Negative Evidence for Sex-Linked Heteroplasmy in the Nemertean Worm Notospermus geniculatus (Delle Chiaje, 1822)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Preparation, and Conservation
2.2. DNA Extraction and Purification
2.3. Amplification and Sequencing
2.4. Data Analysis
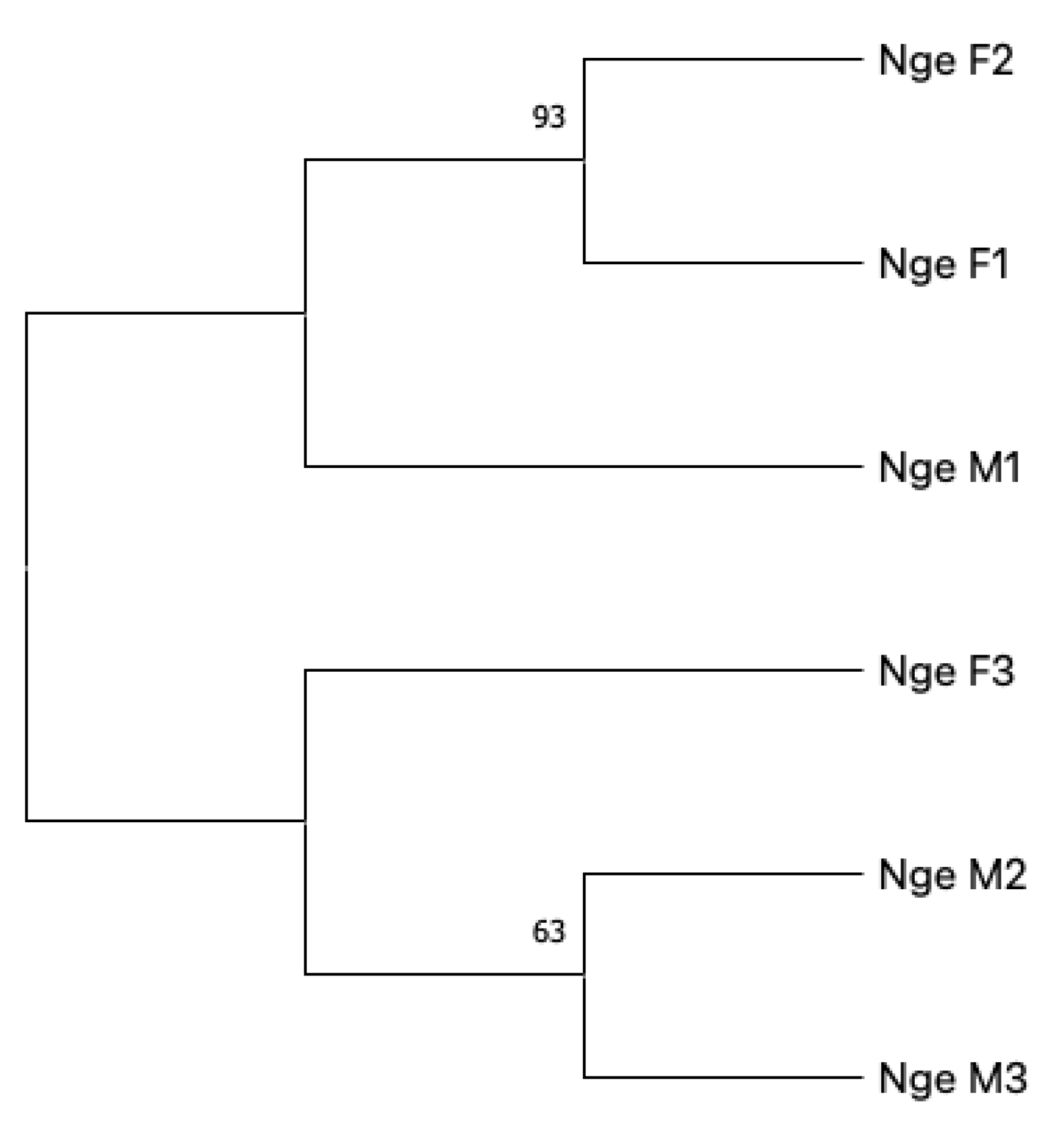
3. Results
| Gene | cox1 | rrnL | rrnS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Specimens’ Positions | 327 | 862 | 946 | 1050 | 384 | 459 | 725 | 951 | 326 | 536 |
| F1 | A | C | K | C | T | G | C | A | A | - |
| F2 | A | C | K | C | T | G | C | A | A | - |
| F3 | G | C | T | T | G | G | T | C | A | - |
| M1 | - | T | T | T | T | G | - | - | A | A |
| M2 | - | T | T | T | T | G | - | - | G | G |
| M3 | - | T | T | T | T | T | - | - | A | G |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tosetto, L.; McNab, J.M.; Hutchings, P.A.; Rodríguez, J.; Williamson, J.E. Fantastic Flatworms and Where to Find Them: Insights into Intertidal Polyclad Flatworm Distribution in Southeastern Australian Boulder Beaches. Diversity 2023, 15, 393. [Google Scholar] [CrossRef]
- Brusca, R.C.; Moore, W.; Shuster, F.M. Invertebrates, 3rd ed.; Sinauer Associates: Oxford, UK, 2016; pp. 435–452. [Google Scholar]
- Carwardine, M. The Guinness Book of Animal Records, 1st ed.; Guinness Publishing: Milan, Italy, 1995. [Google Scholar]
- Moen, F.E.; Svensen, E. Marine Fish & Invertebrates of Northern Europe; Kom: Kristiansund, Norway, 2004. [Google Scholar]
- Chernyshev, A.V. CLSM analysis of phallodin-stained muscle system of the nemertean proboscis and rhynchocoel. Zoolog. Sci. 2015, 32, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Norenburg, J.; Gibson, R.; Herrera Bachiller, A.; Strand, M. World Nemertea Data-base. Notospermus geniculatus (Delle Chiaje, 1828). Accessed through: World Register of Marine Species. 2019. Available online: http://www.marinespecies.org/aphia.php?p=tax-details&id=122586 (accessed on 12 May 2023).
- Helmkampf, M.; Bruchhaus, I.; Hausdorf, B. Phylogenomic analyses of lophophorates (brachiopods, phoronids and bryozoans) confirm the Lophotrochozoa concept. Proc. Biol. Sci. 2008, 275, 1927–1933. [Google Scholar] [CrossRef]
- Struck, T.H.; Fisse, F. Phylogenetic Position of Nemertea Derived from Phylogenomic Data. Mol. Biol. Evol. 2008, 25, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Nesnidal, M.P.; Helmkampf, M.; Meyer, A.; Witek, A.; Bruchhaus, I.; Ebersberger, I.; Hankeln, T.; Lieb, B.; Struck, T.H.; Hausdorf, B. New phylogenomic data support the monophyly of Lophophorata and an Ectoproct-Phoronid clade and indicate that Polyzoa and Kryptrochozoa are caused by systematic bias. BMC Evol. Biol. 2013, 13, 253. [Google Scholar] [CrossRef]
- Weigert, A.; Helm, C.; Meyer, M.; Nickel, B.; Arendt, D.; Hausdorf, B.; Santos, S.R.; Halanych, K.M.; Purschke, G.; Bleidorn, C.; et al. Illuminating the Base of the Annelid Tree Using Transcriptomics. Mol. Biol. Evol. 2014, 31, 1391–1401. [Google Scholar] [CrossRef]
- Kocot, K.M.; Struck, T.H.; Merkel, J.; Waits, D.S.; Todt, C.; Brannock, P.M.; Weese, D.A.; Cannon, J.T.; Moroz, L.L.; Lieb, B.; et al. Phylogenomics of Lophotrochozoa with Consideration of Systematic Error. Syst. Biol. 2016, 66, 256–282. [Google Scholar] [CrossRef]
- Peterson, K.J.; Eernisse, D.J. Animal phylogeny and the ancestry of bilaterians: Inferences from morphology and 18s rDNA gene sequences. Evol. Dev. 2001, 3, 170–205. [Google Scholar] [CrossRef]
- Chan, C.X.; Vaysberg, P.; Price, D.C.; Pelletreau, K.N.; Rumpho, M.E.; Bhattacharya, D. Active Host Response to Algal Symbionts in the Sea Slug Elysia chlorotica. Mol. Biol. Evol. 2018, 35, 1706–1711. [Google Scholar] [CrossRef]
- Struck, T.H.; Schult, N.; Kusen, T.; Hickman, E.; Bleidorn, C.; McHugh, D.; Halanych, K.M. Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evol. Biol. 2007, 7, 57. [Google Scholar] [CrossRef]
- Shen, X.; Ma, X.; Ren, J.; Zhao, F. A close phylogenetic relationship between Sipuncula and Annelida evidenced from the complete mitochondrial genome sequence of Phascolosoma esculenta. BMC Genom. 2009, 10, 136. [Google Scholar] [CrossRef]
- Zrzavý, J.; Říha, P.; Piálek, L.; Janouškovec, J. Phylogeny of Annelida (Lophotrochozoa): Total-evidence analysis of morphology and six genes. BMC Evol. Biol. 2009, 9, 189. [Google Scholar] [CrossRef]
- Struck, T.H.; Paul, C.; Hill, N.; Hartmann, S.; Hösel, C.; Kube, M.; Lieb, B.; Meyer, A.; Tiedemann, R.; Purschke, G.; et al. Phylogenomic analyses unravel annelid evolution. Nature 2011, 471, 95–98. [Google Scholar] [CrossRef]
- Davies, O.K.; Dorey, J.B.; Stevens, M.I.; Gardner, M.G.; Bradford, T.M.; Schwarz, M.P. Unparalleled mitochondrial heteroplasmy and Wolbachia co-infection in the non-model bee, Amphylaeus morosus. Curr. Res. Insect Sci. 2022, 2, 100036. [Google Scholar] [CrossRef]
- Ye, Z.; Zhao, C.; Raborn, R.T.; Lin, M.; Wei, W.; Hao, Y.; Lynch, M. Genetic Diversity, Heteroplasmy, and Recombination in Mitochondrial Genomes of Daphnia pulex, Daphnia pulicaria, and Daphnia obtusa. Mol. Biol. Evol. 2022, 39, msac059. [Google Scholar] [CrossRef]
- Radojičic, J.M.; Krizmanić, I.; Kasapidis, P.; Zouros, E. Extensive mitochondrial heteroplasmy in hybrid water frog (Pelophylax spp.) populations from Southeast Europe. Ecol. Evol. 2015, 5, 4529. [Google Scholar]
- Pizzirani, C.; Viola, P.; Gabbianelli, F.; Fagotti, A.; Simoncelli, F.; Di Rosa, I.; Salvi, P.; Amici, A.; Lucentini, L. First evidence of heteroplasmy in Grey Partridge (Perdix perdix). Avian Res. 2020, 11, 27. [Google Scholar] [CrossRef]
- Parakatselaki, M.E.; Ladoukakis, E.D. mtDNA Heteroplasmy: Origin, Detection, Significance, and Evolutionary Consequences. Life 2021, 11, 633. [Google Scholar] [CrossRef]
- Skibinski, D.O.F.; Gallagher, C.; Beynon, C.M. Mitochondrial DNA inheritance. Nature 1994, 368, 817–818. [Google Scholar] [CrossRef]
- Skibinski, D.O.F.; Gallagher, C.; Beynon, C.M. Sex-limited mitochondrial DNA transmission in the marine mussel Mytilus edulis. Genetics 1994, 138, 801–809. [Google Scholar] [CrossRef]
- Zouros, E.; Oberhauser Ball, A.; Saavedra, C.; Freeman, K.R. An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc. Natl. Acad. Sci. USA 1994, 91, 7463–7467. [Google Scholar] [CrossRef]
- Zouros, E.; Oberhauser Ball, A.; Saavedra, C.; Freeman, K.R. Mitochondrial DNA inheritance. Nature 1994, 368, 818. [Google Scholar] [CrossRef] [PubMed]
- Breton, S.; Beaupré, H.D.; Stewart, D.T.; Hoeh, W.R.; Blier, P.U. The unusual system of doubly uniparental inheritance of mtDNA: Isn’t one enough? Trends Genet. 2007, 23, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Passamonti, M.; Ghiselli, F. Doubly Uniparental Inheritance: Two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 2009, 28, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Zouros, E. Biparental inheritance through uniparental transmission: The doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol. Biol. 2013, 40, 1–31. [Google Scholar] [CrossRef]
- Zouros, E.; Rodakis, G.C. Doubly Uniparental Inheritance of mtDNA: An Unappreciated Defiance of a General Rule. Adv. Anat. Embryol. Cell Biol. 2019, 231, 25–49. [Google Scholar]
- Wang, R.; Li, X.; Qi, J. The complete paternally inherited mitochondrial genomes of three clam species in genus Macridiscus (Bivalvia: Veneridae): A TDRL model of dimer-mitogenome rearrangement of doubly uniparental inheritance. Front. Mar. Sci. 2022, 9, 1016779. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A.; Stewart, D.T.; Sutherland, B.W.; Zouros, E. The distribution of male-transmitted and female-transmitted mitochondrial DNA types in somatic tissues of blue mussels: Implications for the operation of doubly uniparental inheritance of mitochondrial DNA. Genome 1998, 41, 818–824. [Google Scholar] [CrossRef]
- Chakrabarti, R.; Walker, J.M.; Chapman, E.G.; Shepardson, S.P.; Trdan, R.J.; Curole, J.P.; Watters, G.T.; Stewart, D.T.; Vijayaraghavan, S.; Hoeh, W.R. Reproductive Function for a C-terminus Extended, Male-Transmitted Cytochrome c Oxidase Subunit II Protein Expressed in Both Spermatozoa and Eggs. FEBS Lett. 2007, 581, 5213–5219. [Google Scholar] [CrossRef]
- Kyriakou, E.; Zouros, E.; Rodakis, G.C. The atypical presence of the paternal mitochondrial DNA in somatic tissues of male and female individuals of the blue mussel species Mytilus galloprovincialis. BMC Res. Notes 2010, 3, 222. [Google Scholar] [CrossRef]
- Batista, F.M.; Lallias, D.; Taris, N.; Guerdes-Pinto, H.; Beaumont, A.R. Relative quantification of the M and F mitochondrial DNA types in the blue mussel Mytilus edulis by real-time PCR. J. Molluscan Stud. 2011, 77, 24–29. [Google Scholar] [CrossRef]
- Ghiselli, F.; Milani, L.; Passamonti, M. Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae). Mol. Biol. Evol. 2011, 28, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Obata, M.; Sano, N.; Komaru, A. Different transcriptional ratios of male and female transmitted mitochondrial DNA and tissue-specific expression patterns in the blue mussel, Mytilus galloprovincialis. Dev. Growth Diff. 2011, 53, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Brannock, P.M.; Roberts, M.A.; Hilbish, T.J. Ubiquitous heteroplasmy in Mytilus spp. resulting from disruption in doubly uniparental inheritance regulation. Mar. Ecol. Prog. Ser. 2013, 480, 131–143. [Google Scholar] [CrossRef]
- Lucentini, L.; Plazzi, F.; Sfriso, A.A.; Pizzirani, C.; Sfriso, A.; Chiesa, S. Additional taxonomic coverage of the doubly uniparental inheritance in bivalves: Evidence of sex-linked heteroplasmy in the razor clam Solen marginatus Pulteney, 1799, but not in the lagoon cockle Cerastoderma glaucum (Bruguière, 1789). J. Zool. Syst. Evol. Res. 2020, 58, 561–570. [Google Scholar] [CrossRef]
- Gusman, A.; Lecomte, S.; Stewart, D.T.; Passamonti, M.; Breton, S. Pursuing the quest for better understanding the taxonomic distribution of the system of doubly uniparental inheritance of mtDNA. PeerJ 2016, 4, e2760. [Google Scholar] [CrossRef] [PubMed]
- Lubośny, M.; Przyłucka, A.; Śmietanka, B.; Burzyński, A. Semimytilus algosus: First known hermaphroditic mussel with doubly uniparental inheritance of mitochondrial DNA. Sci. Rep. 2020, 10, 11256. [Google Scholar] [CrossRef]
- Milani, L.; Ghiselli, F.; Guerra, D.; Breton, S.; Passamonti, M. A comparative analysis of mitochondrial ORFans: New clues on their origin and role in species with doubly uniparental inheritance of mitochondria. Genome Biol. Evol. 2013, 5, 1408–1434. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.T.; Breton, S.; Chase, E.E.; Robicheau, B.M.; Bettinazzi, S.; Pante, E.; Youssef, N.; Garrido-Ramos, M.A. An unusual evolutionary strategy: The origins, genetic repertoire, and implications of doubly uniparental inheritance of mitochondrial DNA in bivalves. In Evolutionary Biology—A Transdisciplinary Approach; Pontarotti, P., Ed.; Springer International Publishing: New York, NY, USA, 2020; pp. 301–323. [Google Scholar]
- Milani, L.; Ghiselli, F.; Passamonti, M. Mitochondrial selfish elements and the evolution of biological novelties. Curr. Zool. 2016, 62, 687–697. [Google Scholar] [CrossRef]
- Pozzi, A.; Plazzi, F.; Milani, L.; Ghiselli, F.; Passamonti, M. SmithRNAs: Could mitochondria “bend” nuclear regulation? Mol. Biol. Evol. 2017, 34, 1960–1973. [Google Scholar] [CrossRef]
- Bettinazzi, S.; Plazzi, F.; Passamonti, M. The Complete Female- and Male-Transmitted Mitochondrial Genome of Meretrix lamarckii. PLoS ONE 2016, 11, e0153631. [Google Scholar] [CrossRef]
- Parakatselaki, M.E.; Saavedra, C.; Ladoukakis, E.D. Searching for doubly uniparental inheritance of mtDNA in the apple snail Pomacea diffusa. Mitochondrial DNA Part A 2016, 27, 4000–4002. [Google Scholar] [CrossRef] [PubMed]
- Gusman, A.; Azuelos, C.; Breton, S. No evidence of sex-linked heteroplasmy or doubly-uniparental inheritance of mtDNA in five gastropod species. J. Molluscan Stud. 2017, 83, 119–122. [Google Scholar] [CrossRef]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 545–579. [Google Scholar] [CrossRef]
- Palumbi, S.R.; Martin, A.; Romano, S.; McMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR, 1st ed.; University of Hawaii: Honolulu, HI, USA, 1996. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Soroka, M.; Burzyński, A. Doubly uniparental inheritance and highly divergent mitochondrial genomes of the freshwater mussel Unio tumidus (Bivalvia: Unionidae). Hydrobiologia 2018, 810, 239–254. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Theologidis, I.; Fodelianakis, S.; Gaspar, M.B.; Zouros, E. Doubly Uniparental Inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in bivalvia. Evolution 2008, 62, 959–970. [Google Scholar] [CrossRef]
- Breton, S.; Milani, L.; Ghiselli, F.; Guerra, D.; Stewart, D.T.; Passamonti, M. A resourceful genome: Updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 2014, 30, 555–564. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Ancient sex-specific extension of the cytochrome c oxidase II gene in bivalves and the fidelity of doubly-uniparental inheritance. Mol. Biol. Evol. 2002, 19, 1323–1328. [Google Scholar] [CrossRef]
- Breton, S.; Stewart, D.T.; Shepardson, S.; Trdan, R.J.; Bogan, A.E.; Chapman, E.G.; Ruminas, A.J.; Piontkivska, H.; Hoeh, W.R. Novel protein genes in animal mtDNA: A new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol. Biol. Evol. 2011, 28, 1645–1659. [Google Scholar] [CrossRef] [PubMed]
- Obata, M.; Shimizu, M.; Sano, N.; Komaru, A. Maternal inheritance of mitochondrial DNA (mtDNA) in the Pacific oyster (Crassostrea gigas): A preliminary study using mtDNA sequence analysis with evidence of random distribution of MitoTracker-stained sperm mitochondria in fertilized eggs. Zool. Sci. 2008, 25, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Salvi, D.; Mariottini, P. Molecular taxonomy in 2D: A novel ITS2 rRNA sequence-structure approach guides the description of the oysters’ subfamily Saccostreinae and the genus Magallana (Bivalvia: Ostreidae). Zool. J. Linn. Soc.-Lond. 2017, 179, 263–276. [Google Scholar] [CrossRef]
| Target Locus | Primer Name | Sequence (5′-3′) | Primer Length (bp) | Ref. |
|---|---|---|---|---|
| coxI | COX1_F674 (F) | GATCCTATTTTGTATCAGCAT T | 22 | 1 |
| COX1_R1420 (R) | CTCTCCCAAACAATAAACATAA | 22 | 1 | |
| C1-J1709 (F) | AATTGGGGGGTTYGGTAAYTG | 21 | 2 | |
| C1-N2776 (R) | GATAGTCAGAATAACGWCGNGG | 22 | 2 | |
| rrnL | RRNL_F189 (F) | ACCTTTTGTATCATGGTTTA | 20 | 1 |
| RRNL_R917 (R) | AAATGATTATGCTACCTTTG | 20 | 1 | |
| RRNL_F832 (F) | NTTTTATAANAAGTANTTTCTGCCC | 25 | 1 | |
| RRNL_R1405 (R) | ACGTANNATTTTAAAGGTCGAA | 22 | 1 | |
| SbrH(32) (R) | CCGGTCTGAACTCAGATCACGT | 22 | 3 | |
| Sar(34) (F) | CGCCTGTTTAACAAAAACAT | 20 | 3 mod | |
| rrnS | SR-J14197 (F) | GTACAYCTATGTTACGACTT | 20 | 2 |
| SR-N14745 (R) | GTCCCAGCAGYYGCGGTTANAC | 22 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santovito, D.; Brustenga, L.; Lucentini, L.; Plazzi, F.; Chiesa, S.; Passamonti, M. Negative Evidence for Sex-Linked Heteroplasmy in the Nemertean Worm Notospermus geniculatus (Delle Chiaje, 1822). Sustainability 2023, 15, 10212. https://doi.org/10.3390/su151310212
Santovito D, Brustenga L, Lucentini L, Plazzi F, Chiesa S, Passamonti M. Negative Evidence for Sex-Linked Heteroplasmy in the Nemertean Worm Notospermus geniculatus (Delle Chiaje, 1822). Sustainability. 2023; 15(13):10212. https://doi.org/10.3390/su151310212
Chicago/Turabian StyleSantovito, Diletta, Leonardo Brustenga, Livia Lucentini, Federico Plazzi, Stefania Chiesa, and Marco Passamonti. 2023. "Negative Evidence for Sex-Linked Heteroplasmy in the Nemertean Worm Notospermus geniculatus (Delle Chiaje, 1822)" Sustainability 15, no. 13: 10212. https://doi.org/10.3390/su151310212
APA StyleSantovito, D., Brustenga, L., Lucentini, L., Plazzi, F., Chiesa, S., & Passamonti, M. (2023). Negative Evidence for Sex-Linked Heteroplasmy in the Nemertean Worm Notospermus geniculatus (Delle Chiaje, 1822). Sustainability, 15(13), 10212. https://doi.org/10.3390/su151310212










