M.E.; van Groenigen, K.J.; Keenan, T.F.; Sulman, B.N.; Stocker, B.D.; et al. A trade-off between plant and soil carbon storage under elevated CO2. Nature 2021, 591, 599–603. [Google Scholar] [CrossRef]
- Fanin, N.; Hättenschwiler, S.; Barantal, S.; Schimann, H.; Fromin, N. Does variability in litter quality determine soil microbial respiration in an amazonian rainforest? Soil Biol. Biochem. 2011, 43, 1014–1022. [Google Scholar] [CrossRef]
- Lange, M.; Eisenhauer, N.; Sierra, C.A.; Bessler, H.; Engels, C.; Griffiths, R.I.; Mellado-Vázquez, P.G.; Malik, A.A.; Roy, J.; Scheu, S.; et al. Plant diversity increases soil microbial activity and soil carbon storage. Nat. Commun. 2015, 6, 6707. [Google Scholar] [CrossRef]
- Steinauer, K.; Chatzinotas, A.; Eisenhauer, N. Root Exudate Cocktails: The link between plant diversity and soil microorganisms? Ecol. Evol. 2016, 6, 7387–7396. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.Y.H.; Chen, X.; Huang, Z. Meta-analysis shows positive effects of plant diversity on microbial biomass and respiration. Nat. Commun. 2019, 10, 1332. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.Y.H.; Chen, X. Functional diversity enhances, but exploitative traits reduce tree mixture effects on microbial biomass. Funct. Ecol. 2020, 34, 276–286. [Google Scholar] [CrossRef]
- Johnson, D.; Phoenix, G.K.; Grime, J.P. Plant community composition, not diversity, regulates soil respiration in grasslands. Biol. Lett. 2008, 4, 345–348. [Google Scholar] [CrossRef]
- Rottstock, T.; Joshi, J.; Kummer, V.; Fischer, M. Higher plant diversity promotes higher diversity of fungal pathogens, while it decreases pathogen infection per plant. Ecology 2014, 95, 1907–1917. [Google Scholar] [CrossRef]
- Jarzyna, M.A.; Jetz, W. Taxonomic and functional diversity change is scale dependent. Nat. Commun. 2018, 9, 2565. [Google Scholar] [CrossRef]
- Mariotte, P.; Vandenberghe, C.; Meugnier, C.; Rossi, P.; Bardgett, R.D.; Buttler, A. Subordinate plant species impact on soil microbial communities and ecosystem functioning in grasslands: Findings from a removal experiment. Perspect. Plant Ecol. Evol. Syst. 2013, 15, 77–85. [Google Scholar] [CrossRef]
- Eisenhauer, N.; Beßler, H.; Engels, C.; Gleixner, G.; Habekost, M.; Milcu, A.; Partsch, S.; Sabais, A.C.W.; Scherber, C.; Steinbeiss, S.; et al. Plant diversity effects on soil microorganisms support the singular hypothesis. Ecology 2010, 91, 485–496. [Google Scholar] [CrossRef]
- Reich, P.B.; Oleksyn, J.; Modrzynski, J.; Mroziński, P.; Hobbie, S.E.; Eissenstat, D.M.; Chorover, J.; Chadwick, O.A.; Hale, C.M.; Tjoelker, M.G. Linking litter calcium, earthworms and soil properties: A common garden test with 14 tree species. Ecol. Lett. 2005, 8, 811–818. [Google Scholar] [CrossRef]
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; MacE, G.M.; Tilman, D.; Wardle, D.A.; et al. Biodiversity loss and its impact on humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef]
- Frouz, J.; Roubíčková, A.; Heděnec, P.; Tajovský, K. Do Soil Fauna Really Hasten Litter Decomposition? A meta-analysis of enclosure studies. Eur. J. Soil Biol. 2015, 68, 18–24. [Google Scholar] [CrossRef]
- Jagodziński, A.M.; Dyderski, M.K.; Gȩsikiewicz, K.; Horodecki, P. Tree- and stand-level biomass estimation in a Larix decidua Mill. Chronosequence. Forests 2018, 9, 587. [Google Scholar] [CrossRef]
- Kompała-Bąba, A.; Sierka, E.; Dyderski, M.K.; Bierza, W.; Magurno, F.; Besenyei, L.; Błońska, A.; Ryś, K.; Jagodziński, A.M.; Woźniak, G. Do the dominant plant species impact the substrate and vegetation composition of post-coal mining spoil heaps? Ecol. Eng. 2020, 143, 105685. [Google Scholar] [CrossRef]
- Hobbs, R.J.; Higgs, E.S.; Hall, C.M. Novel Ecosystems: Intervening in the New Ecological World Order; Wiley Blackwell: Hoboken, NJ, USA, 2013; ISBN 9781118354186. [Google Scholar]
- Rotherham, I.D. Recombinant Ecology—A Hybrid Future? Springer: Cham, Switzerland, 2017; ISBN 978-3-319-49796-9. [Google Scholar]
- Hobbs, R.J.; Arico, S.; Aronson, J.; Baron, J.S.; Bridgewater, P.; Cramer, V.A.; Epstein, P.R.; Ewel, J.J.; Klink, C.A.; Lugo, A.E.; et al. Novel Ecosystems: Theoretical and management aspects of the new ecological world order. Glob. Ecol. Biogeogr. 2006, 15, 1–7. [Google Scholar] [CrossRef]
- Tropek, R.; Kadlec, T.; Hejda, M.; Kocarek, P.; Skuhrovec, J.; Malenovsky, I.; Vodka, S.; Spitzer, L.; Banar, P.; Konvicka, M. Technical reclamations are wasting the conservation potential of post-mining sites. a case study of black coal spoil dumps. Ecol. Eng. 2012, 43, 13–18. [Google Scholar] [CrossRef]
- Tropek, R.; Kadlec, T.; Karesova, P.; Spitzer, L.; Kocarek, P.; Malenovsky, I.; Banar, P.; Tuf, I.H.; Hejda, M.; Konvicka, M. Spontaneous succession in limestone quarries as an effective restoration tool for endangered arthropods and plants. J. Appl. Ecol. 2010, 47, 139–147. [Google Scholar] [CrossRef]
- Novák, J.; Prach, K. Vegetation succession in basalt quarries: Pattern on a landscape scale. In Applied Vegetation Science; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2003; Volume 6, pp. 111–116. [Google Scholar]
- Řehounková, K.; Čížek, L.; Řehounek, J.; Šebelíková, L.; Tropek, R.; Lencová, K.; Bogusch, P.; Marhoul, P.; Máca, J. Additional disturbances as a beneficial tool for restoration of post-mining sites: A multi-taxa approach. Environ. Sci. Pollut. Res. Int. 2016, 23, 13745–13753. [Google Scholar] [CrossRef]
- Sharma, A.; Guinness, J.; Muyskens, A.; Polizzotto, M.L.; Fuentes, M.; Hesterberg, D. Spatial statistical modeling of arsenic accumulation in microsites of diverse soils. Geoderma 2022, 411, 115697. [Google Scholar] [CrossRef]
- Urbanová, M.; Kopecký, J.; Valášková, V.; Ságová-Marečková, M.; Elhottová, D.; Kyselková, M.; Moënne-Loccoz, Y.; Baldrian, P. Development of bacterial community during spontaneous succession on spoil heaps after brown coal mining. FEMS Microbiol. Ecol. 2011, 78, 59–69. [Google Scholar] [CrossRef]
- Jarosz, J. (Mineral and Energy Economy Research Institute, Polish Academy of Sciences, Kraków, Poland). Personal communication, 2019. [Google Scholar]
- Magurran, A.E. Ecological Diversity and Its Measurement; Springer: Berlin/Heidelberg, Germany, 1988. [Google Scholar]
- Hedberg, P.; Kozub, Ł.; Kotowski, W. Functional diversity analysis helps to identify filters affecting community assembly after fen restoration by top-soil removal and hay transfer. J. Nat. Conserv. 2014, 22, 50–58. [Google Scholar] [CrossRef]
- Laliberte, E.; Legendre, P. A Distance-based framework for measuring functional diversity from multiple traits. Ecology 2010, 91, 299–305. [Google Scholar] [CrossRef]
- Kattge, J.; Bönisch, G.; Díaz, S.; Lavorel, S.; Prentice, I.C.; Leadley, P.; Tautenhahn, S.; Werner, G.D.A.; Aakala, T.; Abedi, M.; et al. TRY Plant Trait Database—Enhanced coverage and open access. Glob. Chang. Biol. 2020, 26, 119–188. [Google Scholar] [CrossRef]
- Kleyer, M.; Bekker, R.M.; Knevel, I.C.; Bakker, J.P.; Thompson, K.; Sonnenschein, M.; Poschlod, P.; Van Groenendael, J.M.; Klimeš, L.; Klimešová, J.; et al. The LEDA traitbase: A database of life-history traits of the Northwest European flora. J. Ecol. 2008, 96, 1266–1274. [Google Scholar] [CrossRef]
- Woźniak, G.; Dyderski, M.K.; Kompała-Bąba, A.; Jagodziński, A.M.; Pasierbiński, A.; Błońska, A.; Bierza, W.; Magurno, F.; Sierka, E. Use of remote sensing to track postindustrial vegetation development. L. Degrad. Dev. 2021, 32, 1426–1439. [Google Scholar] [CrossRef]
- Błońska, A.; Kompała-Bąba, A.; Sierka, E.; Besenyei, L.; Magurno, F.; Frydecka, K.; Bierza, W.; Woźniak, G. Impact of selected plant species on enzymatic activity of soil substratum on post-mining heaps. J. Ecol. Eng. 2019, 20, 138–144. [Google Scholar] [CrossRef]
- Hristov, B.; Filcheva, E.; Ivanov, P. Organic matter content and composition of soils with stagnic properties from Bulgaria. Bulg. J. Soil Sci. 2016, 1, 26–32. [Google Scholar]
- Rutherford, P.M.; McGill, W.B.; Arocena, J.M.; Figueiredo, C.T. Total nitrogen. In Soil Sampling and Methods of Analysis; Carter, M.R., Gregoric, E.G., Eds.; CRC Press: Boca Raton, FL, USA, 2006; pp. 267–278. [Google Scholar]
- Staugaitis, G.; Rutkauskiene, R. Comparison of magnesium determination methods as influenced by soil properties. Zemdirbyste 2010, 97, 105–116. [Google Scholar]
- López-Marcos, D.; Martínez-Ruiz, C.; Turrión, M.B.; Jonard, M.; Titeux, H.; Ponette, Q.; Bravo, F. Soil carbon stocks and exchangeable cations in monospecific and mixed pine forests. Eur. J. For. Res. 2018, 137, 831–847. [Google Scholar] [CrossRef]
- Woźniak, G.; Kamczyc, J.; Bierza, W.; Błońska, A.; Kompała-Bąba, A.; Sierka, E.; Jagodziński, A.M. Functional ecosystem parameters: Soil respiration and diversity of mite (Acari, Mesostigmata) communities after disturbance in a Late Cambrian bedrock environment. L. Degrad. Dev. 2022, 33, 3343–3357. [Google Scholar] [CrossRef]
- Pennanen, T.; Frostegård, Å.; Fritze, H.; Bååth, E. Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal-polluted gradients in coniferous forests. Appl. Environ. Microbiol. 1996, 62, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Frostegård, A.; Bååth, E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertil. Soils 1996, 22, 59–65. [Google Scholar] [CrossRef]
- Bååth, E.; Anderson, T.H. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 2003, 35, 955–963. [Google Scholar] [CrossRef]
- Ushio, M.; Wagai, R.; Balser, T.C.; Kitayama, K. Variations in the soil microbial community composition of a tropical montane forest ecosystem: Does tree species matter? Soil Biol. Biochem. 2008, 40, 2699–2702. [Google Scholar] [CrossRef]
- Chapman, S.K.; Newman, G.S. Biodiversity at the plant-soil interface: Microbial abundance and community structure respond to litter mixing. Oecologia 2010, 162, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, H.Y.H. Global effects of plant litter alterations on soil CO2 to the atmosphere. Glob. Chang. Biol. 2018, 24, 3462–3471. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Bowman, W.D.; Kaufmann, R.; Schmidt, S.K. A Temporal approach to linking aboveground and belowground ecology. Trends Ecol. Evol. 2005, 20, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Oelmann, Y.; Lange, M.; Leimer, S.; Roscher, C.; Aburto, F.; Alt, F.; Bange, N.; Berner, D.; Boch, S.; Boeddinghaus, R.S.; et al. Above- and belowground biodiversity jointly tighten the P cycle in agricultural grasslands. Nat. Commun. 2021, 12, 4431. [Google Scholar] [CrossRef] [PubMed]
- Naeem, S.; Tjossem, S.F.; Byers, D.; Bristow, C.; Li, S. Plant neighborhood diversity and production. Ecoscience 1999, 6, 355–365. [Google Scholar] [CrossRef]
- Díaz, S.; Cabido, M. Vive La Différence: Plant functional diversity matters to ecosystem processes. Trends Ecol. Evol. 2001, 16, 646–655. [Google Scholar] [CrossRef]
- Lefcheck, J.S.; Bastazini, V.A.G.; Griffin, J.N. Choosing and using multiple traits in functional diversity research. Environ. Conserv. 2015, 42, 104–107. [Google Scholar] [CrossRef]
- Mlambo, M.C. Not all traits are “functional”: Insights from taxonomy and biodiversity-ecosystem functioning research. Biodivers. Conserv. 2014, 23, 781–790. [Google Scholar] [CrossRef]
- Petchey, O.L.; Gaston, K.J. Functional diversity (FD), species richness and community composition. Ecol. Lett. 2002, 5, 402–411. [Google Scholar] [CrossRef]
- Petchey, O.L.; Gaston, K.J. Functional diversity: Back to basics and looking forward. Ecol. Lett. 2006, 9, 741–758. [Google Scholar] [CrossRef]
- Guerrero-Ramírez, N.R.; Mommer, L.; Freschet, G.T.; Iversen, C.M.; McCormack, M.L.; Kattge, J.; Poorter, H.; van der Plas, F.; Bergmann, J.; Kuyper, T.W.; et al. Global root traits (GRooT) database. Glob. Ecol. Biogeogr. 2021, 30, 25–37. [Google Scholar] [CrossRef]
- Enquist, B.J.; Condit, R.; Peet, R.K.; Schildhauer, M.; Thiers, B. The Botanical Information and Ecology Network (BIEN): Cyberinfrastructure for an integrated botanical information network to investigate the ecological impacts of global climate change on plant biodiversity. PeerJ Preprints. 2016, e2615v2. [Google Scholar] [CrossRef]
- de Bello, F.; Carmona, C.P.; Dias, A.T.C.; Götzenberger, L.; Moretti, M.; Berg, M.P. Handbook of Trait-Based Ecology; Cambridge University Press: Cambridge, UK, 2021; ISBN 9781108628426. [Google Scholar]
- Wozniak, G.; Markowicz, A.; Borymski, S.; Piotrowska-Seget, Z.; Chmura, D.; Besenyei, L. The relationship between successional vascular plant assemblages and associated microbial communities on coal mine spoil heaps. Community Ecol. 2015, 16, 23–32. [Google Scholar] [CrossRef]
- Hu, Y.-K.; Pan, X.; Liu, X.-Y.; Fu, Z.-X.; Zhang, M.-Y. Above- and belowground plant functional composition show similar changes during temperate forest swamp succession. Front. Plant Sci. 2021, 12, 658883. [Google Scholar] [CrossRef]
- Gilliam, F.S.; Roberts, M.R. The Herbaceous Layer in Forests of Eastern North America, 2nd ed.; Gilliam, F.S., Ed.; Oxford University Press: New York, NY, USA, 2003. [Google Scholar]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Noll, M.; Wellinger, M. Changes of the soil ecosystem along a receding glacier: Testing the correlation between environmental factors and bacterial community structure. Soil Biol. Biochem. 2008, 40, 2611–2619. [Google Scholar] [CrossRef]
- Chodak, M.; Pietrzykowski, M.; Niklińska, M. Development of microbial properties in a chronosequence of sandy mine soils. Appl. Soil Ecol. 2009, 41, 259–268. [Google Scholar] [CrossRef]
- Hinsinger, P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. In Plant and Soil; Springer: Berlin/Heidelberg, Germany, 2001; Volume 237, pp. 173–195. [Google Scholar]
- Treseder, K.K. A Meta-analysis of mycorrhizal responses to nitrogen, phosphorus, and atmospheric CO2 in field studies. N. Phytol. 2004, 164, 347–355. [Google Scholar] [CrossRef]
- Smith, S.; Read, D. Mycorrhizal Symbiosis; Elsevier Ltd.: Amsterdam, The Netherlands, 2008; ISBN 9780123705266. [Google Scholar]
- Stefanowicz, A.M.; Zubek, S.; Stanek, M.; Grześ, I.M.; Rożej-Pabijan, E.; Błaszkowski, J.; Woch, M.W. Invasion of Rosa rugosa induced changes in soil nutrients and microbial communities of coastal sand dunes. Sci. Total Environ. 2019, 677, 340–349. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, S.; Wang, R.; Xu, Z.; Feng, K.; Feng, X.; Li, T.; Liu, H.; Ma, R.; Li, H.; et al. Compositional and functional responses of soil microbial communities to long-term nitrogen and phosphorus addition in a calcareous grassland. Pedobiologia 2020, 78, 150612. [Google Scholar] [CrossRef]
- Treseder, K.K.; Allen, M.F. Direct nitrogen and phosphorus limitation of arbuscular mycorrhizal fungi: A model and field test. New Phytol. 2002, 155, 507–515. [Google Scholar] [CrossRef]
- Odebiri, O.; Mutanga, O.; Odindi, J. Deep learning-based national scale soil organic carbon mapping with Sentinel-3 data. Geoderma 2022, 411, 115695. [Google Scholar] [CrossRef]
- Kompała-Bąba, A.; Bierza, W.; Sierka, E.; Błońska, A.; Besenyei, L.; Woźniak, G. The role of plants and soil properties in the enzyme activities of substrates on hard coal mine spoil heaps. Sci. Rep. 2021, 11, 5155. [Google Scholar] [CrossRef]
- Stefanowicz, A.M.; Kapusta, P.; Błońska, A.; Kompała-Bąba, A.; Woźniak, G. Effects of Calamagrostis epigejos, Chamaenerion palustre and Tussilago farfara on nutrient availability and microbial activity in the surface layer of spoil heaps after hard coal mining. Ecol. Eng. 2015, 83, 328–337. [Google Scholar] [CrossRef]
- Borymski, S.; Cycoń, M.; Beckmann, M.; Mur, L.A.J.; Piotrowska-Seget, Z. Plant species and heavy metals affect biodiversity of microbial communities associated with metal-tolerant plants in metalliferous soils. Front. Microbiol. 2018, 9, 1425. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płociniczak, T.; Yu, D.; Kurola, J.M.; Sinkkonen, A.; Piotrowska-Seget, Z.; Romantschuk, M. Effect of Silene vulgaris and heavy metal pollution on soil microbial diversity in long-term contaminated soil. Water. Air. Soil Pollut. 2018, 229, 13. [Google Scholar] [CrossRef]
- Kozdrój, J.; Van Elsas, J.D. Response of the bacterial community to root exudates in soil polluted with heavy metals assessed by molecular and cultural approaches. Soil Biol. Biochem. 2000, 32, 1405–1417. [Google Scholar] [CrossRef]
- Eisenhauer, N.; Milcu, A.; Sabais, A.C.W.; Bessler, H.; Brenner, J.; Engels, C.; Klarner, B.; Maraun, M.; Partsch, S.; Roscher, C.; et al. Plant diversity surpasses plant functional groups and plant productivity as driver of soil biota in the long term. PLoS One 2011, 6, e16055. [Google Scholar] [CrossRef]
- Zhang, C.B.; Ke, S.S.; Wang, J.; Ge, Y.; Chang, S.X.; Zhu, S.X.; Chang, J. Responses of microbial activity and community metabolic profiles to plant functional group diversity in a full-scale constructed wetland. Geoderma 2011, 160, 503–508. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, S.; Chen, X.; Chen, H.Y.H. Plant diversity increases the abundance and diversity of soil fauna: A meta-analysis. Geoderma 2022, 411, 115694. [Google Scholar] [CrossRef]
- Shafi, M.I.; Yarranton, G.A. Vegetational heterogeneity during a secondary (postfire) succession. Can. J. Bot. 1973, 51, 73–90. [Google Scholar] [CrossRef]
- Prach, K. Succession of vegetation on dumps from strip coal mining, N.W. Bohemia, Czechoslovakia. Folia Geobot. Phytotaxon. 1987, 22, 339–354. [Google Scholar] [CrossRef]
- Prach, K. Spontaneous Succession in Central-European man-made habitats: What information can be used in restoration practice. In Applied Vegetation Science; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2003; Volume 6, pp. 125–129. [Google Scholar]
- Horn, H.S.; MacArthur, R.H. Competition among fugitive species in a harlequin environment. Ecology 1972, 53, 749–752. [Google Scholar] [CrossRef]
- Horn, H.S. The ecology of secondary succession. Annu. Rev. Ecol. Syst. 1974, 5, 25–37. [Google Scholar] [CrossRef]
- Baldrian, P.; Trögl, J.; Frouz, J.; Šnajdr, J.; Valášková, V.; Merhautová, V.; Cajthaml, T.; Herinková, J. Enzyme activities and microbial biomass in topsoil layer during spontaneous succession in spoil heaps after brown coal mining. Soil Biol. Biochem. 2008, 40, 2107–2115. [Google Scholar] [CrossRef]
- Józefowska, A.; Pietrzykowski, M.; Woś, B.; Cajthaml, T.; Frouz, J. The effects of tree species and substrate on carbon sequestration and chemical and biological properties in reforested post-mining soils. Geoderma 2017, 292, 9–16. [Google Scholar] [CrossRef]
- Mastrogianni, A.; Papatheodorou, E.M.; Monokrousos, N.; Menkissoglu-Spiroudi, U.; Stamou, G.P. Reclamation of lignite mine areas with Triticum aestivum: The dynamics of soil functions and microbial communities. Appl. Soil Ecol. 2014, 80, 51–59. [Google Scholar] [CrossRef]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 2011, 43, 1621–1625. [Google Scholar] [CrossRef]
- Zhang, B.; Li, Y.; Ren, T.; Tian, Z.; Wang, G.; He, X.; Tian, C. Short-term effect of tillage and crop rotation on microbial community structure and enzyme activities of a clay loam soil. Biol. Fertil. Soils 2014, 50, 1077–1085. [Google Scholar] [CrossRef]
- Muhammad, N.; Dai, Z.; Xiao, K.; Meng, J.; Brookes, P.C.; Liu, X.; Wang, H.; Wu, J.; Xu, J. Changes in microbial community structure due to biochars generated from different feedstocks and their relationships with soil chemical properties. Geoderma 2014, 226–227, 270–278. [Google Scholar] [CrossRef]
- Doran, J.W.; Parkin, T.B. Defining and assessing soil quality. In Defining Soil Quality for a Sustainable Environment; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 3–21. ISBN 9780891189305. [Google Scholar]
- Handa, I.T.; Aerts, R.; Berendse, F.; Berg, M.P.; Bruder, A.; Butenschoen, O.; Chauvet, E.; Gessner, M.O.; Jabiol, J.; Makkonen, M.; et al. Consequences of biodiversity loss for litter decomposition across biomes. Nature 2014, 509, 218–221. [Google Scholar] [CrossRef]
- Liang, J.; Crowther, T.W.; Picard, N.; Wiser, S.; Zhou, M.; Alberti, G.; Schulze, E.D.; McGuire, A.D.; Bozzato, F.; Pretzsch, H.; et al. Positive biodiversity-productivity relationship predominant in global forests. Science 2016, 354, aaf8957. [Google Scholar] [CrossRef]
- Sitaula, B.K.; Bakken, L.R.; Abrahamsen, G. N-fertilization and soil acidification effects on N2O and CO2 emission from temperate pine forest soil. Soil Biol. Biochem. 1995, 27, 1401–1408. [Google Scholar] [CrossRef]



| H′ | FDis | Number of Specimens | |
|---|---|---|---|
| HH | 1.76 ± 0.07 a | 0.34 ± 0.00 bc | 49.67 ± 6.94 |
| HL | 1.53 ± 0.03 ab | 0.29 ± 0.00 b | 40.00 ± 5.29 |
| LH | 1.12 ± 0.07 bc | 0.33 ± 0.01 ac | 33.00 ± 4.58 |
| LL | 0.81 ± 0.09 c | 0.22 ± 0.03 b | 28.67 ± 3.53 |
| R | 1.17 ± 0.03 bc | 0.46 ± 0.01 ac | 12.27 ± 6.62 |
| HH | HL | LH | LL | R | |
|---|---|---|---|---|---|
| pH H2O | 7.51 ± 0.17 a | 6.86 ± 0.19 b | 6.38 ± 0.57 b | 5.06 ± 0.37 c | 6.31 ± 0.12 bc |
| pH KCl | 7.20 ± 0.18 a | 6.41 ± 0.19 b | 5.77 ± 0.56 b | 4.49 ± 0.44 c | 5.41 ± 0.12 c |
| EC (mS cm−1) | 0.62 ± 0.19 a | 0.70 ± 0.27 a | 0.32 ± 0.07 a | 0.33 ± 0.08 a | 0.21 ± 0.02 a |
| NT (%) | 0.33 ± 0.06 b | 0.47 ± 0.03 a | 0.37 ± 0.02 b | 0.39 ± 0.01 b | 0.21 ± 0.01 c |
| SOM (%) | 16.31 ± 3.18 b | 23.59 ± 0.92 a | 20.07 ± 1.14 ab | 19.46 ± 0.91 ab | 8.32 ± 0.82 c |
| Mg ava. (mg kg−1) | 223.33 ± 29.10 d | 337.5 ± 9.02 b | 370.42 ± 18.25 a | 300.39 ± 19.96 c | 367.83 ± 3.14 a |
| P ava. (mg kg−1) | 31.03 ± 8.64 a | 2.97 ± 069 c | 5.57 ± 0.62 b | 4.51 ± 1.89 bc | 24.33 ± 2.76 a |
| Na+ (mg kg−1) | 127.69 ± 6.94 bc | 132.36 ± 7.95 bc | 143.69 ± 7.22 b | 119.56 ± 3.65 c | 232.18 ± 20.71 a |
| K+(mg kg−1) | 158.31 ± 15.17 bc | 160.31 ± 5.38 bc | 182.8 ± 15.19 b | 144.13 ± 10.16 c | 227.24 ± 3.55 a |
| Moisture (%) | 12.49 ± 1.08 a | 6.56 ± 0.80 b | 8.03 ± 1.01 b | 8.72 ± 1.24 b | 7.99 ± 0.49 b |
| HH | HL | LH | LL | R | |
|---|---|---|---|---|---|
| G+/G− | 1.22 ± 0.18 a | 1.42 ± 0.20 a | 1.30 ± 0.21 a | 1.57 ± 0.27 a | 1.13 ± 0.03 a |
| B/F | 1.56 ± 0.08 a | 2.01 ± 0.25 a | 1.84 ± 0.13 a | 1.96 ± 0.19 a | 1.74 ± 0.05 a |
| Simpson dominance index (D) | 0.11 ± 0.00 a | 0.11 ± 0.01 a | 0.11 ± 0.01 a | 0.10 ± 0.00 a | 0.10 ± 0.00 a |
| Shannon-Wiener diversity index (H′) | 2.38 ± 0.03 a | 2.42 ± 0.04 a | 2.42 ± 0.03 a | 2.45 ± 0.02 a | 2.45 ± 0.01 a |
| Evenness (E) | 0.77 ± 0.02 a | 0.80 ± 0.02 a | 0.76 ± 0.02 a | 0.81 ± 0.02 a | 0.77 ± 0.01 a |
| Margalef richness index (DS) | 5.01 ± 0.70 a | 5.27 ± 0.75 a | 4.66 ± 0.30 a | 5.62 ± 0.86 a | 3.23 ± 0.03 b |
| i15:0 + i17:0/a15:0 + a17:0 | 0.62 ± 0.06 c | 0.68 ± 0.09 c | 0.88 ± 0.04 b | 0.75 ± 0.06 bc | 1.29 ± 0.03 a |
| iso/anteiso | 1.49 ± 0.05 c | 1.56 ± 0.10 c | 1.80 ± 0.05 b | 1.78 ± 0.10 b | 2.03 ± 0.03 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bierza, W.; Woźniak, G.; Kompała-Bąba, A.; Magurno, F.; Malicka, M.; Chmura, D.; Błońska, A.; Jagodziński, A.M.; Piotrowska-Seget, Z. The Effect of Plant Diversity and Soil Properties on Soil Microbial Biomass and Activity in a Novel Ecosystem. Sustainability 2023, 15, 4880. https://doi.org/10.3390/su15064880
Bierza W, Woźniak G, Kompała-Bąba A, Magurno F, Malicka M, Chmura D, Błońska A, Jagodziński AM, Piotrowska-Seget Z. The Effect of Plant Diversity and Soil Properties on Soil Microbial Biomass and Activity in a Novel Ecosystem. Sustainability. 2023; 15(6):4880. https://doi.org/10.3390/su15064880
Chicago/Turabian StyleBierza, Wojciech, Gabriela Woźniak, Agnieszka Kompała-Bąba, Franco Magurno, Monika Malicka, Damian Chmura, Agnieszka Błońska, Andrzej M. Jagodziński, and Zofia Piotrowska-Seget. 2023. "The Effect of Plant Diversity and Soil Properties on Soil Microbial Biomass and Activity in a Novel Ecosystem" Sustainability 15, no. 6: 4880. https://doi.org/10.3390/su15064880
APA StyleBierza, W., Woźniak, G., Kompała-Bąba, A., Magurno, F., Malicka, M., Chmura, D., Błońska, A., Jagodziński, A. M., & Piotrowska-Seget, Z. (2023). The Effect of Plant Diversity and Soil Properties on Soil Microbial Biomass and Activity in a Novel Ecosystem. Sustainability, 15(6), 4880. https://doi.org/10.3390/su15064880










