Climate-Resilient Microbial Biotechnology: A Perspective on Sustainable Agriculture
Abstract
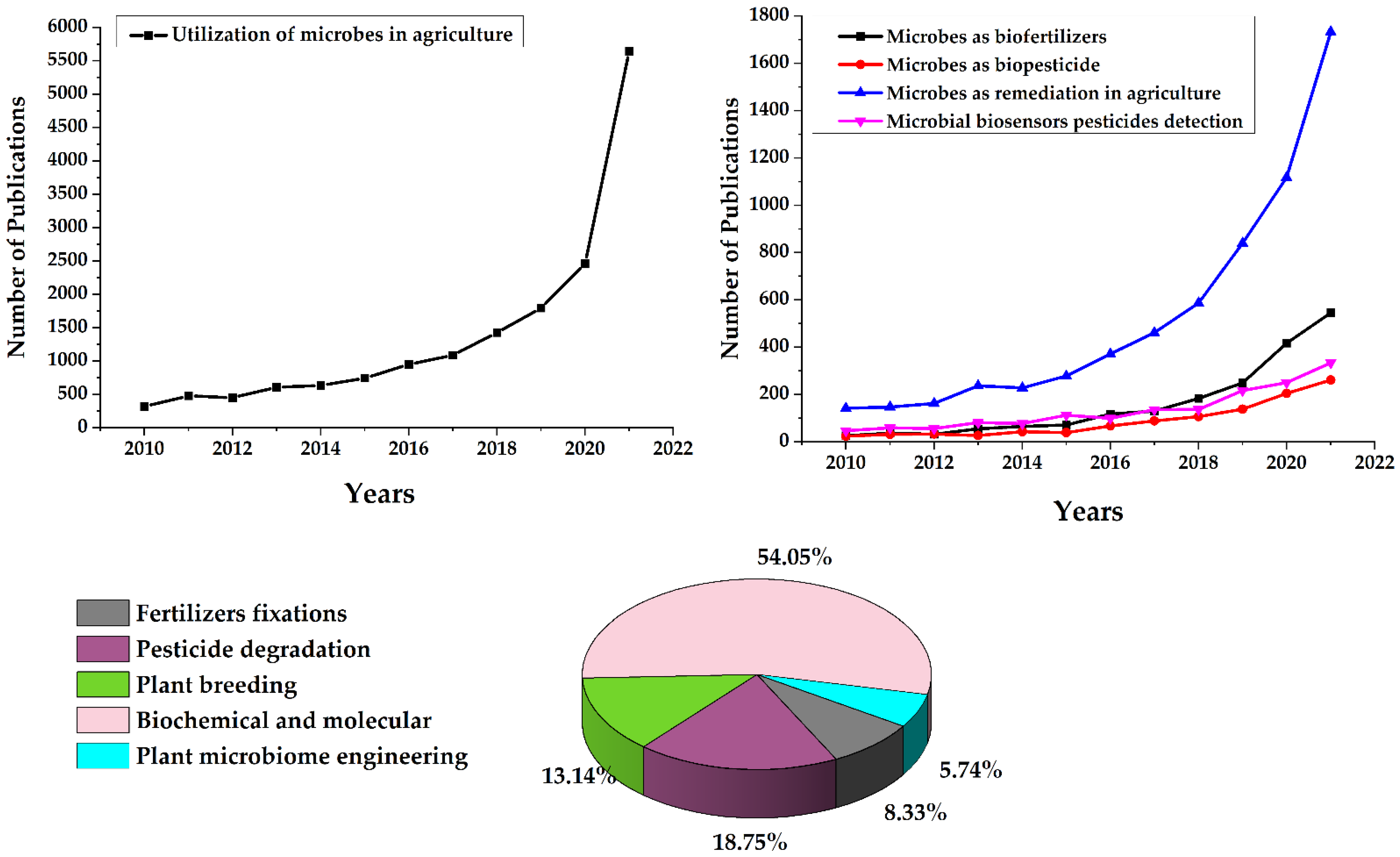
1. Introduction
2. Methodology
3. Climate Change
3.1. Harnessing Resilient Microbes for Climate Change
3.2. Climate-Smart Agriculture
4. Plant Pathogens, Biocontrol Strategies, and Metabolomic Applications
4.1. Plant Pathogens and Control Mechanisms
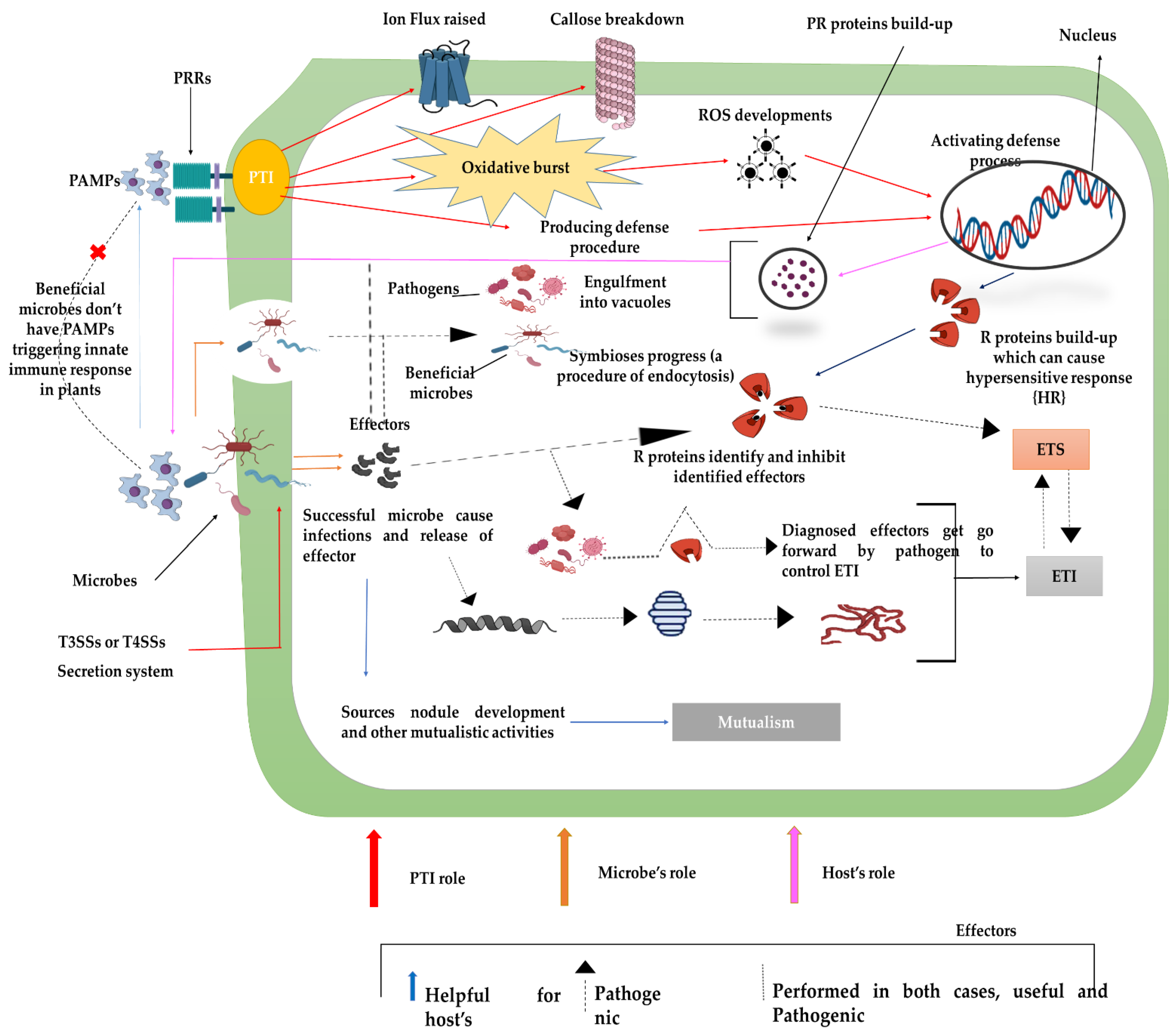
4.2. Plant–Microbes and Metabolome Variations
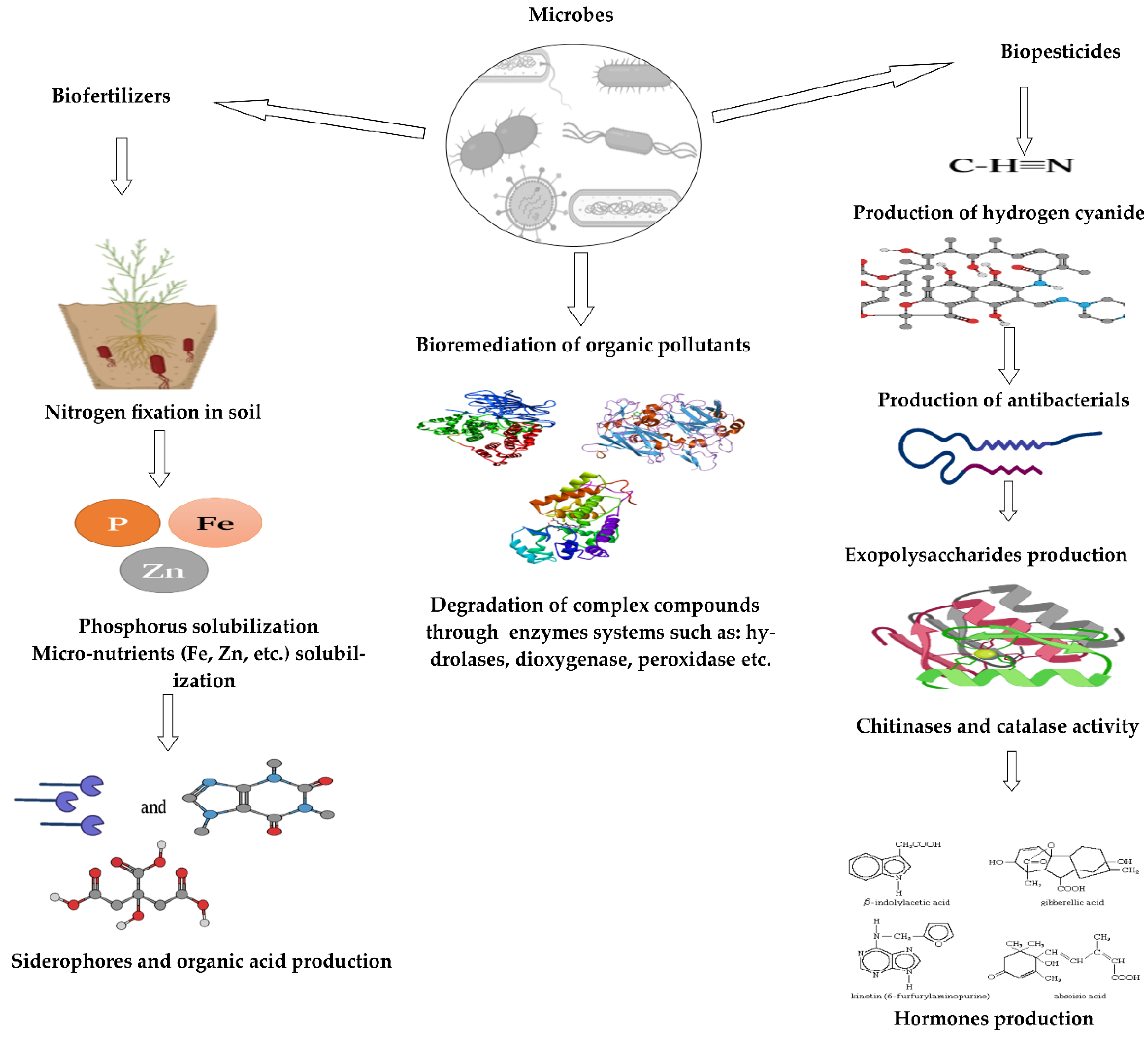
5. Plant–Microbes Symbiosis: Bio-Fertilizer
Application of Bio-Fertilizers to Control Replanting Disorders
6. Microbiome Engineering
6.1. Modification in Soil
6.2. Synthetic Microbial Consortia
6.3. Handling and Inoculation of Microbiome to Plants
7. Microbial Pesticides
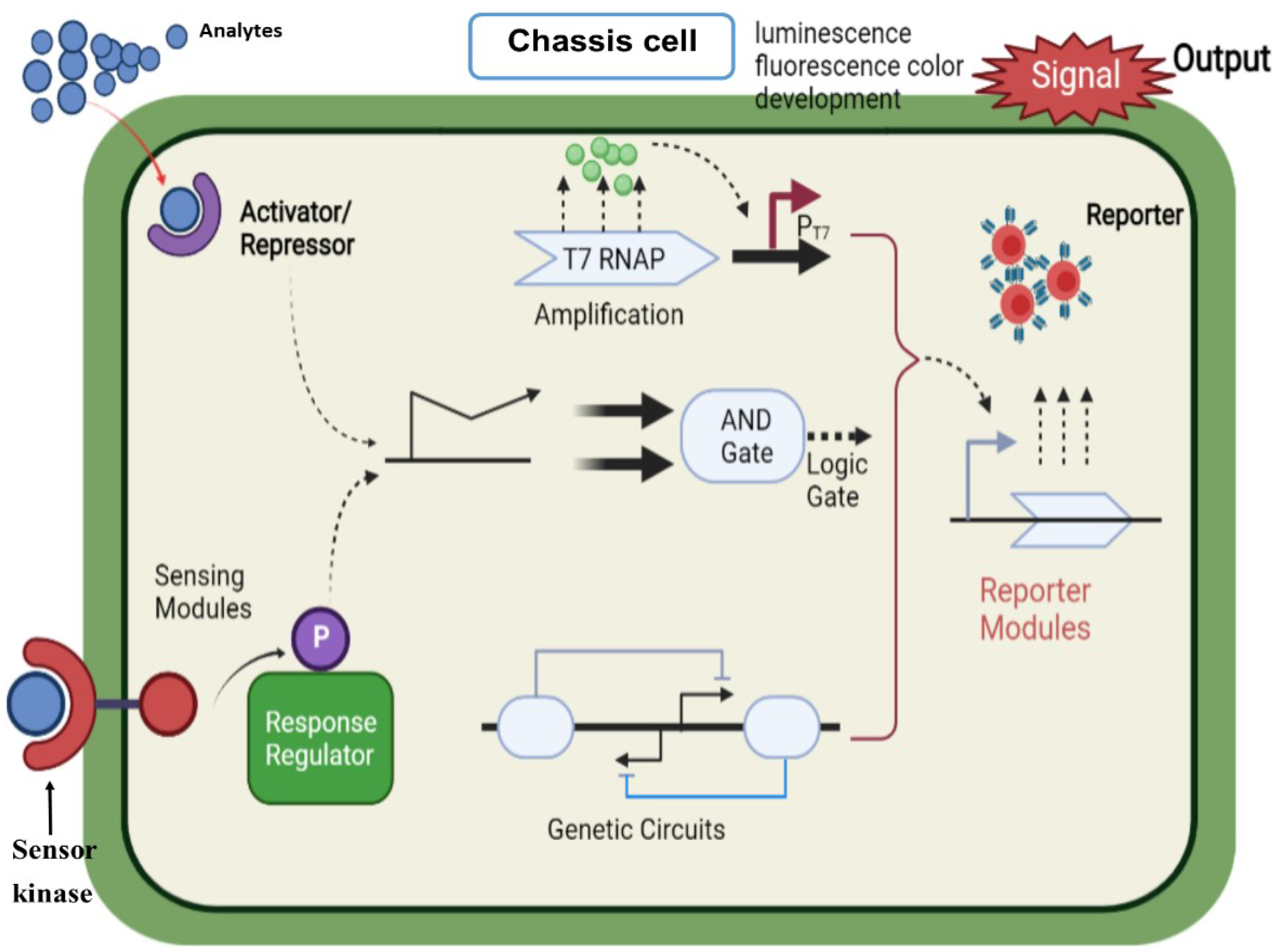
7.1. Application of Reporter Genes
7.2. Microbial Sensors and Gene Promoter
7.3. Advancements in Biosensors through Nanotechnology Application
8. Application of Microbes in Pesticide Degradation
8.1. Microbial Biosensors and Applications in Sensing Pesticide Residues
8.2. Microbes in Biochemical Degradation of Pesticides
8.3. Pesticide Degradation through Microbial Engineering
9. Conclusions and Future Prospectus
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghose, B. Food security and food self-sufficiency in China: From past to 2050. Food Energy Secur. 2014, 3, 86–95. [Google Scholar] [CrossRef]
- Alexandratos, N.; Bruinsma, J. World Agriculture towards 2030/2050: The 2012 Revision; Food and Agriculture Organization of the United Nations: Rome, Italy, 2012. [Google Scholar]
- Bruinsma, J. The Resource Outlook to 2050: By How Much Do Land, Water and Crop Yields Need to increase by 2050? In How to Feed the World in 2050, Proceedings of the a Technical Meeting of Experts, Rome, Italy, 24–26 June 2009; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2009; pp. 1–33. [Google Scholar]
- Wei, Z.; Jousset, A. Plant breeding goes microbial. Trends Plant Sci. 2017, 22, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.; Sharma, S.; Srivastava, A.K.; Penna, S. Halotolerant microbes and their applications in sustainable agriculture. In Physiological and Biotechnological Aspects of Extremophiles; Elsevier: Amsterdam, The Netherlands, 2020; pp. 39–49. [Google Scholar]
- Rohr, J.R.; Barrett, C.B.; Civitello, D.J.; Craft, M.E.; Delius, B.; DeLeo, G.A.; Hudson, P.J.; Jouanard, N.; Nguyen, K.H.; Ostfeld, R.S. Emerging human infectious diseases and the links to global food production. Nat. Sustain. 2019, 2, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Kukal, M.S.; Irmak, S. Climate-driven crop yield and yield variability and climate change impacts on the US Great Plains agricultural production. Sci. Rep. 2018, 8, 3450. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef]
- Kalhoro, M.T.; Zhang, H.; Kalhoro, G.M.; Wang, F.; Chen, T.; Faqir, Y.; Nabi, F. Fungicidal properties of ginger (Zingiber officinale) essential oils against Phytophthora colocasiae. Sci. Rep. 2022, 12, 2191. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K. Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Jayne, T.S.; Mason, N.M.; Burke, W.J.; Ariga, J. Taking stock of Africa’s second-generation agricultural input subsidy programs. Food Policy 2018, 75, 1–14. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, C. Natural and Human Factors Affect the Distribution of Soil Heavy Metal Pollution: A Review. Water Air Soil Pollut. 2020, 231, 350. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Fritschi, F.B.; Mittler, R. Global warming, climate change, and environmental pollution: Recipe for a multifactorial stress combination disaster. Trends Plant Sci. 2021, 26, 588–599. [Google Scholar] [CrossRef]
- Karakaya, A.; Dikilitas, M. Biochemical, physiological and molecular defence mechanisms of tea plants against pathogenic agents under changing climate conditions. In Stress Physiology of Tea in the Face of Climate Change; Springer: Singapore, 2018; pp. 241–268. [Google Scholar]
- Newton, A.C.; Fitt, B.D.; Atkins, S.D.; Walters, D.R.; Daniell, T.J. Pathogenesis, parasitism and mutualism in the trophic space of microbe–plant interactions. Trends Microbiol. 2010, 18, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.S.; Gupta, V.K. Soil microbial biomass: A key soil driver in management of ecosystem functioning. Sci. Total Environ. 2018, 634, 497–500. [Google Scholar] [CrossRef]
- Niu, B.; Wang, W.; Yuan, Z.; Sederoff, R.R.; Sederoff, H.; Chiang, V.L.; Borriss, R. Microbial interactions within multiple-strain biological control agents impact soil-borne plant disease. Front. Microbiol. 2020, 11, 2452. [Google Scholar] [CrossRef] [PubMed]
- Berlanga-Clavero, M.V.; Molina-Santiago, C.; de Vicente, A.; Romero, D. More than words: The chemistry behind the interactions in the plant holobiont. Environ. Microbiol. 2020, 22, 4532–4544. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Yang, F.; Tang, C.; Antonietti, M. Natural and artificial humic substances to manage minerals, ions, water, and soil microorganisms. Chem. Soc. Rev. 2021, 50, 6221–6239. [Google Scholar] [CrossRef]
- Kumar, V. Rhizomicrobiome Dynamics in Bioremediation; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2021. [Google Scholar]
- Basu, S.; Kumar, G. Plant microbe interaction for changing endophytic colonization to improve plant productivity. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2021; pp. 137–147. [Google Scholar]
- Itelima, J.; Bang, W.; Onyimba, I.; Oj, E. A review: Biofertilizer; a key player in enhancing soil fertility and crop productivity. J. Microbiol. Biotechnol. Rep. 2018, 2, 22–28. [Google Scholar]
- Soumare, A.; Boubekri, K.; Lyamlouli, K.; Hafidi, M.; Ouhdouch, Y.; Kouisni, L. From isolation of phosphate solubilizing microbes to their formulation and use as biofertilizers: Status and needs. Front. Bioeng. Biotechnol. 2019, 7, 425. [Google Scholar] [CrossRef]
- Patil, H.J.; Solanki, M.K. Microbial inoculant: Modern era of fertilizers and pesticides. In Microbial Inoculants in Sustainable Agricultural Productivity; Springer: New Delhi, India, 2016; pp. 319–343. [Google Scholar]
- Singh, R. Microorganism as a tool of bioremediation technology for cleaning environment: A review. Proc. Int. Acad. Ecol. Environ. Sci. 2014, 4, 1. [Google Scholar]
- Dykes, G.E. Soil Microbial Ecology and Biogeochemical Cycling of Arsenic and Iron in Flooded Rice Paddies; University of Delaware: Newark, DE, USA, 2021. [Google Scholar]
- Tscharntke, T.; Grass, I.; Wanger, T.C.; Westphal, C.; Batáry, P. Beyond organic farming–harnessing biodiversity-friendly landscapes. Trends Ecol. Evol. 2021, 36, 919–930. [Google Scholar] [CrossRef]
- Abo Nouh, F.A. Endophytic fungi for sustainable agriculture. Microb. Biosyst. 2019, 4, 31–44. [Google Scholar]
- Parasuraman, P.; Pattnaik, S.; Busi, S. Plant-Microbe Interactions in Ecosystems Functioning and Sustainability. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2019; pp. 255–266. [Google Scholar]
- Papik, J.; Folkmanova, M.; Polivkova, M.; Suman, J.; Uhlik, O. The invisible life inside plants: Deciphering the riddles of endophytic bacterial diversity. Biotechnol. Adv. 2020, 44, 107614. [Google Scholar] [CrossRef] [PubMed]
- Hirakue, A.; Sugiyama, S. Relationship between foliar endophytes and apple cultivar disease resistance in an organic orchard. Biol. Control 2018, 127, 139–144. [Google Scholar] [CrossRef]
- Verma, S.K.; Kingsley, K.; Bergen, M.; English, C.; Elmore, M.; Kharwar, R.N.; White, J.F. Bacterial endophytes from rice cut grass (Leersia oryzoides L.) increase growth, promote root gravitropic response, stimulate root hair formation, and protect rice seedlings from disease. Plant Soil 2018, 422, 223–238. [Google Scholar] [CrossRef]
- Barra-Bucarei, L.; González, M.G.; Iglesias, A.F.; Aguayo, G.S.; Peñalosa, M.G.; Vera, P.V. Beauveria bassiana multifunction as an endophyte: Growth promotion and biologic control of Trialeurodes vaporariorum, (Westwood) (Hemiptera: Aleyrodidae) in tomato. Insects 2020, 11, 591. [Google Scholar] [CrossRef]
- Sahu, P.K.; Singh, S.; Gupta, A.; Singh, U.B.; Brahmaprakash, G.; Saxena, A.K. Antagonistic potential of bacterial endophytes and induction of systemic resistance against collar rot pathogen Sclerotium rolfsii in tomato. Biol. Control 2019, 137, 104014. [Google Scholar] [CrossRef]
- Mei, C.; Flinn, B.S. The use of beneficial microbial endophytes for plant biomass and stress tolerance improvement. Recent Pat. Biotechnol. 2010, 4, 81–95. [Google Scholar] [CrossRef]
- Dastogeer, K.M.; Wylie, S.J. Plant–fungi association: Role of fungal endophytes in improving plant tolerance to water stress. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Singapore, 2017; pp. 143–159. [Google Scholar]
- Egamberdieva, D.; Wirth, S.; Behrendt, U.; Ahmad, P.; Berg, G. Antimicrobial activity of medicinal plants correlates with the proportion of antagonistic endophytes. Front. Microbiol. 2017, 8, 199. [Google Scholar] [CrossRef]
- Godson-ibeji, C.C.; Chikaire, J.U. Consequences of environmental pollution on agricultural productivity in developing countries: A case of Nigeria. Int. J. Agric. Res. 2016, 5, 1–12. [Google Scholar] [CrossRef]
- Guetsky, R.; Shtienberg, D.; Elad, Y.; Dinoor, A. Combining biocontrol agents to reduce the variability of biological control. Phytopathology 2001, 91, 621–627. [Google Scholar] [CrossRef]
- Olfs, H.W.; Blankenau, K.; Brentrup, F.; Jasper, J.; Link, A.; Lammel, J. Soil-and plant-based nitrogen-fertilizer recommendations in arable farming. J. Plant. Nutr. Soil Sci. 2005, 168, 414–431. [Google Scholar] [CrossRef]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Hasanuzzaman, M.; Alam, M.; Ullah, H.; Saeed, M.; Khan, I.A.; Adnan, M. Environment, Climate, Plant and Vegetation Growth; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Igiehon, N.O.; Babalola, O.O. Rhizosphere microbiome modulators: Contributions of nitrogen fixing bacteria towards sustainable agriculture. Int. J. Environ. Res. Public Health 2018, 15, 574. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, P.L.; Srivastava, A.K.; Tiwari, S.P.; Kumar, S. Microbes for Climate Resilient Agriculture; John Wiley & Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Hagagy, N.I.; Saddiq, A.A.; Hamedo, H.A.; Selim, S.A. Extremophiles Inhabiting Unique Ecosystems in Egypt. In Extreme Environments; CRC Press: Boca Raton, FL, USA, 2021; pp. 252–262. [Google Scholar]
- Khan, I.; Khan, F.; Ahmad, S.; Pandey, P.; Khan, M.M. Microbes and Climate: A Tangled Relation. In Microbiomes and the Global Climate Change; Springer: Cham, Switzerland, 2021; pp. 3–15. [Google Scholar]
- Meena, R.P.; Jha, A. Conservation agriculture for climate change resilience: A microbiological perspective. In Microbes for Climate Resilient Agriculture; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 165–190. [Google Scholar]
- El Chami, D.; Daccache, A.; El Moujabber, M. How can sustainable agriculture increase climate resilience? A systematic review. Sustainability 2020, 12, 3119. [Google Scholar] [CrossRef]
- Yadav, A.N. Biodiversity and bioprospecting of extremophilic microbiomes for agro-environmental sustainability. J. Appl. Biol. 2021, 9, 1–6. [Google Scholar]
- De Souza, R.S.C.; Armanhi, J.S.L.; Arruda, P. From microbiome to traits: Designing synthetic microbial communities for improved crop resiliency. Front. Plant Sci. 2020, 11, 1179. [Google Scholar] [CrossRef]
- Lyu, D.; Backer, R.; Subramanian, S.; Smith, D.L. Phytomicrobiome coordination signals hold potential for climate change-resilient agriculture. Front. Plant Sci. 2020, 11, 634. [Google Scholar] [CrossRef]
- Mohanty, S.; Swain, C.K. Role of microbes in climate smart agriculture. In Microorganisms for Green Revolution; Springer: Singapore, 2018; pp. 129–140. [Google Scholar]
- Sethi, S. Utilization of beneficial fungal strain/bacterial strains in climate-resilient agriculture. In Microbiome under Changing Climate; Elsevier: Amsterdam, The Netherlands, 2022; pp. 313–331. [Google Scholar]
- Waaswa, A.; Oywaya Nkurumwa, A.; Mwangi Kibe, A.; Ngeno Kipkemoi, J. Climate-Smart agriculture and potato production in Kenya: Review of the determinants of practice. Clim. Dev. 2022, 14, 75–90. [Google Scholar] [CrossRef]
- Bungau, S.; Behl, T.; Aleya, L.; Bourgeade, P.; Aloui-Sossé, B.; Purza, A.L.; Abid, A.; Samuel, A.D. Expatiating the impact of anthropogenic aspects and climatic factors on long-term soil monitoring and management. Environ. Sci. Pollut. Res. 2021, 28, 30528–30550. [Google Scholar] [CrossRef]
- Singh, B.K.; Trivedi, P.; Singh, S.; Macdonald, C.A.; Verma, J.P. Emerging microbiome technologies for sustainable increase in farm productivity and environmental security. Microbiol. Aust. 2018, 39, 17–23. [Google Scholar] [CrossRef]
- Hamilton, C.E.; Bever, J.D.; Labbé, J.; Yang, X.; Yin, H. Mitigating climate change through managing constructed-microbial communities in agriculture. Agric. Ecosyst. Environ. 2016, 216, 304–308. [Google Scholar] [CrossRef]
- Dhankher, O.P.; Foyer, C.H. Climate resilient crops for improving global food security and safety. Wiley Online Libr. 2018, 41, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, M. Climatic resilient agriculture for root, tuber, and banana crops using plant growth-promoting microbes. In Climate Change and Agricultural Ecosystems; Elsevier: Amsterdam, The Netherlands, 2019; pp. 307–329. [Google Scholar]
- Wong, C.K.F. Application of Microbes in Climate-Resilient Crops. In Application of Microbes in Environmental and Microbial Biotechnology; Springer: Singapore, 2022; pp. 93–112. [Google Scholar]
- Dastogeer, K.M.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Sarkar, B.; Jat, H.S.; Sharma, P.C.; Bolan, N.S. Soil salinity under climate change: Challenges for sustainable agriculture and food security. J. Environ. Manag. 2021, 280, 111736. [Google Scholar] [CrossRef] [PubMed]
- Gorguner, M.; Kavvas, M.L. Modeling impacts of future climate change on reservoir storages and irrigation water demands in a Mediterranean basin. Sci. Total Environ. 2020, 748, 141246. [Google Scholar] [CrossRef]
- Etesami, H.; Glick, B.R. Halotolerant plant growth–promoting bacteria: Prospects for alleviating salinity stress in plants. Environ. Exp. Bot. 2020, 178, 104124. [Google Scholar] [CrossRef]
- Saini, R.; Sharma, S. Climate resilient microbes in sustainable crop production. In Contaminants in Agriculture and Environment: Health Risks and Remediation; Agriculture and Environmental Science Academy: Haridwar, India, 2019; Volume 1, p. 264. [Google Scholar]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022. [Google Scholar] [CrossRef]
- Passarini, M.R.Z.; Duarte, A.W.F.; Rosa, L.H.; de Oliveira, V.M.; Ottoni, J.R. Extremofuels: Production of biofuels by extremophile microbes as an alternative to avoid climate change effects. In Microbiome under Changing Climate; Elsevier: Amsterdam, The Netherlands, 2022; pp. 237–256. [Google Scholar]
- Al-Shaibani, M.M.; Radin Mohamed, R.M.S.; Sidik, N.M.; Enshasy, H.A.E.; Al-Gheethi, A.; Noman, E.; Al-Mekhlafi, N.A.; Zin, N.M. Biodiversity of secondary metabolites compounds isolated from phylum actinobacteria and its therapeutic applications. Molecules 2021, 26, 4504. [Google Scholar] [CrossRef]
- Rizvi, A.; Ahmed, B.; Khan, M.S.; Umar, S.; Lee, J. Psychrophilic Bacterial Phosphate-Biofertilizers: A Novel Extremophile for Sustainable Crop Production under Cold Environment. Microorganisms 2021, 9, 2451. [Google Scholar] [CrossRef]
- Pouvreau, B.; Vanhercke, T.; Singh, S. From plant metabolic engineering to plant synthetic biology: The evolution of the design/build/test/learn cycle. Plant Sci. 2018, 273, 3–12. [Google Scholar] [CrossRef]
- Song, C.; Jin, K.; Raaijmakers, J.M. Designing a home for beneficial plant microbiomes. Curr. Opin. Plant Biol. 2021, 62, 102025. [Google Scholar] [CrossRef] [PubMed]
- Dey, R.; Pal, K.; Thomas, M.; Sherathia, D.; Mandaliya, V.; Bhadania, R.; Patel, M.; Maida, P.; Mehta, D.; Nawade, B. Endophytic microorganisms: Future tools for climate resilient agriculture. In Microbes for Climate Resilient Agriculture; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 235–253. [Google Scholar]
- Lone, S.A.; Malik, A. Microbiomes and the Global Climate Change; Springer: Singapore, 2021. [Google Scholar]
- Omomowo, O.I.; Babalola, O.O. Bacterial and fungal endophytes: Tiny giants with immense beneficial potential for plant growth and sustainable agricultural productivity. Microorganisms 2019, 7, 481. [Google Scholar] [CrossRef] [PubMed]
- Hanvi, D.M.; Lawson-Evi, P.; De Boevre, M.; Goto, C.; De Saeger, S.; Eklu-Gadegbeku, K. Natural occurrence of mycotoxins in maize and sorghum in Togo. Mycotoxin Res. 2019, 35, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.T.; Petta, T.; Rottinghaus, G.E.; Bordin, K.; Gomes, G.A.; Oliveira, C.A. Co-occurrence of mycotoxins in maize food and maize-based feed from small-scale farms in Brazil: A pilot study. Mycotoxin Res. 2019, 35, 65–73. [Google Scholar] [CrossRef]
- Chilaka, C.A.; De Boevre, M.; Atanda, O.O.; De Saeger, S. Occurrence of Fusarium mycotoxins in cereal crops and processed products (Ogi) from Nigeria. Toxins 2016, 8, 342. [Google Scholar] [CrossRef]
- Lee, H.J.; Ryu, D. Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: Public health perspectives of their co-occurrence. J. Agric. Food Chem. 2017, 65, 7034–7051. [Google Scholar] [CrossRef]
- Abdallah, M.F.; De Boevre, M.; Audenaert, K.; Haesaert, G.; De Saeger, S. Highlight report: Mycotoxins as food contaminants in Africa—challenges and perspectives. Arch Toxicol 2018, 92, 2151–2152. [Google Scholar] [CrossRef]
- Ghorbanpour, M.; Omidvari, M.; Abbaszadeh-Dahaji, P.; Omidvar, R.; Kariman, K. Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 2018, 117, 147–157. [Google Scholar] [CrossRef]
- Kagot, V.; Okoth, S.; De Boevre, M.; De Saeger, S. Biocontrol of Aspergillus and Fusarium mycotoxins in Africa: Benefits and limitations. Toxins 2019, 11, 109. [Google Scholar] [CrossRef]
- Nafuka, S.N.; Misihairabgwi, J.M.; Bock, R.; Ishola, A.; Sulyok, M.; Krska, R. Variation of fungal metabolites in sorghum malts used to prepare Namibian traditional fermented beverages Omalodu and Otombo. Toxins 2019, 11, 165. [Google Scholar] [CrossRef]
- Babar, M.M.; Khan, S.F.; Zargaham, M.K.; Gul, A. Plant-microbe interactions: A molecular approach. In Plant, Soil and Microbes; Springer: Cham, Switzerland, 2016; pp. 1–22. [Google Scholar]
- Egamberdieva, D.; Shrivastava, S.; Varma, A. Plant-Growth-Promoting Rhizobacteria (PGPR) and Medicinal Plants; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Patel, P.; Shah, R.; Joshi, B.; Ramar, K.; Natarajan, A. Molecular identification and biocontrol activity of sugarcane rhizosphere bacteria against red rot pathogen Colletotrichum falcatum. Biotechnol. Rep. 2019, 21, e00317. [Google Scholar] [CrossRef] [PubMed]
- Nookongbut, P.; Kantachote, D.; Khuong, N.Q.; Sukhoom, A.; Tantirungkij, M.; Limtong, S. Selection of acid-resistant purple nonsulfur bacteria from peat swamp forests to apply as biofertilizers and biocontrol agents. J. Soil Sci. Plant Nutr. 2019, 19, 488–500. [Google Scholar] [CrossRef]
- Venieraki, A.; Chorianopoulou, S.N.; Katinakis, P.; Bouranis, D.L. Multi-Trait Wheat Rhizobacteria from Calcareous Soil with Biocontrol Activity Promote Plant Growth and Mitigate Salinity Stress. Microorganisms 2021, 9, 1588. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Verma, H.; Singh, V.K.; Singh, P.P.; Singh, S.K.; Ansari, W.A.; Yadav, A.; Singh, P.; Pandey, K. Role of Pseudomonas sp. in sustainable agriculture and disease management. In Agriculturally Important Microbes for Sustainable Agriculture; Springer: Singapore, 2017; pp. 195–215. [Google Scholar]
- Bakker, P.A.; Ran, L.; Mercado-Blanco, J. Rhizobacterial salicylate production provokes headaches! Plant Soil 2014, 382, 1–16. [Google Scholar] [CrossRef]
- Mirzaee, H.; Shuey, L.; Schenk, P.M. Transcriptomics of plants interacting with pathogens and beneficial microbes. In Genomics, Proteomics and Metabolomics in Nutraceuticals and Functional Foods; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 525–536. [Google Scholar]
- Darvill, A.G.; Albersheim, P. Phytoalexins and their elicitors-a defense against microbial infection in plants. Annu. Rev. Plant Physiol. 1984, 35, 243–275. [Google Scholar] [CrossRef]
- D’Antuono, A.L.; Ott, T.; Krusell, L.; Voroshilova, V.; Ugalde, R.A.; Udvardi, M.; Lepek, V.C. Defects in rhizobial cyclic glucan and lipopolysaccharide synthesis alter legume gene expression during nodule development. Mol. Plant-Microbe Interact. 2008, 21, 50–60. [Google Scholar] [CrossRef][Green Version]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Singh, P.P.; Kujur, A.; Yadav, A.; Kumar, A.; Singh, S.K.; Prakash, B. Mechanisms of plant-microbe interactions and its significance for sustainable agriculture. In PGPR Amelioration in Sustainable Agriculture; Elsevier: Amsterdam, The Netherlands, 2019; pp. 17–39. [Google Scholar]
- Emwas, A.-H.M.; Salek, R.M.; Griffin, J.L.; Merzaban, J. NMR-based metabolomics in human disease diagnosis: Applications, limitations, and recommendations. Metabolomics 2013, 9, 1048–1072. [Google Scholar] [CrossRef]
- Zamora-Ros, R.; Touillaud, M.; Rothwell, J.A.; Romieu, I.; Scalbert, A. Measuring exposure to the polyphenol metabolome in observational epidemiologic studies: Current tools and applications and their limits. Am. J. Clin. Nutr. 2014, 100, 11–26. [Google Scholar] [CrossRef]
- Arbona, V.; Gomez-Cadenas, A. Metabolomics of disease resistance in crops. Curr. Issues Mol. Biol. 2016, 19, 13–30. [Google Scholar] [PubMed]
- Adeniji, A.A.; Babalola, O.O. Metabolomic applications for understanding complex tripartite plant-microbes interactions: Strategies and perspectives. Biotechnol. Rep. 2020, 25, e00425. [Google Scholar] [CrossRef] [PubMed]
- Azzollini, A.; Boggia, L.; Boccard, J.; Sgorbini, B.; Lecoultre, N.; Allard, P.-M.; Rubiolo, P.; Rudaz, S.; Gindro, K.; Bicchi, C. Dynamics of metabolite induction in fungal co-cultures by metabolomics at both volatile and non-volatile levels. Front. Microbiol. 2018, 9, 72. [Google Scholar] [CrossRef]
- Sevastos, A.; Kalampokis, I.; Panagiotopoulou, A.; Pelecanou, M.; Aliferis, K. Implication of Fusarium graminearum primary metabolism in its resistance to benzimidazole fungicides as revealed by 1H NMR metabolomics. Pestic. Biochem. Phys. 2018, 148, 50–61. [Google Scholar] [CrossRef]
- Bucher, R.; Veyel, D.; Willmitzer, L.; Krattinger, S.; Keller, B.; Bigler, L. Combined GC-and UHPLC-HR-MS based metabolomics to analyze durable anti-fungal resistance processes in cereals. Chim. Int. J. Chem. 2017, 71, 156–159. [Google Scholar] [CrossRef]
- Sun, J.; Duan, Z.; Zhang, Y.; Cao, S.; Tang, Z.; Abozeid, A. Metabolite Profiles Provide Insights into Underlying Mechanism in Bupleurum (Apiaceae) in Response to Three Levels of Phosphorus Fertilization. Plants 2022, 11, 752. [Google Scholar] [CrossRef] [PubMed]
- Aliferis, K.A.; Faubert, D.; Jabaji, S. A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS ONE 2014, 9, e111930. [Google Scholar] [CrossRef] [PubMed]
- Dhayalan, V.; Sudalaimuthu, K. Plant growth promoting rhizobacteria in promoting sustainable agriculture. Glob. J. Environ. Sci. Manag. 2021, 7, 401–418. [Google Scholar]
- Berrada, H.; Fikri-Benbrahim, K. Taxonomy of the rhizobia: Current perspectives. Microbiol. Res. Int. 2014, 616–639. [Google Scholar] [CrossRef]
- Prasad, R.; Kumar, M.; Varma, A. Role of PGPR in soil fertility and plant health. In Plant-Growth-Promoting Rhizobacteria (PGPR) and Medicinal Plants; Springer: Cham, Switzerland, 2015; pp. 247–260. [Google Scholar]
- Ji, H.; Dong, H. Key steps in type III secretion system (T3SS) towards translocon assembly with potential sensor at plant plasma membrane. Mol. Plant Pathol. 2015, 16, 762–773. [Google Scholar] [CrossRef]
- Almario, J.; Gobbin, D.; Défago, G.; Moënne-Loccoz, Y.; Rezzonico, F. Prevalence of type III secretion system in effective biocontrol pseudomonads. Res. Microbiol. 2014, 165, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Pandey, P.; Maheshwari, D. Reduction in dose of chemical fertilizers and growth enhancement of sesame (Sesamum indicum L.) with application of rhizospheric competent Pseudomonas aeruginosa LES4. Eur. J. Soil Biol. 2009, 45, 334–340. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Dey, U. Biofertilizer, a way towards organic agriculture: A review. Afr. J. Microbiol. Res. 2014, 8, 2332–2343. [Google Scholar]
- Wu, H.; Zhang, Z.; Wang, J.; Qin, X.; Chen, J.; Wu, L.; Lin, S.; Rensing, C.; Lin, W. Bio-fertilizer Amendment Alleviates the Replanting Disease under Consecutive Monoculture Regimes by Reshaping Leaf and Root Microbiome. Microb. Ecol. 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Li, Y.; Xu, J.; Yang, J.; Wei, G.; Shen, L.; Ding, W.; Chen, S. Biofertilizers regulate the soil microbial community and enhance Panax ginseng yields. Chin. Med. 2019, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Samuel, A.D.; Bungau, S.; Tit, D.M.; Melinte, C.E.; Purza, L.; Badea, G.E. Effects of long term application of organic and mineral fertilizers on soil enzymes. Rev. Chim. 2018, 69, 2608–2612. [Google Scholar] [CrossRef]
- Ganesh, K.S.; Sundaramoorthy, P.; Nagarajan, M.; Xavier, R.L. Role of organic amendments in sustainable agriculture. In Sustainable Agriculture towards Food Security; Springer: Singapore, 2017; pp. 111–124. [Google Scholar]
- Wang, S.; Tan, Y.; Fan, H.; Ruan, H.; Zheng, A. Responses of soil microarthropods to inorganic and organic fertilizers in a poplar plantation in a coastal area of eastern China. Appl. Soil Ecol. 2015, 89, 69–75. [Google Scholar] [CrossRef]
- Saeid, A.; Chojnacka, K. Fertlizers: Need for new strategies. In Organic Farming; Elsevier: Amsterdam, The Netherlands, 2019; pp. 91–116. [Google Scholar]
- Elouear, Z.; Bouhamed, F.; Boujelben, N.; Bouzid, J. Application of sheep manure and potassium fertilizer to contaminated soil and its effect on zinc, cadmium and lead accumulation by alfalfa plants. Sustain. Environ. Res. 2016, 26, 131–135. [Google Scholar] [CrossRef]
- Usmani, Z.; Kumar, V.; Mritunjay, S.K. Vermicomposting of coal fly ash using epigeic and epi-endogeic earthworm species: Nutrient dynamics and metal remediation. RSC Adv. 2017, 7, 4876–4890. [Google Scholar] [CrossRef]
- Hellequin, E.; Monard, C.; Quaiser, A.; Henriot, M.; Klarzynski, O.; Binet, F. Specific recruitment of soil bacteria and fungi decomposers following a biostimulant application increased crop residues mineralization. PLoS ONE 2018, 13, e0209089. [Google Scholar] [CrossRef]
- De Corato, U. Disease-suppressive compost enhances natural soil suppressiveness against soil-borne plant pathogens: A critical review. Rhizosphere 2020, 13, 100192. [Google Scholar] [CrossRef]
- Testen, A.L.; Miller, S.A. Carbon source and soil origin shape soil microbiomes and tomato soilborne pathogen populations during anaerobic soil disinfestation. Phytobiomes 2018, 2, 138–150. [Google Scholar] [CrossRef]
- Hart, M.M.; Antunes, P.M.; Chaudhary, V.B.; Abbott, L.K. Fungal inoculants in the field: Is the reward greater than the risk? Funct. Ecol. 2018, 32, 126–135. [Google Scholar] [CrossRef]
- Kong, Z.; Hart, M.; Liu, H. Paving the way from the lab to the field: Using synthetic microbial consortia to produce high-quality crops. Front. Plant Sci. 2018, 9, 1467. [Google Scholar] [CrossRef] [PubMed]
- Baas, P.; Bell, C.; Mancini, L.M.; Lee, M.N.; Conant, R.T.; Wallenstein, M.D. Phosphorus mobilizing consortium Mammoth P™ enhances plant growth. PeerJ 2016, 4, e2121. [Google Scholar] [CrossRef] [PubMed]
- Dal Cortivo, C.; Barion, G.; Ferrari, M.; Visioli, G.; Dramis, L.; Panozzo, A.; Vamerali, T. Effects of field inoculation with VAM and bacteria consortia on root growth and nutrients uptake in common wheat. Sustainability 2018, 10, 3286. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Yu, K.; Feussner, K.; De Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.; Feussner, I.; Pieterse, C.M. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef]
- Tsolakidou, M.-D.; Stringlis, I.A.; Fanega-Sleziak, N.; Papageorgiou, S.; Tsalakou, A.; Pantelides, I.S. Rhizosphere-enriched microbes as a pool to design synthetic communities for reproducible beneficial outputs. FEMS Microbiol. Ecol. 2019, 95, fiz138. [Google Scholar] [CrossRef]
- Vitullo, D.; Di Pietro, A.; Romano, A.; Lanzotti, V.; Lima, G. Role of new bacterial surfactins in the antifungal interaction between Bacillus amyloliquefaciens and Fusarium oxysporum. Plant Pathol. 2012, 61, 689–699. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Dubey, S.; Sharma, S. Storage of soil microbiome for application in sustainable agriculture: Prospects and challenges. Environ. Sci. Pollut. Res. 2022, 29, 3171–3183. [Google Scholar] [CrossRef]
- Koskey, G.; Mburu, S.W.; Awino, R.; Njeru, E.M.; Maingi, J.M. Potential use of beneficial microorganisms for soil amelioration, phytopathogen biocontrol, and sustainable crop production in smallholder agroecosystems. Front. Sustain. Food Syst. 2021, 5, 130. [Google Scholar] [CrossRef]
- Azevedo, J.L.; Quecine, M.C. Biodiversity and biotechnological applications of microorganisms associated with tropical plants. In Microbiome in Plant Health and Disease; Springer: Singapore, 2019; pp. 293–313. [Google Scholar]
- Tambo, J.A.; Romney, D.; Mugambi, I.; Mbugua, F.; Bundi, M.; Uzayisenga, B.; Matimelo, M.; Ndhlovu, M. Can plant clinics enhance judicious use of pesticides? Evidence from Rwanda and Zambia. Food Policy 2021, 101, 102073. [Google Scholar] [CrossRef]
- Magnoli, K.; Carranza, C.S.; Aluffi, M.E.; Magnoli, C.E.; Barberis, C.L. Herbicides based on 2, 4-D: Its behavior in agricultural environments and microbial biodegradation aspects. A review. Environ. Sci. Pollut. Res. 2020, 27, 38501–38512. [Google Scholar] [CrossRef] [PubMed]
- Chaín, J.M.; Tubert, E.; Graciano, C.; Castagno, L.N.; Recchi, M.; Pieckenstain, F.L.; Estrella, M.J.; Gudesblat, G.; Amodeo, G.; Baroli, I. Growth promotion and protection from drought in Eucalyptus grandis seedlings inoculated with beneficial bacteria embedded in a superabsorbent polymer. Sci. Rep. 2020, 10, 18221. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Salwan, R.; Al-Ani, L.K.T. Molecular Aspects of Plant Beneficial Microbes in Agriculture; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Kalra, A.; Chandra, M.; Awasthi, A.; Singh, A.K.; Khanuja, S.P.S. Natural compounds enhancing growth and survival of rhizobial inoculants in vermicompost-based formulations. Biol. Fertil. Soils 2010, 46, 521–524. [Google Scholar] [CrossRef]
- Herrmann, L.; Atieno, M.; Brau, L.; Lesueur, D. Microbial quality of commercial inoculants to increase BNF and nutrient use efficiency. In Biological Nitrogen Fixation; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1031–1040. [Google Scholar]
- Young, C.-C.; Shen, F.-T.; Singh, S. Strategies for the Exploration and Development of Biofertilizer. In Bacteria in Agrobiology: Plant Probiotics; Springer: Berlin/Heidelberg, Germany, 2012; pp. 127–139. [Google Scholar]
- Sanap, D.; Satpute, T.; Dudhate, D.; Babar, A.; Nagure, D. Constraints and suggestions in soybean crop under organic and inorganic system. Int. J. Agric. Sci. 2009, 5, 608–610. [Google Scholar]
- Bharti, N.; Sharma, S.K.; Saini, S.; Verma, A.; Nimonkar, Y.; Prakash, O. Microbial plant probiotics: Problems in application and formulation. In Probiotics and Plant Health; Springer: Singapore, 2017; pp. 317–335. [Google Scholar]
- Luna, G.M.; Quero, G.M.; Kokou, F.; Kormas, K. Time to integrate biotechnological approaches into fish gut microbiome research. Curr. Opin. Biotechnol. 2022, 73, 121–127. [Google Scholar] [CrossRef]
- Aguilar, C.N.; Hernández-Almanza, A.; Sabu, A.; Loredo, A.; Sudhakaran, S.; Davila-Medina, D.; Morales-Martínez, T.K.; Sepúlveda-Torre, L.; Sugathan, S.; Teixeira, J.A. Microbial Technology: Advances and Challenges. In Advances in Food Bioproducts and Bioprocessing Technologies; CRC Press: Boca Raton, FL, USA, 2019; pp. 3–24. [Google Scholar]
- Bisht, N.; Chauhan, P.S. Excessive and Disproportionate Use of Chemicals Cause Soil Contamination and Nutritional Stress. In Soil Contamination; IntechOpen: London, UK, 2020. [Google Scholar]
- Arora, S.; Arora, S.; Sahni, D.; Sehgal, M.; Srivastava, D.; Singh, A. Pesticides use and its effect on soil bacteria and fungal populations, microbial biomass carbon and enzymatic activity. Curr. Sci. 2019, 116, 643–649. [Google Scholar] [CrossRef]
- Zhang, L.; Li, X.; Yu, J.; Yao, X. Toward cleaner production: What drives farmers to adopt eco-friendly agricultural production? J. Clean. Prod. 2018, 184, 550–558. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Wang, T.; Li, C.; Wu, Z. Effects of ozone treatment on pesticide residues in food: A review. Int. J. Food Sci. 2019, 54, 301–312. [Google Scholar] [CrossRef]
- Costa, J.A.V.; Freitas, B.C.B.; Cruz, C.G.; Silveira, J.; Morais, M.G. Potential of microalgae as biopesticides to contribute to sustainable agriculture and environmental development. J. Environ. Sci. Health B 2019, 54, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Parte, S.G.; Mohekar, A.D.; Kharat, A.S. Microbial degradation of pesticide: A review. Afr. J. Microbiol. Res. 2017, 11, 992–1012. [Google Scholar]
- Ye, X.; Dong, F.; Lei, X. Microbial resources and ecology-microbial degradation of pesticides. Nat. Resour. Res. 2018, 1, 22–28. [Google Scholar] [CrossRef]
- Liu, X.; Cao, A.; Yan, D.; Ouyang, C.; Wang, Q.; Li, Y. Overview of mechanisms and uses of biopesticides. Int. J. Pest Manag. 2021, 67, 65–72. [Google Scholar] [CrossRef]
- Meena, R.K.; Mishra, P. Bio-pesticides for Agriculture and Environment Sustainability. In Resources Use Efficiency in Agriculture; Springer: Singapore, 2020; pp. 85–107. [Google Scholar]
- Butu, M.; Stef, R.; Grozea, I.; Corneanu, M.; Butnariu, M. Biopesticides: Clean and Viable Technology for Healthy Environment. In Bioremediation and Biotechnology; Springer: Cham, Switzerland, 2020; pp. 107–151. [Google Scholar]
- Jan, S.; Singh, R.; Bhardwaj, R.; Ahmad, P.; Kapoor, D. Plant growth regulators: A sustainable approach to combat pesticide toxicity. 3 Biotech 2020, 10, 466. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, V.; Dhanjal, D.S.; Singh, J. Herbicides and plant growth regulators: Current developments and future challenges. In Natural Bioactive Products in Sustainable Agriculture; Springer: Singapore, 2020; pp. 67–81. [Google Scholar]
- Zhang, W. Global pesticide use: Profile, trend, cost/benefit and more. Proc. Int. Acad. Ecol. Environ. Sci. 2018, 8, 1. [Google Scholar]
- Köberl, M.; Ramadan, E.M.; Adam, M.; Cardinale, M.; Hallmann, J.; Heuer, H.; Smalla, K.; Berg, G. Bacillus and Streptomyces were selected as broad-spectrum antagonists against soilborne pathogens from arid areas in Egypt. FEMS Microbiol. Lett. 2013, 342, 168–178. [Google Scholar] [CrossRef]
- Rasul, M.; Yasmin, S.; Zubair, M.; Mahreen, N.; Yousaf, S.; Arif, M.; Sajid, Z.I.; Mirza, M.S. Phosphate solubilizers as antagonists for bacterial leaf blight with improved rice growth in phosphorus deficit soil. Biol. Control 2019, 136, 103997. [Google Scholar] [CrossRef]
- Nagendran, K.; Karthikeyan, G.; Mohammed Faisal, P.; Kalaiselvi, P.; Raveendran, M.; Prabakar, K.; Raguchander, T. Exploiting endophytic bacteria for the management of sheath blight disease in rice. Biol. Agric. Hortic. 2014, 30, 8–23. [Google Scholar] [CrossRef]
- Bashan, Y. Alternative strategies for controlling plant diseases caused by Pseudomonas syringae. In Pseudomonas syringae Pathovars and Related Pathogens; Springer: Dordrecht, The Netherlands, 1997; pp. 575–583. [Google Scholar]
- Crone, S.; Vives-Flórez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martínez-García, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H. The environmental occurrence of Pseudomonas aeruginosa. Apmis 2020, 128, 220–231. [Google Scholar] [CrossRef]
- Almoneafy, A.A.; Moustafa-Farag, M.; Mohamed, H.I. The Auspicious Role of Plant Growth-Promoting Rhizobacteria in the Sustainable Management of Plant Diseases. In Plant Growth-Promoting Microbes for Sustainable Biotic and Abiotic Stress Management; Springer: Cham, Switzerland, 2021; pp. 251–283. [Google Scholar]
- Kunda, P.; Mukherjee, A.; Dhal, P.K. Bacterial Biological Control Agents for Soilborne Diseases Management in Pulses: Present Status and Future Prospects. In Microbial Mitigation of Stress Response of Food Legumes; CRC Press: Boca Raton, FL, USA, 2020; pp. 231–243. [Google Scholar]
- Välitalo, P.; Kruglova, A.; Mikola, A.; Vahala, R. Toxicological impacts of antibiotics on aquatic micro-organisms: A mini-review. Int. J. Hyg. Environ. Health 2017, 220, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Chandra, N.; Kumar, S. Antibiotics producing soil microorganisms. In Antibiotics and Antibiotics Resistance Genes in Soils; Springer: Cham, Switzerland, 2017; pp. 1–18. [Google Scholar]
- Stockwell, V.; Duffy, B. Use of antibiotics in plant agriculture. Rev. Sci. Et Tech.-Off. Int. Des Epizoot. 2012, 31, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Huygens, J.; Daeseleire, E.; Mahillon, J.; Van Elst, D.; Decrop, J.; Meirlaen, J.; Dewulf, J.; Heyndrickx, M.; Rasschaert, G. Presence of Antibiotic Residues and Antibiotic Resistant Bacteria in Cattle Manure Intended for Fertilization of Agricultural Fields: A One Health Perspective. Antibiotics 2021, 10, 410. [Google Scholar] [CrossRef] [PubMed]
- Bian, C.; Duan, Y.; Xiu, Q.; Wang, J.; Tao, X.; Zhou, M. Mechanism of validamycin A inhibiting DON biosynthesis and synergizing with DMI fungicides against Fusarium graminearum. Mol. Plant Pathol. 2021, 22, 769–785. [Google Scholar] [CrossRef]
- Kochansky, J.; Knox, D.A.; Feldlaufer, M.; Pettis, J.S. Screening alternative antibiotics against oxytetracycline-susceptible and-resistant Paenibacillus larvae. Apidologie 2001, 32, 215–222. [Google Scholar] [CrossRef][Green Version]
- Roggo, C.; van der Meer, J.R. Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol. 2017, 45, 24–33. [Google Scholar] [CrossRef]
- Park, M.; Tsai, S.-L.; Chen, W. Microbial biosensors: Engineered microorganisms as the sensing machinery. Sensors 2013, 13, 5777–5795. [Google Scholar] [CrossRef]
- Daunert, S.; Barrett, G.; Feliciano, J.S.; Shetty, R.S.; Shrestha, S.; Smith-Spencer, W. Genetically engineered whole-cell sensing systems: Coupling biological recognition with reporter genes. Chem. Rev. 2000, 100, 2705–2738. [Google Scholar] [CrossRef]
- Daniel, R.; Almog, R.; Ron, A.; Belkin, S.; Diamand, Y.S. Modeling and measurement of a whole-cell bioluminescent biosensor based on a single photon avalanche diode. Biosens. Bioelectron. 2008, 24, 882–887. [Google Scholar] [CrossRef]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 2003, 6, 206–212. [Google Scholar] [CrossRef]
- Bazin, I.; Seo, H.B.; Suehs, C.M.; Ramuz, M.; De Waard, M.; Gu, M.B. Profiling the biological effects of wastewater samples via bioluminescent bacterial biosensors combined with estrogenic assays. Environ. Sci. Pollut. Res. 2017, 24, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Ooi, L.; Heng, L.Y.; Mori, I.C. A high-throughput oxidative stress biosensor based on Escherichia coli roGFP2 cells immobilized in a k-carrageenan matrix. Sensors 2015, 15, 2354–2368. [Google Scholar] [CrossRef] [PubMed]
- Ikeno, S.; Ogino, C.; Ito, T.; Shimizu, N. Detection of benzene derivatives by recombinant E. coli with Ps promoter and GFP as a reporter protein. Biochem. Eng. J. 2003, 15, 193–197. [Google Scholar] [CrossRef]
- Farré, M.; Gonçalves, C.; Lacorte, S.; Barceló, D.; Alpendurada, M. Pesticide toxicity assessment using an electrochemical biosensor with Pseudomonas putida and a bioluminescence inhibition assay with Vibrio fischeri. Anal. Bioanal. Chem. 2002, 373, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Chinalia, F.; Paton, G.I.; Killham, K. Physiological and toxicological characterization of an engineered whole-cell biosensor. Bioresour. Technol. 2008, 99, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Abadian, P.N.; Kelley, C.P.; Goluch, E.D. Cellular analysis and detection using surface plasmon resonance techniques. Anal. Chem. 2014, 86, 2799–2812. [Google Scholar] [CrossRef]
- Sørensen, S.J.; Burmølle, M.; Hansen, L.H. Making bio-sense of toxicity: New developments in whole-cell biosensors. Curr. Opin. Biotechnol. 2006, 17, 11–16. [Google Scholar] [CrossRef]
- Liu, X.; Germaine, K.J.; Ryan, D.; Dowling, D.N. Whole-cell fluorescent biosensors for bioavailability and biodegradation of polychlorinated biphenyls. Sensors 2010, 10, 1377–1398. [Google Scholar] [CrossRef]
- Cases, I.; De Lorenzo, V. Promoters in the environment: Transcriptional regulation in its natural context. Nat. Rev. Microbiol. 2005, 3, 105–118. [Google Scholar] [CrossRef]
- Rosi, N.L.; Mirkin, C.A. Nanostructures in biodiagnostics. Chem. Rev. 2005, 105, 1547–1562. [Google Scholar] [CrossRef]
- Kaittanis, C.; Santra, S.; Perez, J.M. Emerging nanotechnology-based strategies for the identification of microbial pathogenesis. Adv. Drug Deliv. Rev. 2010, 62, 408–423. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Q.; Luo, K.; Li, L.; Zheng, J. Direct electrochemistry of glucose oxidase immobilized on NdPO4 nanoparticles/chitosan composite film on glassy carbon electrodes and its biosensing application. Bioelectrochemistry 2009, 74, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Guo, S.; Zhou, M.; Liu, L.; Liu, C.; Dong, S. A silk derived carbon fiber mat modified with Au@ Pt urchilike nanoparticles: A new platform as electrochemical microbial biosensor. Biosens. Bioelectron. 2010, 25, 2189–2193. [Google Scholar] [CrossRef] [PubMed]
- Tuncagil, S.; Ozdemir, C.; Demirkol, D.O.; Timur, S.; Toppare, L. Gold nanoparticle modified conducting polymer of 4-(2, 5-di (thiophen-2-yl)-1H-pyrrole-1-l) benzenamine for potential use as a biosensing material. Food Chem. 2011, 127, 1317–1322. [Google Scholar] [CrossRef]
- Maas, M.B.; Maybery, G.H.; Perold, W.J.; Neveling, D.P.; Dicks, L.M. Borosilicate glass fiber-optic biosensor for the detection of Escherichia coli. Curr. Microbiol. 2018, 75, 150–155. [Google Scholar] [CrossRef]
- Eltzov, E.; Pavluchkov, V.; Burstin, M.; Marks, R.S. Creation of a fiber optic based biosensor for air toxicity monitoring. Sens. Actuators B Chem. 2011, 155, 859–867. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, I.; Sehgal, A.; Singh, S.; Sharma, N.; Bhatia, S.; Al-Harrasi, A.; Bungau, S. The dichotomy of nanotechnology as the cutting edge of agriculture: Nano-farming as an asset versus nanotoxicity. Chemosphere 2022, 288, 132533. [Google Scholar] [CrossRef]
- Bhatt, P.; Pathak, V.M.; Joshi, S.; Bisht, T.S.; Singh, K.; Chandra, D. Major metabolites after degradation of xenobiotics and enzymes involved in these pathways. In Smart Bioremediation Technologies; Elsevier: Amsterdam, The Netherlands, 2019; pp. 205–215. [Google Scholar]
- Copley, S.D. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat. Chem. Biol. 2009, 5, 559–566. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques–classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef]
- Villegas, L.B.; Martínez, M.A.; Rodríguez, A.; Amoroso, M.J. Microbial consortia, a viable alternative for cleanup of contaminated soils. In Bioremediation in Latin America; Springer: Cham, Switzerland, 2014; pp. 135–148. [Google Scholar]
- Negi, G.; Gangola, S.; Khati, P.; Kumar, G.; Srivastava, A.; Sharma, A. Differential expression and characterization of cypermethrin-degrading potential proteins in Bacillus thuringiensis strain, SG4. 3 Biotech 2016, 6, 225. [Google Scholar]
- Bhatt, P. Smart Bioremediation Technologies: Microbial Enzymes; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Lin, Z.; Zhang, W.; Pang, S.; Huang, Y.; Mishra, S.; Bhatt, P.; Chen, S. Current approaches to and future perspectives on methomyl degradation in contaminated soil/water environments. Molecules 2020, 25, 738. [Google Scholar] [CrossRef] [PubMed]
- Solomon, K.R.; Baker, D.B.; Richards, R.P.; Dixon, K.R.; Klaine, S.J.; La Point, T.W.; Kendall, R.J.; Weisskopf, C.P.; Giddings, J.M.; Giesy, J.P. Ecological risk assessment of atrazine in North American surface waters. Environ. Toxicol. Chem. Int. J. Res. 1996, 15, 31–76. [Google Scholar] [CrossRef]
- Bhatt, P.; Huang, Y.; Rene, E.R.; Kumar, A.J.; Chen, S. Mechanism of allethrin biodegradation by a newly isolated Sphingomonas trueperi strain CW3 from wastewater sludge. Bioresour. Technol. 2020, 305, 123074. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhan, H.; Bhatt, P.; Chen, S. Paraquat degradation from contaminated environments: Current achievements and perspectives. Front. Microbiol. 2019, 10, 1754. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Zhang, W.; Lin, Z.; Pang, S.; Huang, Y.; Bhatt, P.; Chen, S. Carbofuran toxicity and its microbial degradation in contaminated environments. Chemosphere 2020, 259, 127419. [Google Scholar] [CrossRef]
- Agmas, B.; Adugna, M. Attitudes and practices of farmers with regard to pesticide use in NorthWest Ethiopia. Cogent Environ. Sci. 2020, 6, 1791462. [Google Scholar] [CrossRef]
- Tadesse, A. Increasing Crop Production through Improved Plant Protection; Plant Protection Society of Ethiopia (PPSE): Addis Ababa, Ethiopia, 2008. [Google Scholar]
- Liu, S.; Zheng, Z.; Li, X. Advances in pesticide biosensors: Current status, challenges, and future perspectives. Anal. Bioanal. Chem. 2013, 405, 63–90. [Google Scholar] [CrossRef]
- Negatu, B.; Kromhout, H.; Mekonnen, Y.; Vermeulen, R. Use of chemical pesticides in Ethiopia: A cross-sectional comparative study on knowledge, attitude and practice of farmers and farm workers in three farming systems. Ann. Occup. Hyg. 2016, 60, 551–566. [Google Scholar] [CrossRef]
- Asghar, U.; Malik, M.; Javed, A. Pesticide exposure and human health: A review. J. Ecosys. Ecograph. S 2016, 5, 2. [Google Scholar]
- Rose, M.T.; Cavagnaro, T.R.; Scanlan, C.A.; Rose, T.J.; Vancov, T.; Kimber, S.; Kennedy, I.R.; Kookana, R.S.; Van Zwieten, L. Impact of herbicides on soil biology and function. Adv. Agron. 2016, 136, 133–220. [Google Scholar]
- Mulchandani, A. Microbial biosensors for organophosphate pesticides. Appl. Biochem. Biotechnol. 2011, 165, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Upadhyay, N.; Kumar, V.; Sharma, S. A review on sample preparation and chromatographic determination of acephate and methamidophos in different samples. Arab. J. Chem. 2015, 8, 624–631. [Google Scholar] [CrossRef]
- Sporring, S.; Bøwadt, S.; Svensmark, B.; Björklund, E. Comprehensive comparison of classic Soxhlet extraction with Soxtec extraction, ultrasonication extraction, supercritical fluid extraction, microwave assisted extraction and accelerated solvent extraction for the determination of polychlorinated biphenyls in soil. J. Chromatogr. A 2005, 1090, 1–9. [Google Scholar] [PubMed]
- Balootaki, P.A.; Hassanshahian, M. Microbial biosensor for marine environments. Bull. Environ. Pharmacol. Life Sci. 2014, 3, 1–13. [Google Scholar]
- Adetunji, C.O.; Nwankwo, W.; Ukhurebor, K.E.; Olayinka, A.S.; Makinde, A.S. Application of Biosensor for the Identification of Various Pathogens and Pests Mitigating Against the Agricultural Production: Recent Advances. In Biosensors in Agriculture: Recent Trends and Future Perspectives; Springer: Cham, Switzerland, 2021; pp. 169–189. [Google Scholar]
- Malhotra, B.D.; Chaubey, A.; Singh, S. Prospects of conducting polymers in biosensors. Anal. Chim. Acta 2006, 578, 59–74. [Google Scholar] [CrossRef]
- Karim, F.; Fakhruddin, A. Recent advances in the development of biosensor for phenol: A review. Rev. Environ. Sci. Biotechnol. 2012, 11, 261–274. [Google Scholar] [CrossRef]
- Lindemann, S.R.; Bernstein, H.C.; Song, H.-S.; Fredrickson, J.K.; Fields, M.W.; Shou, W.; Johnson, D.R.; Beliaev, A.S. Engineering microbial consortia for controllable outputs. ISME J. 2016, 10, 2077–2084. [Google Scholar] [CrossRef]
- Bhatt, P.; Zhou, X.; Huang, Y.; Zhang, W.; Chen, S. Characterization of the role of esterases in the biodegradation of organophosphate, carbamate, and pyrethroid pesticides. J. Hazard. Mater. 2021, 411, 125026. [Google Scholar] [CrossRef]
- Chakraborty, R.; Wu, C.H.; Hazen, T.C. Systems biology approach to bioremediation. Curr. Opin. Biotechnol. 2012, 23, 483–490. [Google Scholar] [CrossRef]
- Jariyal, M.; Jindal, V.; Mandal, K.; Gupta, V.K.; Singh, B. Bioremediation of organophosphorus pesticide phorate in soil by microbial consortia. Ecotoxicol. Environ. Saf. 2018, 159, 310–316. [Google Scholar] [CrossRef]
- Irving, S.E.; Choudhury, N.R.; Corrigan, R.M. The stringent response and physiological roles of (pp) pGpp. in bacteria. Nat. Rev. Microbiol. 2021, 19, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Pang, S.; Zhang, W.; Lin, Z.; Bhatt, P.; Chen, S. Insights into the microbial degradation and biochemical mechanisms of carbamates. Chemosphere 2021, 279, 130500. [Google Scholar] [CrossRef] [PubMed]
- LaPara, T.M.; Zakharova, T.; Nakatsu, C.H.; Konopka, A. Functional and structural adaptations of bacterial communities growing on particulate substrates under stringent nutrient limitation. Microb. Ecol. 2002, 44, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Arenas, F.; Sánchez, I.; Hawkins, S.J.; Jenkins, S.R. The invasibility of marine algal assemblages: Role of functional diversity and identity. Ecology 2006, 87, 2851–2861. [Google Scholar] [CrossRef]
- Briones, A.; Raskin, L. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr. Opin. Biotechnol. 2003, 14, 270–276. [Google Scholar] [CrossRef]
- Gangola, S.; Sharma, A.; Bhatt, P.; Khati, P.; Chaudhary, P. Presence of esterase and laccase in Bacillus subtilis facilitates biodegradation and detoxification of cypermethrin. Sci. Rep. 2018, 8, 12755. [Google Scholar] [CrossRef]
- Adebami, G.E.; Kuila, A.; Ajunwa, O.M.; Fasiku, S.A.; Asemoloye, M.D. Genetics and metabolic engineering of yeast strains for efficient ethanol production. J. Food Process Eng. 2021, e13798. [Google Scholar] [CrossRef]
- Lyon, G.J.; Novick, R.P. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 2004, 25, 1389–1403. [Google Scholar] [CrossRef]
- Slamti, L.; Lereclus, D. The oligopeptide ABC-importers are essential communication channels in Gram-positive bacteria. Res. Microbiol. 2019, 170, 338–344. [Google Scholar] [CrossRef]
- Gao, B.; Sabnis, R.; Costantini, T.; Jinkerson, R.; Sun, Q. A peek in the micro-sized world: A review of design principles, engineering tools, and applications of engineered microbial community. Biochem. Soc. Trans. 2020, 48, 399–409. [Google Scholar] [CrossRef]
- Qian, X.; Chen, L.; Sui, Y.; Chen, C.; Zhang, W.; Zhou, J.; Dong, W.; Jiang, M.; Xin, F.; Ochsenreither, K. Biotechnological potential and applications of microbial consortia. Biotechnol. Adv. 2020, 40, 107500. [Google Scholar] [CrossRef] [PubMed]
- Brenner, K.; Karig, D.K.; Weiss, R.; Arnold, F.H. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc. Natl. Acad. Sci. USA 2007, 104, 17300–17304. [Google Scholar] [CrossRef]
- Bhatt, P.; Bhatt, K.; Sharma, A.; Zhang, W.; Mishra, S.; Chen, S. Biotechnological basis of microbial consortia for the removal of pesticides from the environment. Crit. Rev. Biotechnol. 2021, 41, 317–338. [Google Scholar] [CrossRef] [PubMed]
- Hawley, A.K.; Brewer, H.M.; Norbeck, A.D.; Paša-Tolić, L.; Hallam, S.J. Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc. Natl. Acad. Sci. USA 2014, 111, 11395–11400. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, C.; Kalhoro, M.T.; Faqir, Y.; Ma, J.; Osei, M.D.; Khaliq, G. Climate-Resilient Microbial Biotechnology: A Perspective on Sustainable Agriculture. Sustainability 2022, 14, 5574. https://doi.org/10.3390/su14095574
Tan C, Kalhoro MT, Faqir Y, Ma J, Osei MD, Khaliq G. Climate-Resilient Microbial Biotechnology: A Perspective on Sustainable Agriculture. Sustainability. 2022; 14(9):5574. https://doi.org/10.3390/su14095574
Chicago/Turabian StyleTan, Chengjia, Mohammad Talib Kalhoro, Yahya Faqir, Jiahua Ma, Matthew Duah Osei, and Ghulam Khaliq. 2022. "Climate-Resilient Microbial Biotechnology: A Perspective on Sustainable Agriculture" Sustainability 14, no. 9: 5574. https://doi.org/10.3390/su14095574
APA StyleTan, C., Kalhoro, M. T., Faqir, Y., Ma, J., Osei, M. D., & Khaliq, G. (2022). Climate-Resilient Microbial Biotechnology: A Perspective on Sustainable Agriculture. Sustainability, 14(9), 5574. https://doi.org/10.3390/su14095574








