Selective Isolation of Bioactive-Pigmented Bacteria from Saline Agricultural Soil and Assessment of Their Antimicrobial Potential against Plant Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Bioactive Pigment-Producing Bacteria
Selective Isolation of Bioactive Pigment-Producing Bacteria
2.3. Screening Isolates for Antimicrobial Activity
2.3.1. Phytopathogenic Microorganisms Tested
2.3.2. Screening Bioactive Pigment-Producing Bacteria for Antifungal Activity
2.3.3. Screening Bioactive Pigment-Producing Bacteria for Antibacterial Activity
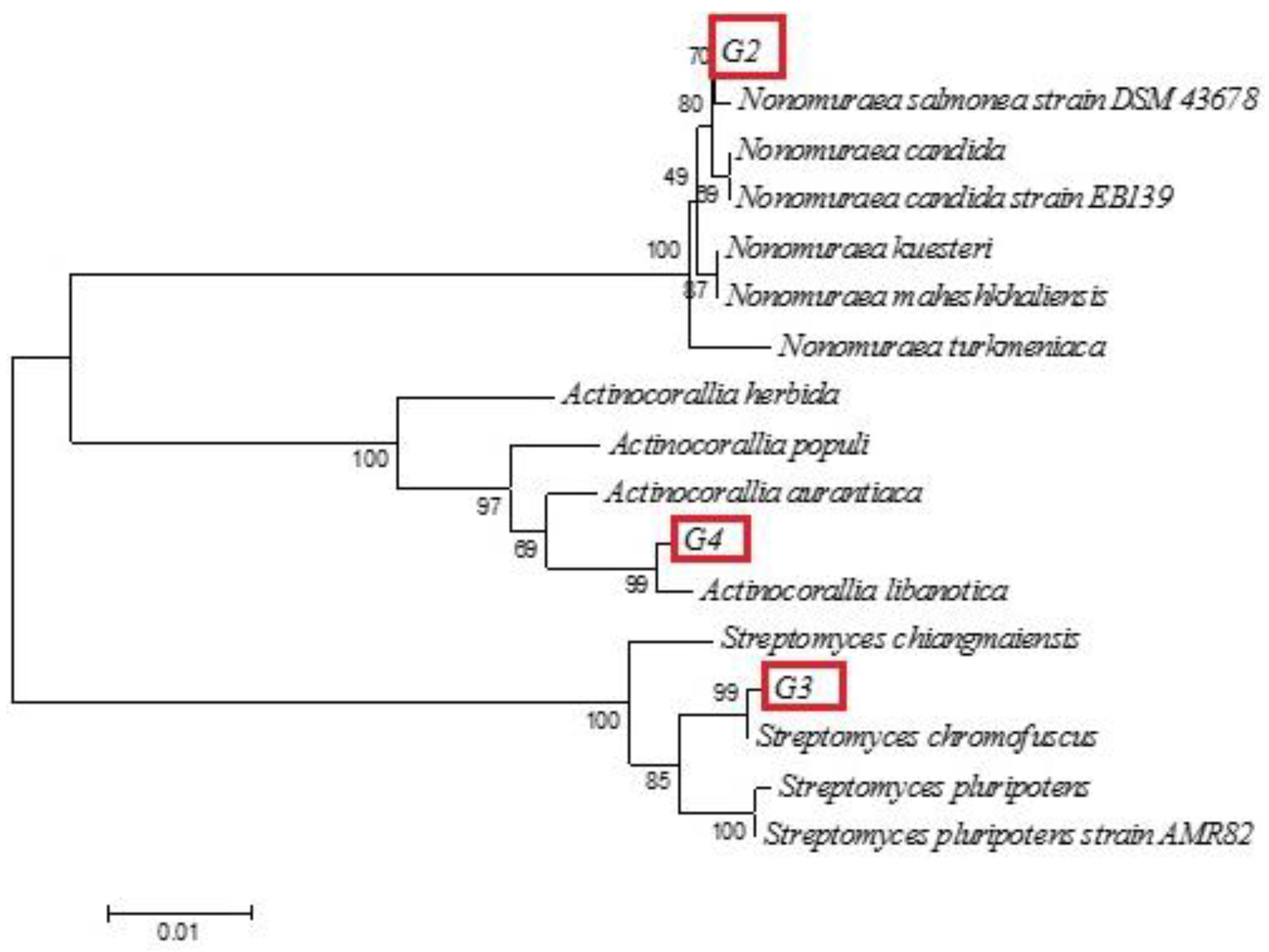
2.4. Strain Identification
2.5. Crude Bioactive Pigment Extract Preparation
2.6. Antifungal Activity of Crude Bioactive Pigment Extract
Determination of Minimum Inhibitory Concentration (MIC) against Fungal Phytopathogens
2.7. Antifungal Activity on Mycelial Radial Growth (MRG)
2.8. Antibacterial Activity of Crude Bioactive Pigment Extract against Bacterial Phytopathogens
Determination of MIC of Crude Bioactive Pigment Extract against Phytopathogenic Bacteria
2.9. Statistical Analysis
3. Results
3.1. Isolation and Identification of Bioactive Pigment-Producing Bacteria
3.2. Screening Bioactive Pigment-Producing Bacteria for Antimicrobial Activity
3.3. Antifungal Activity of Crude Bioactive Pigment Extract and Determination of MIC
3.4. Antifungal Activity on Mycelial Radial Growth (MRG)
3.5. Antibacterial Activity and MIC Determination
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ajmal, A.W.; Saroosh, S.; Mulk, S.; Hassan, M.N.; Yasmin, H.; Jabeen, Z.; Nosheen, A.; Shah, S.M.U.; Naz, R.; Hasnain, Z. Bacteria isolated from wastewater irrigated agricultural soils adapt to heavy metal toxicity while maintaining their plant growth promoting traits. Sustainability 2021, 13, 7792. [Google Scholar] [CrossRef]
- Zhou, L.; Song, C.; Li, Z.; Kuipers, O.P. Antimicrobial activity screening of rhizosphere soil bacteria from tomato and genome-based analysis of their antimicrobial biosynthetic potential. BMC Genom. 2021, 22, 29. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Radwan, M.M.; Taráwneh, A.H.; Gao, J.; Wedge, D.E.; Rosa, L.H.; Cutler, H.G.; Cutler, S.J. Antifungal activity against plant pathogens of metabolites from the endophytic fungus Cladosporium cladosporioides. J. Agric. Food Chem. 2013, 61, 4551–4555. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Dessaux, Y.; Llamas, I. Saline environments as a source of potential quorum sensing disruptors to control bacterial infections: A review. Mar. Drugs 2019, 17, 191. [Google Scholar] [CrossRef]
- Rashid, M.; Fakruddin, M.; Mazumdar, R.M.; Kaniz, F.; Chowdhury, M. Anti-Bacterial Activity of Pigments Isolated From Pigment-Forming Soil Bacteria. J. Pharm. Res. Int. 2014, 4, 880–894. [Google Scholar] [CrossRef]
- Ramesh, C.; Vinithkumar, N.V.; Kirubagaran, R.; Venil, C.K.; Dufossé, L. Multifaceted applications of microbial pigments: Current knowledge, challenges and future directions for public health implications. Microorganisms 2019, 7, 186. [Google Scholar] [CrossRef] [PubMed]
- Sekurova, O.N.; Schneider, O.; Zotchev, S.B. Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering. Microb. Biotechnol. 2019, 12, 828–844. [Google Scholar] [CrossRef]
- Venil, C.K.; Dufossé, L.; Renuka Devi, P. Bacterial Pigments: Sustainable Compounds With Market Potential for Pharma and Food Industry. Front. Sustain. Food Syst. 2020, 4, 1–17. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, R.; Bashir, S.; Numan, M.; Shinwari, Z.K.; Ali, M. Pigments from Soil Bacteria and Their Therapeutic Properties: A Mini Review. Curr. Microbiol. 2018, 76, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.; Chatterjee, S.; Banerjee, U.; Guha, A.K.; Ray, L. Green Pigment from Bacillus cereus M 1 16 (MTCC 5521): Production Parameters and Antibacterial Activity. Appl. Biochem. Biotechnol. 2011, 164, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Drewnowska, J.M.; Zambrzycka, M.; Kalska-Szostko, B.; Fiedoruk, K.; Swiecicka, I. Melanin-like pigment synthesis by soil Bacillus weihenstephanensis isolates from Northeastern Poland. PLoS ONE 2015, 10, e0125428. [Google Scholar] [CrossRef]
- Kim, H.; Han, S.; Lee, C.; Lee, K.; Park, S.; Kim, Y. Use of Prodigiosin for Treating Diabetes Mellitus. US 6,638,968 B1, 28 October 2003. [Google Scholar]
- Sakthivel, N.; Gnanamanickam, S. Toxicity of Pseudomonas fluorescens towards rice sheath-rot pathogen Acrocylindrium oryzae Saw. Curr. Sci. 1986, 55, 106–107. [Google Scholar]
- Chen, Y.; Zhou, D.; Qi, D.; Gao, Z.; Xie, J.; Luo, Y. Growth promotion and disease suppression ability of a Streptomyces sp. CB-75 from banana rhizosphere soil. Front. Microbiol. 2018, 8, 2704. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Chandraker, S.; Patel, V.; Ramteke, P. Antibacterial activity of medicinal plants against pathogens causing complicated urinary tract infections. Indian J. Pharm. Sci. 2009, 71, 136. [Google Scholar] [CrossRef]
- Chen, L.; Luo, S.; Xiao, X.; Guo, H.; Chen, J.; Wan, Y.; Li, B.; Xu, T.; Xi, Q.; Rao, C. Application of plant growth-promoting endophytes (PGPE) isolated from Solanum nigrum L. for phytoextraction of Cd-polluted soils. Appl. Soil Ecol. 2010, 46, 383–389. [Google Scholar] [CrossRef]
- Kim, O.-S.; Cho, Y.-J.; Lee, K.; Yoon, S.-H.; Kim, M.; Na, H.; Park, S.-C.; Jeon, Y.S.; Lee, J.-H.; Yi, H. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 3, 716–721. [Google Scholar] [CrossRef]
- Fatima, H.; Khan, K.; Zia, M.; Ur-Rehman, T.; Mirza, B.; Haq, I.-U. Extraction optimization of medicinally important metabolites from Datura innoxia Mill.: An in vitro biological and phytochemical investigation. BMC Complement. Altern. Med. 2015, 15, 376. [Google Scholar] [CrossRef]
- Sharma, P.; Kalita, M.C.; Thakur, D. Broad Spectrum Antimicrobial Activity of Forest-Derived Soil Actinomycete, Nocardia sp. PB-52. Front. Microbiol. 2016, 7, 347. [Google Scholar] [CrossRef] [PubMed]
- Nimaichand, S.; Devi, A.M.; Tamreihao, K.; Ningthoujam, D.S.; Li, W.-J. Actinobacterial diversity in limestone deposit sites in Hundung, Manipur (India) and their antimicrobial activities. Front. Microbiol. 2015, 6, 413. [Google Scholar] [CrossRef] [PubMed]
- Ahameethunisa, A.R.; Hopper, W. Antibacterial activity of Artemisia nilagirica leaf extracts against clinical and phytopathogenic bacteria. BMC Complement. Altern. Med. 2010, 10, 6. [Google Scholar] [CrossRef]
- Qi, D.; Zou, L.; Zhou, D.; Chen, Y.; Gao, Z.; Feng, R.; Zhang, M.; Li, K.; Xie, J.; Wang, W. Taxonomy and broad-spectrum antifungal activity of Streptomyces sp. SCA3-4 isolated from rhizosphere soil of Opuntia stricta. Front. Microbiol. 2019, 10, 1390. [Google Scholar] [CrossRef] [PubMed]
- Human, Z.R.; Moon, K.; Bae, M.; De Beer, Z.W.; Cha, S.; Wingfield, M.J.; Slippers, B.; Oh, D.-C.; Venter, S.N. Antifungal Streptomyces spp. associated with the infructescences of Protea spp. in South Africa. Front. Microbiol. 2016, 7, 1657. [Google Scholar] [CrossRef]
- Sapkota, A.; Thapa, A.; Budhathoki, A.; Sainju, M.; Shrestha, P.; Aryal, S. Isolation, characterization, and screening of antimicrobial-producing actinomycetes from soil samples. Int. J. Microbiol. 2020, 2020, 2716584. [Google Scholar] [CrossRef] [PubMed]
- Ryandini, D.; Radjasa, O. Bioactive compounds derived from Streptomyces sp. SA32: Antibacterial activity, chemical profile, and their related genes. IOP Conference Series: Earth and Environmental Science. In Proceedings of the 4th International Conference on Biosciences (ICoBio 2021), Bogor, Indonesia, 11–12 August 2021; p. 012062. [Google Scholar]
- Bundale, S.; Singh, J.; Begde, D.; Nashikkar, N.; Upadhyay, A. Rare actinobacteria: A potential source of bioactive polyketides and peptides. World J. Microbiol. Biotechnol. 2019, 35, 92. [Google Scholar] [CrossRef] [PubMed]
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary metabolites and biodiversity of actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Yang, L.J.; Zhang, W.D.; Shen, Y.H. The secondary metabolites of rare actinomycetes: Chemistry and bioactivity. RSC Adv. 2019, 9, 21964–21988. [Google Scholar] [CrossRef] [PubMed]
- Dezfully, N.K.; Ramanayaka, J.G. Isolation, identification and evaluation of antimicrobial activity of Streptomyces flavogriseus, strain ACTK2 from soil sample of Kodagu, Karnataka State (India). Jundishapur J. Microbiol. 2015, 8, e15107. [Google Scholar]
- Pacios-Michelena, S.; Aguilar, C.N.; Alvarez-Perez, O.B.; Rodriguez-Herrera, R.; Chávez-González, M.L.; Arredondo Valdés, R.; Ascacio Valdes, J.A.; Govea Salas, M.; Ilyina, A. Application of Streptomyces antimicrobial compounds for the control of phytopathogens. Front. Sustain. Food Syst. 2021, 310, 1–8. [Google Scholar] [CrossRef]
- Amin, D.H.; Abdallah, N.A.; Abolmaaty, A.; Tolba, S.; Wellington, E.M. Microbiological and molecular insights on rare Actinobacteria harboring bioactive prospective. Bull. Natl. Res. Cent. 2020, 44, 1–12. [Google Scholar] [CrossRef]
- Messaoudi, O.; Wink, J.; Bendahou, M. Diversity of actinobacteria isolated from date palms rhizosphere and saline environments: Isolation, identification and biological activity evaluation. Microorganisms 2020, 8, 1853. [Google Scholar] [CrossRef] [PubMed]
- Axenov-Gribanov, D.V.; Kostka, D.V.; Vasilieva, U.A.; Shatilina, Z.M.; Krasnova, M.E.; Pereliaeva, E.V.; Zolotovskaya, E.D.; Morgunova, M.M.; Rusanovskaya, O.O.; Timofeyev, M.A. Cultivable actinobacteria first found in baikal endemic algae is a new source of natural products with antibiotic activity. Int. J. Microbiol. 2020, 2020, 5359816. [Google Scholar] [CrossRef] [PubMed]
- Sungthong, R.; Nakaew, N. The genus Nonomuraea: A review of a rare actinomycete taxon for novel metabolites. J. Basic Microbiol. 2015, 55, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Han, K.-Y.; Wu, X.; Jiang, C.; Huang, R.; Li, Z.-H.; Feng, T.; Chen, H.-P.; Liu, J.-K. Three new compounds from the actinomycete Actinocorallia aurantiaca. Nat. Prod. Bioprospect. 2019, 9, 351–354. [Google Scholar] [CrossRef] [PubMed]




| Bacterial Isolates | Color | Shape | Margin | Elevation | Texture | Opacity | Size | Spore Color |
|---|---|---|---|---|---|---|---|---|
| G2 | Dark purple/magenta | Irregular | Irregular | Raised | Hard, dry, breaks apart | Opaque | 3 mm | White with very slight purplish tinch |
| G3 | Light green | Irregular | Undulate | Raised | Hard | Opaque | 5–7 mm | Blackish |
| G4 | Dark green | Round | Entire | Convex | Very hard, cannot be easily picked | Opaque | 2 mm | White |
| Bacterial IDs | Accession No. | Closet Match in NCBI with Accession No. | Link to NCBI Closet Match | Similarity % |
|---|---|---|---|---|
| G2 | MT071630 | Nonomurae salmonae strain JCM 3324 (MT760449) | https://www.ncbi.nlm.nih.gov/nucleotide/MT760449.1?report=genbank&log$=nuclalign&blast_rank=7&RID=G8YD90N001R (accessed on 30 July 2020) | 100 |
| G3 | MT071631 | Streptomyces chromofuscus strain AS-2 (JX442508) | https://www.ncbi.nlm.nih.gov/nucleotide/JX442508.1?report=genbank&log$=nucltop&blast_rank=2&RID=G8YNXKZ0013 (accessed on 30 July 2020) | 100 |
| G4 | MT071632 | Actinocorallia libanotica strain 11-2 (KJ571069) | https://www.ncbi.nlm.nih.gov/nucleotide/KJ571069.1?report=genbank&log$=nucltop&blast_rank=5&RID=G8YWRK2S016 (accessed on 30 July 2020) | 99.64 |
| Bacterial Pathogens | Nonomurae salmonae (G2) | Streptomyces chromofuscus (G3) | Actinocorallia libanotica (G4) | Kruskal Wallis Test Summary |
|---|---|---|---|---|
| P. syringae | 14.33 ± 0.58 | 17.66 ± 0.58 | 0 ± 0 | P = 0.0006 Kruskal–Wallis statistics = 10.73 |
| X. axonopodis | 0 ± 0 | 9 ± 1 | 0 ± 0 | P = 0.0182, Kruskal–Wallis statistics = 10.73 |
| Dunn’s Multiple Comparisons | P. syringae | X. axonopodis | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean Rank Difference | Significant | Summary | Adjusted p Value | Mean Rank Difference | Significant | Summary | Adjusted p Value | |
| NC to N. salmonae (G2) | 0 | No | ns | >0.9999 | −4.5 | No | ns | 0.3041 |
| NC to S. chromofuscus (G3) | −6 | Yes | * | 0.0224 | −7.5 | Yes | * | 0.0190 |
| NC to A. libanotica (G4) | 0 | No | ns | >0.9999 | 0 | No | ns | >0.9999 |
| Nonomurae salmonae (G2) | Streptomyces chromofuscus (G3) | Actinocorallia libanotica (G4) | ANOVA Summary | ||||
|---|---|---|---|---|---|---|---|
| Inhibition Zone (mm) | MIC (μg/mL) | Inhibition Zone (mm) | MIC (μg/mL) | Inhibition Zone (mm) | MIC (μg/mL) | ||
| F. oxysporum | 7 ± 1 a | <50 | 11 ± 1 a | <50 | 0.67 ± 0.57 c | Not in range | F = 142, P < 0.0001, DF = 11 |
| F. solani | 6.67 ± 1.5 b | <25 | 16.7 ± 0.4 a | <25 | 13.3 ± 1.5 a | <100 | F = 128.5, P < 0.0001, DF = 11 |
| A. flavus | 0 ± 0 c | No activity | 28 ± 1 a | <12.5 | 0 ± 0 c | No activity | F = 2352, P < 0.0001, DF = 11 |
| A. alternata | 11.3 ± 1.15 a | <50 | 22.67 ± 0.5 a | <12.5 | 11 ± 1 a | <100 | F = 385.5, P < 0.0001, DF = 11 |
| A. niger | 19.33 ± 1.15 a | <50 | 26.33 ± 1.5 a | <6.25 | 4 ± 0.7 b | <100 | F = 436.5, P < 0.0001, DF = 11 |
| Pathogenic Fungi | Nonomurae salmonae (G2) | Streptomyces chromofuscus (G3) | Actinocorallia libanotica(G4) | ANOVA Summary |
|---|---|---|---|---|
| F. oxysporum | 15.49 ± 0.81 a | 24.88 ± 0.81 a | 0.94 ± 0.81 c | F = 610.6, P < 0.0001, df = 11 |
| F. solani | 13.14 ± 0.7 a | 30.98 ± 0.8 a | 22.07 ± 0.8 a | F = 812, P < 0.0001, df = 11 |
| A. flavus | 2.82 ± 1.4 c | 36.62 ± 0.8 a | 0.94 ± 0.8 c | F = 1078, P < 0.0001, df = 11 |
| A. alternata | 14.56 ± 0.7 a | 30.99 ± 1.07 a | 17.37 ± 0.8 a | F = 591, P < 0.0001, df = 11 |
| A. niger | 23.01 ± 0.8 a | 32.86 ± 1.2 a | 7.98 ± 0.8 a | F = 718, P < 0.0001, df = 11 |
| Pathogenic Bacteria | Nonomurae salmonae | Streptomyces chromofuscus | Streptomycin |
|---|---|---|---|
| P. syringae | 15.49 | 24.88 | 3.906 |
| X. axonopodis | NA | 30.98 | 1.953 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashir, S.; Numan, M.; Shinwari, Z.K. Selective Isolation of Bioactive-Pigmented Bacteria from Saline Agricultural Soil and Assessment of Their Antimicrobial Potential against Plant Pathogens. Sustainability 2022, 14, 13574. https://doi.org/10.3390/su142013574
Bashir S, Numan M, Shinwari ZK. Selective Isolation of Bioactive-Pigmented Bacteria from Saline Agricultural Soil and Assessment of Their Antimicrobial Potential against Plant Pathogens. Sustainability. 2022; 14(20):13574. https://doi.org/10.3390/su142013574
Chicago/Turabian StyleBashir, Samina, Muhammad Numan, and Zabta Khan Shinwari. 2022. "Selective Isolation of Bioactive-Pigmented Bacteria from Saline Agricultural Soil and Assessment of Their Antimicrobial Potential against Plant Pathogens" Sustainability 14, no. 20: 13574. https://doi.org/10.3390/su142013574
APA StyleBashir, S., Numan, M., & Shinwari, Z. K. (2022). Selective Isolation of Bioactive-Pigmented Bacteria from Saline Agricultural Soil and Assessment of Their Antimicrobial Potential against Plant Pathogens. Sustainability, 14(20), 13574. https://doi.org/10.3390/su142013574







