DNA Barcoding Revealing the Parrotfish (Perciformes: Scaridae) Diversity of the Coral Reef Ecosystem of the South China Sea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Morphological Identification
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Molecular Data Analysis
3. Results
3.1. Morphology-Based Species Identification
3.2. DNA Barcodes Identification
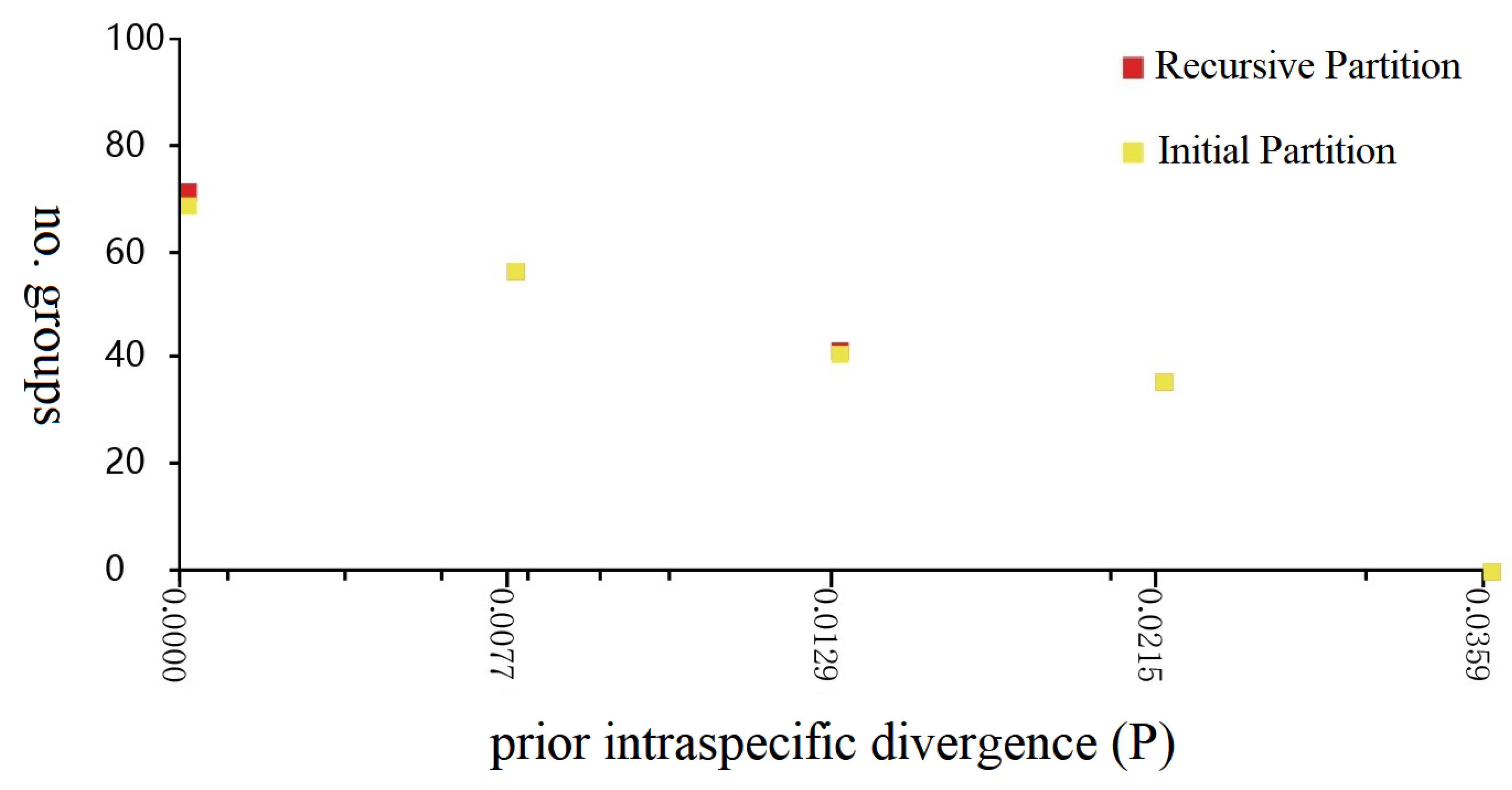
3.3. ABGD Analyses, BIN Analyses and Phylogenetic Tree Delimitation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilkinson, C.R. Status of Coral Reefs of the World: 2008; Australian Institute of Marine Science: Townsville, Australia, 2008. [Google Scholar]
- Hong, X.; Guoning, S.; Baolin, L.; Gang, C.; Shuyi, D.; Longwei, Q.; Qi, L.; Jinxiong, L.; Jian, S.; Xiaoqi, Z. Coral-coral reefs in China seas: The biodiversity characteristics of coral-coral reefs in the central South China Sea. J. Palaeogeogr. Chin. Ed. 2021, 23, 771–788. [Google Scholar]
- Hughes, T.P.; Huang, H.U.I.; Young, M.A.L. The wicked problem of China’s disappearing coral reefs. Conserv. Biol. 2013, 27, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Z.; Shi, Y.R.; Ai, H.; Dong, L.N.; Li, N.N.; Li, X.; Gao, T.X. Large scale distribution patterns of taxonomic diversity of fish in coral reef waters, South China Sea. J. Fish. Sci. China 2011, 18, 619–628. [Google Scholar] [CrossRef]
- Wang, Z.M.; Kong, X.Y.; Huang, L.M. Taxonomic revision of parrotfishes (Perciformes: Scaridae) in China seas. J. Trop. Oceanogr. 2013, 32, 22–32. [Google Scholar]
- Quan, Q.M.; Wang, T.; Liu, Y.; Guo, J.T.; Xie, Y.F.; Li, C.R.; Li, C.H. The community structure of parrotfish in typical islands and reefs of the South China Sea. Chin. J. Ecol. 2021, 40, 2133–2145. [Google Scholar]
- Mumby, P.J. Herbivory versus corallivory: Are parrotfish good or bad for Caribbean coral reefs? Coral Reefs 2009, 28, 683–690. [Google Scholar] [CrossRef]
- Rotjan, R.D.; Lewis, S.M. Parrotfish abundance and selective corallivory on a Belizean coral reef. J. Exp. Mar. Biol. Ecol. 2006, 335, 292–301. [Google Scholar] [CrossRef]
- Sánchez, J.A.; Gil, M.F.; Chasqui, L.H.; Alvarado, E.M. Grazing dynamics on a Caribbean reef-building coral. Coral Reefs 2004, 23, 578–583. [Google Scholar]
- Bruckner, A.W.; Bruckner, R.J.; Sollins, P. Parrotfish predation on live coral: “spot biting” and “focused biting”. Coral Reefs 2000, 19, 50. [Google Scholar] [CrossRef]
- Mumby, P.J. The impact of exploiting grazers (Scaridae) on the dynamics of Caribbean coral reefs. Ecol. Appl. 2006, 16, 747–769. [Google Scholar] [CrossRef]
- Gromova, E.S.; Makhotin, V.V. Morphofunctional Features of the Visceral Apparatus in Ember Parrotfish Scarus rubroviolaceus (Scaridae). J. Ichthyol. 2020, 60, 204–229. [Google Scholar] [CrossRef]
- Roos, N.C.; Longo, G.O.; Pennino, M.G.; Francini-Filho, R.B.; Carvalho, A.R. Protecting nursery areas without fisheries management is not enough to conserve the most endangered parrotfish of the Atlantic Ocean. Sci. Rep. 2020, 10, 19143. [Google Scholar] [CrossRef] [PubMed]
- Brock, R.E. An experimental study on the effects of grazing by parrotfishes and role of refuges in benthic community structure. Mar. Biol. 1979, 51, 381–388. [Google Scholar] [CrossRef]
- Szmant, A.M. Nutrient enrichment on coral reefs: Is it a major cause of coral reef decline? Estuaries 2002, 25, 743–766. [Google Scholar] [CrossRef]
- Hou, G.; Chen, W.T.; Lu, H.S.; Cheng, F.; Xie, S.G. Developing a DNA barcode library for perciform fishes in the South China Sea: Species identification, accuracy and cryptic diversity. Mol. Ecol. Resour. 2018, 18, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Choat, J.H.; Robertson, D.R. Protogynous Hermaphroditism in Fishes of the Family Scaridae; Springer: Berlin/Heidelberg, Germany, 1975; pp. 263–283. [Google Scholar]
- Shen, Y.; Hubert, N.; Yan, H.; Wang, X.; He, S. DNA barcoding the ichthyofauna of the Yangtze River: Insights from the molecular inventory of a mega-diverse temperate fauna. Mol. Ecol. Resour. 2018, 19, 1278–1291. [Google Scholar] [CrossRef]
- Ferri, G.; Alu, M.; Corradini, B.; Licata, M.; Beduschi, G. Species Identification Through DNA ‘Barcodes’. Genet. Test. 2009, 13, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. B 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Hebert, P.D.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Chase, M.W.; Salamin, N.; Wilkinson, M.; Dunwell, J.M.; Kesanakurthi, R.P.; Haidar, N.; Savolainen, V. Land plants and DNA barcodes: Short-term and long-term goals. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1889–1895. [Google Scholar] [CrossRef]
- Mir, R.A.; Bhat, K.A.; Rashid, G.; Ebinezer, L.B.; Masi, A.; Rakwal, R.; Shah, A.; Zargar, S.M. DNA barcoding: A way forward to obtain deep insights about the realistic diversity of living organisms. Nucleus 2021, 64, 157–165. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Shadrin, D. DNA Barcoding: Applications. Russ. J. Genet. 2021, 57, 489–497. [Google Scholar] [CrossRef]
- Yancy, H.F.; Zemlak, T.S.; Mason, J.A.; Washington, J.D.; Tenge, B.J.; Nguyen, N.L.T.; Barnett, J.D.; Savary, W.E.; Hill, W.E.; Moore, M.M. Potential use of DNA barcodes in regulatory science: Applications of the Regulatory Fish Encyclopedia. J. Food Prot. 2008, 71, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.S. Fishes of South China Sea; Science Press: Beijing, China, 1962. [Google Scholar]
- Yang, J.J. The Fishes of the Islands in the South China Sea; Science Press: Beijing, China, 1979. [Google Scholar]
- Ward, R.; Zemlak, T.; Innes, B.; Last, P.; Hebert, P. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Shen, Y.J.; Guan, L.H.; Wang, D.Q.; Gan, X.N. DNA barcoding and evaluation of genetic diversity in Cyprinidae fish in the midstream of the Yangtze River. Ecol. Evol. 2016, 6, 2702–2713. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1. 4.0. A Graphical Viewer of Phylogenetic Trees; University of Edinburgh: Edinburgh, UK, 2012. [Google Scholar]
- Meyer, C.P.; Paulay, G. DNA Barcoding: Error Rates Based on Comprehensive Sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef]
- Landi, M.; Dimech, M.; Arculeo, M.; Biondo, G.; Martins, R.; Carneiro, M.; Carvalho, G.R.; Brutto, S.L.; Costa, F.O. DNA barcoding for species assignment: The case of Mediterranean marine fishes. PLoS ONE 2014, 9, e106135. [Google Scholar] [CrossRef] [PubMed]
- Weigt, L.A.; Baldwin, C.C.; Driskell, A.; Smith, D.G.; Ormos, A.; Reyier, E.A. Using DNA barcoding to assess Caribbean reef fish biodiversity: Expanding taxonomic and geographic coverage. PLoS ONE 2012, 7, e41059. [Google Scholar] [CrossRef] [PubMed]
- Walczyńska, K.S.; Mańko, M.K.; Weydmann, A. Arctic Ocean biodiversity and DNA barcoding—A climate change perspective. In YOUMARES 8–Oceans across Boundaries: Learning from Each Other; Springer: Cham, Switzerland, 2018; pp. 145–153. [Google Scholar]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M.; Godfray, C. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, Z.; Liu, M.; Liu, W.; Zhao, W.; Liu, H.; You, F. DNA barcoding of marine fish species from Rongcheng Bay, China. PeerJ 2018, 6, e5013. [Google Scholar] [CrossRef]
- Rock, J.; Costa, F.O.; Walker, D.I.; North, A.W.; Hutchinson, W.F.; Carvalho, G.R. DNA barcodes of fish of the Scotia Sea, Antarctica indicate priority groups for taxonomic and systematics focus. Antarct. Sci. 2008, 20, 253–262. [Google Scholar] [CrossRef]
- Neigel, J.; Domingo, A.; Stake, J. DNA barcoding as a tool for coral reef conservation. Coral Reefs 2007, 26, 487. [Google Scholar] [CrossRef]
- Collins, R.A.; Cruickshank, R.H. The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 2013, 13, 969–975. [Google Scholar] [CrossRef]
- Čandek, K.; Kuntner, M. DNA barcoding gap: Reliable species identification over morphological and geographical scales. Mol. Ecol. Resour. 2015, 15, 268–277. [Google Scholar] [CrossRef]
- Bay, L.K.; Choat, J.H.; Herwerden, L.; Robertson, D.R. High genetic diversities and complex genetic structure in an Indo-Pacific tropical reef fish (Chlorurus sordidus): Evidence of an unstable evolutionary past? Mar. Biol. 2004, 144, 757–767. [Google Scholar] [CrossRef]
- Winters, K.L.; Herwerden, L.V.; Choat, J.H.; Robertson, D.R. Phylogeography of the Indo-Pacific parrotfish Scarus psittacus: Isolation generates distinctive peripheral populations in two oceans. Mar. Biol. 2010, 157, 1679–1691. [Google Scholar] [CrossRef]
- Bezerra, I.M.; Gramacho, K.P.; Barreto, M.A.; Hackradt, C.W.; Leão Feitosa, J.L.; Torres, R.A.; Ferreira, B.P.; González-Wanguemert, M.; Félix-Hackradt, F.C. Genetic diversity and gene flow of the threatened Brazilian endemic parrotfish Scarus trispinosus (Valenciennes, 1840). Mar. Environ. Res. 2018, 142, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Visram, S.; Yang, M.C.; Pillay, R.M.; Said, S.; Henriksson, O.; Grahn, M.; Chen, C.A. Genetic connectivity and historical demography of the blue barred parrotfish (Scarus ghobban) in the western Indian Ocean. Mar. Biol. 2010, 157, 1475–1487. [Google Scholar] [CrossRef]
- Green, A.L.; Maypa, A.P.; Almany, G.R.; Rhodes, K.L.; Weeks, W.; Abesamis, R.A. Larval dispersal and movement patterns of coral reef fishes, and implications for marine reserve network design. Biol. Rev. 2015, 90, 1215–1247. [Google Scholar] [CrossRef]
- Gong, Y.Y.; Zhang, Q.; Cao, Y.; Lv, J.L.; Yang, X.S. Application of Mitochondrial Cytochrome Oxidase Subunit I sequence in DNA barcoding Upeneus in the South China Sea. Mar. Fish. 2016, 38, 113–119. [Google Scholar]
- Bostrom, M.A.; Collette, B.B.; Luckhurst, B.E.; Reece, K.S.; Graves, J.E. Hybridization between two serranids, the coney (Cephalopholis fulva) and the creole-fish (Paranthias furcifer), at Bermuda. Fish. Bull. 2002, 100, 651–661. [Google Scholar]
- Hobbs, J.-P.A.; Frisch, A.J.; Allen, G.R.; Van Herwerden, L. Marine hybrid hotspot at Indo-Pacific biogeographic border. Biol. Lett. 2009, 5, 258–261. [Google Scholar] [CrossRef]
- DiBattista, J.D.; Whitney, J.; Craig, M.T.; Hobbs, J.-P.A.; Rocha, L.A.; Feldheim, K.A.; Berumen, M.L.; Bowen, B.W. Surgeons and suture zones: Hybridization among four surgeonfish species in the Indo-Pacific with variable evolutionary outcomes. Mol. Phylogenetics Evol. 2016, 101, 203–215. [Google Scholar] [CrossRef]
- Carlon, D.B.; Robertson, D.R.; Barron, R.L.; Choat, J.H.; Anderson, D.J.; Schwartz, S.A.; Sánchez-Ortiz, C.A. The origin of the parrotfish species Scarus compressus in the Tropical Eastern Pacific: Region-wide hybridization between ancient species pairs. BMC Ecol. Evol. 2021, 21, 7. [Google Scholar] [CrossRef]
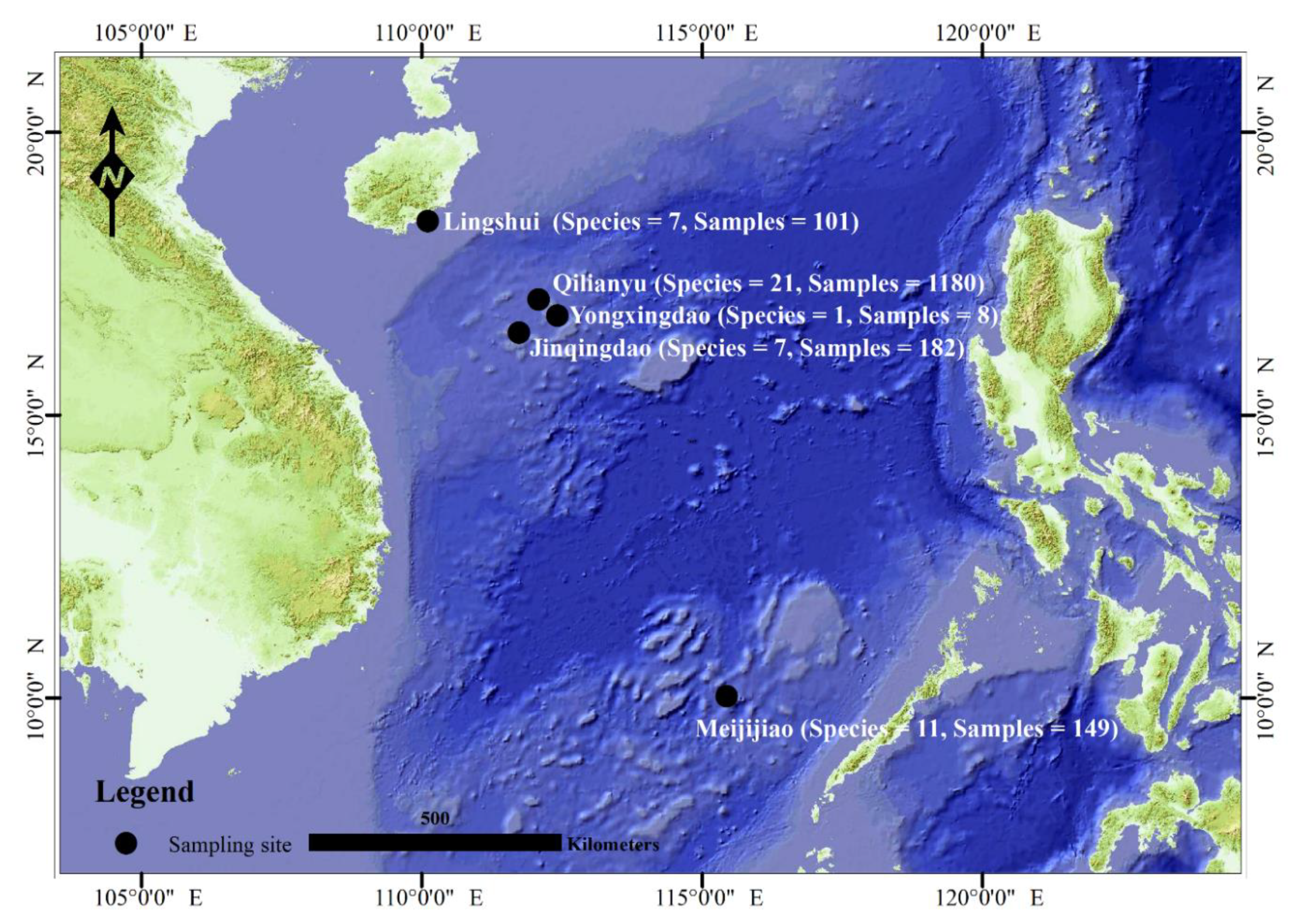


| Genus | Species | Size | Sample Site and Size |
|---|---|---|---|
| Bolbometopon | B. muricatum | 5 | Qilianyu (5) |
| Calotomus | C. carolinus | 41 | Qilianyu (38); Meijijiao (3) |
| Cetoscarus | C. bicolor | 4 | Qilianyu (4) |
| Chlorurus | C. bleekeri | 1 | Meijijiao (1) |
| C. japanensis | 5 | Qilianyu (1); Meijijiao (1); Lingshui (3) | |
| C. microrhinos | 22 | Qilianyu (21); Meijijiao (1) | |
| C. sordidus | 362 | Qilianyu (271); Meijijiao (43); Jinqingdao (48) | |
| Hipposcarus | H. longiceps | 135 | Qilianyu (47); Meijijiao (12); Jinqingdao (75); Lingshui (1) |
| Scarus | S. altipinnis | 1 | Lingshui (1) |
| S. chameleon | 31 | Qilianyu (6); Meijijiao (17); Yongxingdao (8) | |
| S. dimidiatus | 59 | Qilianyu (48); Meijijiao (11) | |
| S. forsteni | 276 | Qilianyu (224); Meijijiao (35); Jinqingdao (14); Lingshui (3) | |
| S. frenatus | 6 | Qilianyu (6) | |
| S. ghobban | 93 | Qilianyu (15); Jinqingdao (2); Lingshui (76) | |
| S. globiceps | 78 | Qilianyu (78) | |
| S. niger | 64 | Qilianyu (59); Meijijiao (4); Lingshui (1) | |
| S. oviceps | 196 | Qilianyu (157); Jinqingdao (39) | |
| S. prasiognathos | 9 | Qilianyu (9) | |
| S. psittacus | 12 | Qilianyu (12) | |
| S. rivulatus | 20 | Qilianyu (20) | |
| S. rubroviolaceus | 32 | Qilianyu (13); Jinqingdao (3); Lingshui (16) | |
| S. schlegeli | 158 | Qilianyu (136); Jinqingdao (1); Meijijiao (21) | |
| S. spinus | 10 | Qilianyu (10) |
| Level | N | Comparisons | Minimum | Mean | Maximum | SE |
|---|---|---|---|---|---|---|
| Within species | 1618 | 157,247 | 0.00 | 0.23 | 12.08 | 0.00 |
| Within genus | 1435 | 473,979 | 0.32 | 8.52 | 21.60 | 0.00 |
| Within family | 1620 | 680,164 | 6.30 | 13.89 | 29.48 | 0.00 |
| Species | Mean Intra-Species (%) | Max Intra-Species (%) | Nearest Species (%) | Distance to NN (%) |
|---|---|---|---|---|
| B. muricatum | 0.1 | 0.16 | C. bicolor | 14.75 |
| C. carolinus | 0.06 | 0.32 | C. bicolor | 16.15 |
| C. bicolor | 0.27 | 0.49 | S. altipinnis | 13.29 |
| C. bleekeri | N/A | 0 | C. microrhinos | 3.16 |
| C. japanensis | 0.86 | 2.15 | C. microrhinos | 6.52 |
| C. microrhinos | 1.3 | 11.92 | C. bleekeri | 3.16 |
| C. sordidus | 0.25 | 9.81 | C. microrhinos | 5.97 |
| H. longiceps | 0.32 | 1.31 | C. sordidus | 14.78 |
| S. altipinnis | N/A | 0 | S. forsteni | 3.34 |
| S. chameleon | 0.08 | 0.49 | S. globiceps | 7.08 |
| S. dimidiatus | 0.11 | 0.65 | S. oviceps | 3.15 |
| S. forsteni | 0.19 | 6.26 | S. niger | 2.48 |
| S. frenatus | 0 | 0 | S. oviceps | 4.01 |
| S. ghobban | 0.18 | 0.98 | S. rubroviolaceus | 3.68 |
| S. globiceps | 0.34 | 6.44 | S. rivulatus | 0.32 |
| S. niger | 0.2 | 1.81 | S. forsteni | 2.48 |
| S. oviceps | 0.09 | 1.47 | S. forsteni | 2.65 |
| S. prasiognathos | 0.33 | 0.66 | S. forsteni | 3.68 |
| S. psittacus | 0.38 | 1.14 | S. schlegeli | 8.35 |
| S. rivulatus | 0.13 | 0.65 | S. globiceps | 0.32 |
| S. rubroviolaceus | 0.27 | 0.98 | S. ghobban | 3.68 |
| S. schlegeli | 0.15 | 5.78 | S. globiceps | 5.58 |
| S. spinus | 0.23 | 0.65 | S. globiceps | 9.55 |
| OTU Information | Dist. (%). Max. Intra. | Dist. (%). Near. Neigh. |
|---|---|---|
| Chlorurus japanensis | 2.15 | 6.52 |
| OTU4 (BIN: ACK7947) | 0.16 | 6.69 |
| OTU5 (BIN: AAE8961) | - | 6.52 |
| Chlorurus microrhinos | 11.92 | 3.16 |
| OTU7 (BIN: AAJ5287) | 9.81 | 3.16 |
| OTU8 (BIN: AAD0850) | - | 10.99 |
| Chlorurus sordidus | 9.81 | 5.97 |
| OTU9 (BIN: AAB6670) | 5.97 | |
| OTU10 (BIN: AEL9035) | - | 14.56 |
| OTU11 (BIN: AEM3067) | - | 13.32 |
| OTU12 (BIN: AAB6670) | - | 8.37 |
| Scarus forsteni | 6.26 | 2.48 |
| OTU18 (BIN: AAE4369) | 1.16 | 2.48 |
| OTU19 (BIN: AEL8860) | 1.31 | 3.32 |
| OTU20 (BIN: AAE4369) | - | 3.60 |
| OTU21 (BIN: AEL8858) | - | 4.37 |
| OTU22 (BIN: AEM1502) | - | 6.44 |
| OTU23 (BIN: AEL8859) | - | 4.36 |
| Scarus globiceps | 6.44 | 0.32 |
| OTU14 (BIN: ADB4663) | 1.97 | 0.32 |
| OTU26 (BIN: ADB4663) | - | 3.31 |
| OTU27 (BIN: AEM1397) | - | 4.37 |
| Scarus schlegeli | 5.78 | 5.58 |
| OTU33 (BIN: ACF2863) | 0.98 | 7.73 |
| OTU34 (BIN: AEL7735) | - | 5.83 |
| OUT35 (BIN: ACF2863) | - | 11.74 |
| OTU36 (BIN: ACF2863) | - | 9.67 |
| OTU Information | Dist. (%). Max. Intra. | Dist. (%). Near. Neigh. | Species |
|---|---|---|---|
| OTU14 | 3.81 | 0.81 | Scarus globiceps Scarus rivulatus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Y.; Li, C.; Wang, T.; Lin, L.; Guo, J.; Quan, Q.; Liu, Y. DNA Barcoding Revealing the Parrotfish (Perciformes: Scaridae) Diversity of the Coral Reef Ecosystem of the South China Sea. Sustainability 2022, 14, 15386. https://doi.org/10.3390/su142215386
Xiao Y, Li C, Wang T, Lin L, Guo J, Quan Q, Liu Y. DNA Barcoding Revealing the Parrotfish (Perciformes: Scaridae) Diversity of the Coral Reef Ecosystem of the South China Sea. Sustainability. 2022; 14(22):15386. https://doi.org/10.3390/su142215386
Chicago/Turabian StyleXiao, Yayuan, Chunhou Li, Teng Wang, Lin Lin, Jiatong Guo, Qiumei Quan, and Yong Liu. 2022. "DNA Barcoding Revealing the Parrotfish (Perciformes: Scaridae) Diversity of the Coral Reef Ecosystem of the South China Sea" Sustainability 14, no. 22: 15386. https://doi.org/10.3390/su142215386
APA StyleXiao, Y., Li, C., Wang, T., Lin, L., Guo, J., Quan, Q., & Liu, Y. (2022). DNA Barcoding Revealing the Parrotfish (Perciformes: Scaridae) Diversity of the Coral Reef Ecosystem of the South China Sea. Sustainability, 14(22), 15386. https://doi.org/10.3390/su142215386






