Pyrolyzed or Composted Sewage Sludge Application Induces Short-Term Changes in the Terra Rossa Soil Bacterial and Fungal Communities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup
2.2. DNA Extraction and Sequencing
2.3. Data Analysis
3. Results
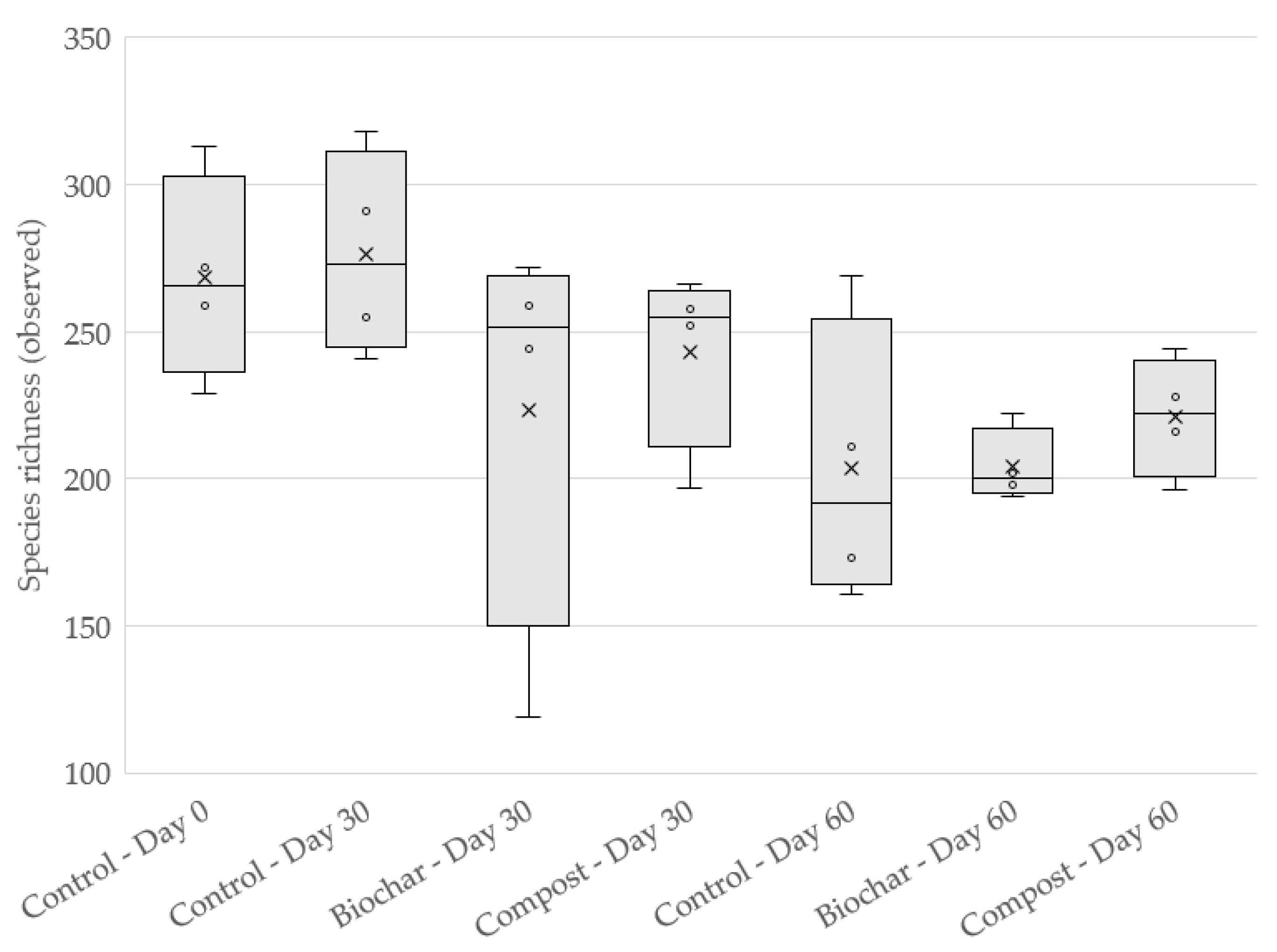
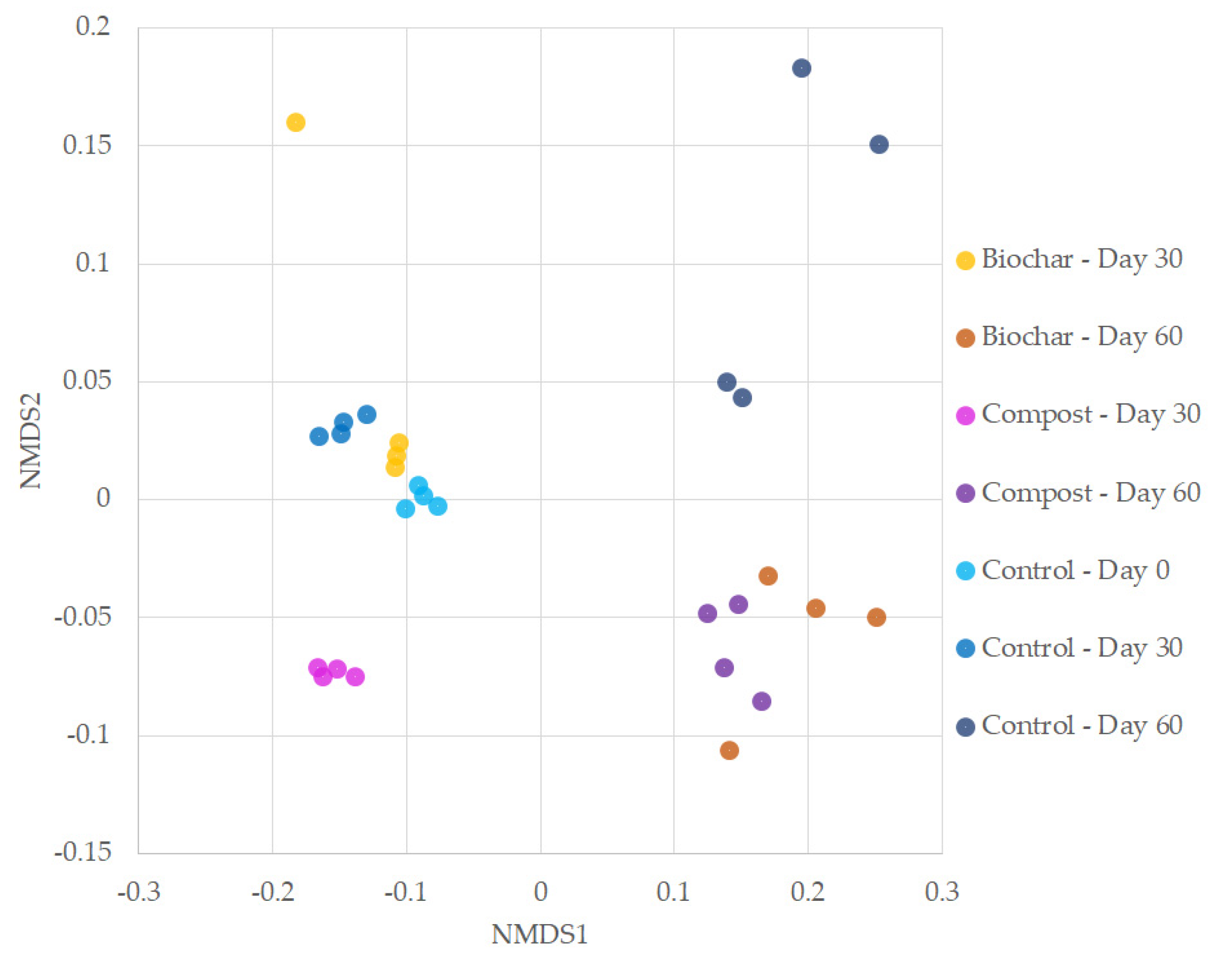
3.1. Species Richness and Community Analysis
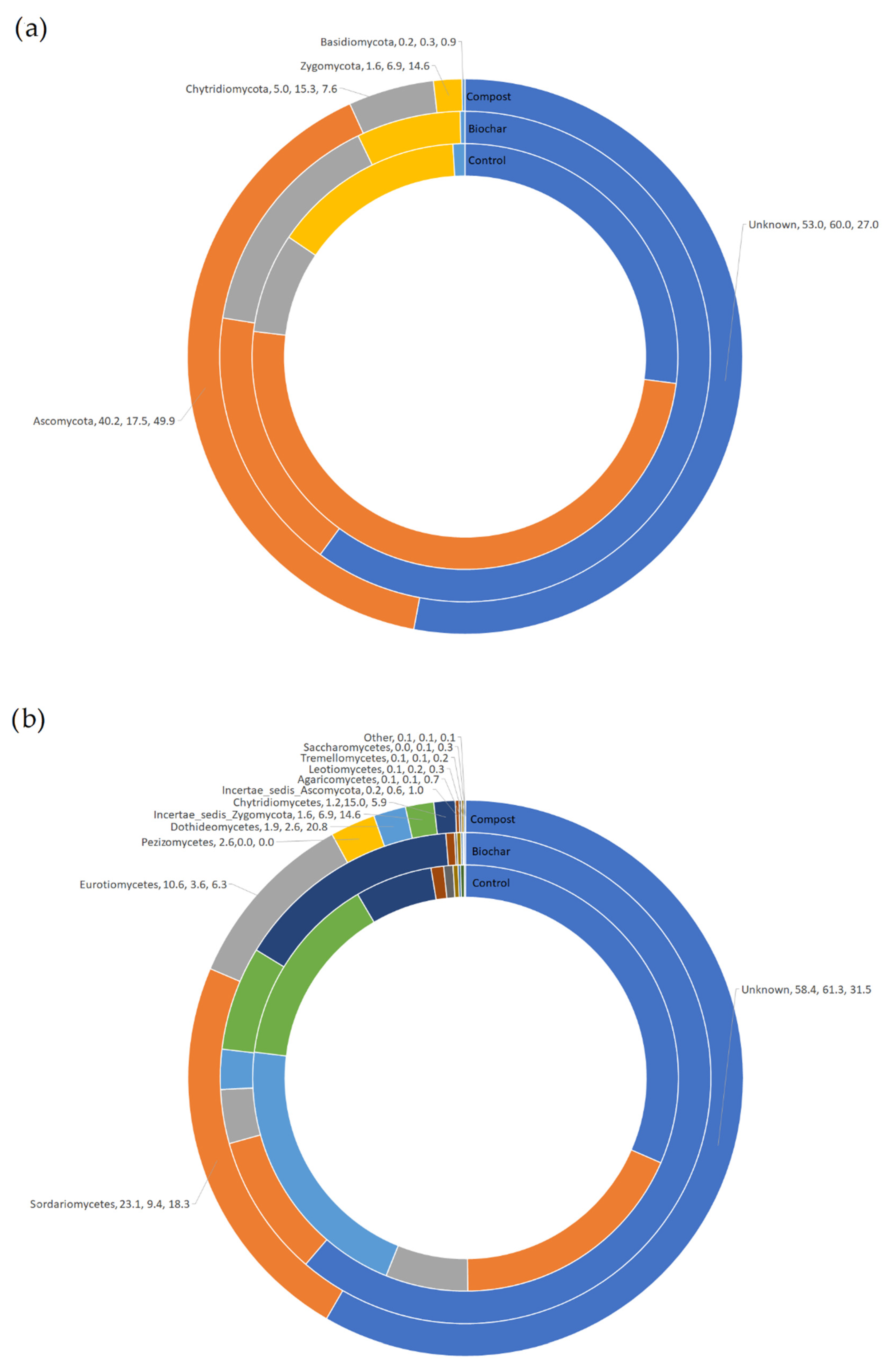
3.2. Taxonomic Classification
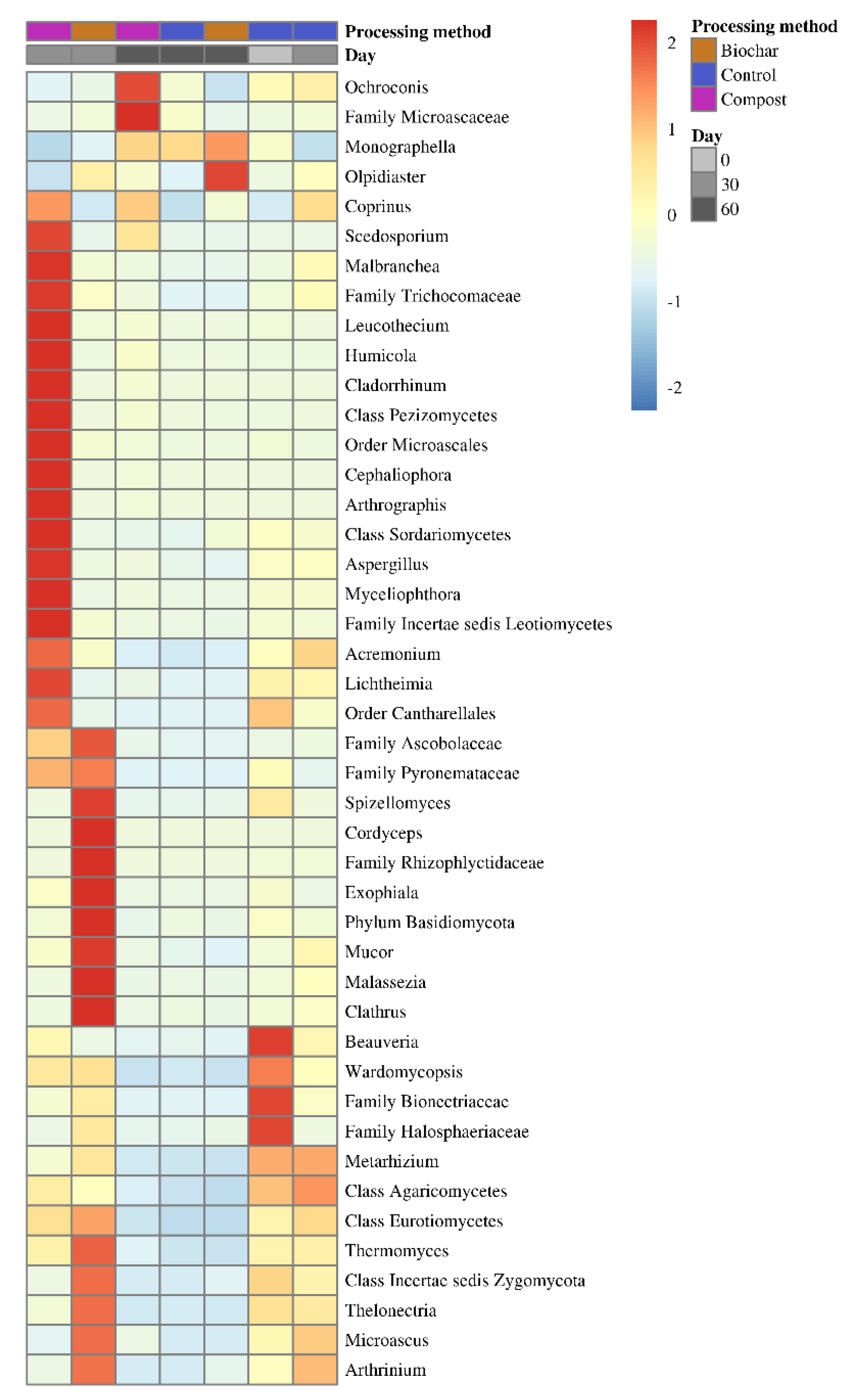
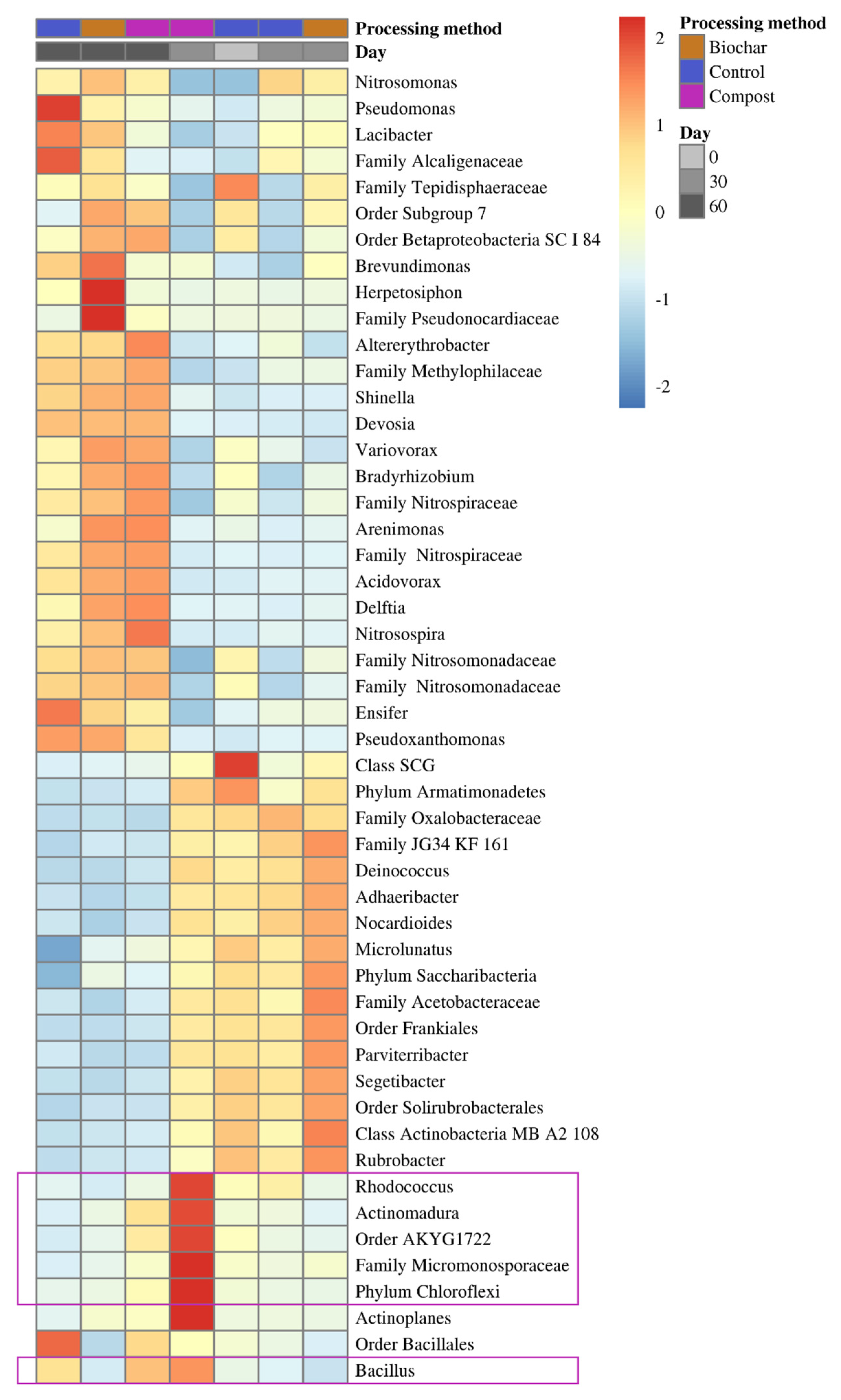
3.3. Differential Abundant Taxa
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the Soil Microbiome to Increase Soil Health and Plant Fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Jackson, R.B. The Diversity and Biogeography of Soil Bacterial Communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.R.; Viger, M.; Arnold, E.C.; Harris, Z.M.; Ventura, M.; Miglietta, F.; Girardin, C.; Edwards, R.J.; Rumpel, C.; Fornasier, F.; et al. Biochar Alters the Soil Microbiome and Soil Function: Results of next-Generation Amplicon Sequencing across Europe. GCB Bioenergy 2017, 9, 591–612. [Google Scholar] [CrossRef]
- Fischer, D.; Glaser, B. Synergisms between Compost and Biochar for Sustainable Soil Amelioration. In Management of Organic Waste; InTech Open: Rijeka, Croatia, 2012. [Google Scholar]
- Lupatini, M.; Korthals, G.W.; de Hollander, M.; Janssens, T.K.S.; Kuramae, E.E. Soil Microbiome Is More Heterogeneous in Organic Than in Conventional Farming System. Front. Microbiol. 2017, 7, 2064. [Google Scholar] [CrossRef]
- Safaei Khorram, M.; Zhang, G.; Fatemi, A.; Kiefer, R.; Maddah, K.; Baqar, M.; Zakaria, M.P.; Li, G. Impact of Biochar and Compost Amendment on Soil Quality, Growth and Yield of a Replanted Apple Orchard in a 4-Year Field Study. J. Sci. Food Agric. 2019, 99, 1862–1869. [Google Scholar] [CrossRef] [PubMed]
- Cesarano, G.; De Filippis, F.; La Storia, A.; Scala, F.; Bonanomi, G. Organic Amendment Type and Application Frequency Affect Crop Yields, Soil Fertility and Microbiome Composition. Appl. Soil Ecol. 2017, 120, 254–264. [Google Scholar] [CrossRef]
- Wong, J.W.C.; Ma, K.K.; Fang, K.M.; Cheung, C. Utilization of a Manure Compost for Organic Farming in Hong Kong. Bioresour. Technol. 1999, 67, 43–46. [Google Scholar] [CrossRef]
- Annabi, M.; Le Bissonnais, Y.; Le Villio-Poitrenaud, M.; Houot, S. Improvement of Soil Aggregate Stability by Repeated Applications of Organic Amendments to a Cultivated Silty Loam Soil. Agric. Ecosyst. Environ. 2011, 144, 382–389. [Google Scholar] [CrossRef]
- Curtis, M.J.; Claassen, V.P. Compost Incorporation Increases Plant Available Water in a Drastically Disturbed Serpentine Soil. Soil Sci. 2005, 170, 939–953. [Google Scholar] [CrossRef]
- Diacono, M.; Montemurro, F. Long-Term Effects of Organic Amendments on Soil Fertility. A Review. Agron. Sustain. Dev. 2012, 30, 401–422. [Google Scholar] [CrossRef]
- Lourenço, K.S.; Suleiman, A.K.A.; Pijl, A.; van Veen, J.A.; Cantarella, H.; Kuramae, E.E. Resilience of the Resident Soil Microbiome to Organic and Inorganic Amendment Disturbances and to Temporary Bacterial Invasion. Microbiome 2018, 6, 142. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Lal, R.; Zimmerman, A.R. Effects of Biochar and Other Amendments on the Physical Properties and Greenhouse Gas Emissions of an Artificially Degraded Soil. Sci. Total Environ. 2014, 487, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Razzaghi, F.; Obour, P.B.; Arthur, E. Does Biochar Improve Soil Water Retention? A Systematic Review and Meta-Analysis. Geoderma 2020, 361, 114055. [Google Scholar] [CrossRef]
- Herath, H.M.S.K.; Camps-Arbestain, M.; Hedley, M. Effect of Biochar on Soil Physical Properties in Two Contrasting Soils: An Alfisol and an Andisol. Geoderma 2013, 209–210, 188–197. [Google Scholar] [CrossRef]
- Major, N.; Schierstaedt, J.; Jechalke, S.; Nesme, J.; Ban, S.G.; Černe, M.; Sørensen, S.J.; Ban, D.; Schikora, A. Composted Sewage Sludge Influences the Microbiome and Persistence of Human Pathogens in Soil. Microorganisms 2020, 8, 1020. [Google Scholar] [CrossRef]
- Jechalke, S.; Schierstaedt, J.; Becker, M.; Flemer, B.; Grosch, R.; Smalla, K.; Schikora, A. Salmonella Establishment in Agricultural Soil and Colonization of Crop Plants Depend on Soil Type and Plant Species. Front. Microbiol. 2019, 10, 967. [Google Scholar] [CrossRef]
- Schierstaedt, J.; Grosch, R.; Schikora, A. Agricultural Production Systems Can Serve as Reservoir for Human Pathogens. FEMS Microbiol. Lett. 2019, 366, 16. [Google Scholar] [CrossRef]
- Bziuk, N.; Maccario, L.; Straube, B.; Wehner, G.; Sørensen, S.J.; Schikora, A.; Smalla, K. The Treasure inside Barley Seeds: Microbial Diversity and Plant Beneficial Bacteria. Environ. Microbiome 2021, 16, 1–21. [Google Scholar] [CrossRef]
- Wang, H.; Liu, R.; You, M.P.; Barbetti, M.J.; Chen, Y. Pathogen Biocontrol Using Plant Growth-Promoting Bacteria (PGPR): Role of Bacterial Diversity. Microorganisms 2021, 9, 1988. [Google Scholar] [CrossRef]
- Schierstaedt, J.; Jechalke, S.; Nesme, J.; Neuhaus, K.; Sørensen, S.J.; Grosch, R.; Smalla, K.; Schikora, A. Salmonella Persistence in Soil Depends on Reciprocal Interactions with Indigenous Microorganisms. Environ. Microbiol. 2020, 22, 2639–2652. [Google Scholar] [CrossRef] [PubMed]
- Kelessidis, A.; Stasinakis, A.S. Comparative Study of the Methods Used for Treatment and Final Disposal of Sewage Sludge in European Countries. Waste Manag. 2012, 32, 1186–1195. [Google Scholar] [CrossRef]
- Mininni, G.; Blanch, A.R.; Lucena, F.; Berselli, S. EU Policy on Sewage Sludge Utilization and Perspectives on New Approaches of Sludge Management. Environ. Sci. Pollut. Res. 2015, 22, 7361–7374. [Google Scholar] [CrossRef]
- Alvarenga, P.; Mourinha, C.; Farto, M.; Santos, T.; Palma, P.; Sengo, J.; Morais, M.-C.; Cunha-Queda, C. Sewage Sludge, Compost and Other Representative Organic Wastes as Agricultural Soil Amendments: Benefits versus Limiting Factors. Waste Manag. 2015, 40, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Casado-Vela, J.; Sellés, S.; Navarro, J.; Bustamante, M.A.; Mataix, J.; Guerrero, C.; Gomez, I. Evaluation of Composted Sewage Sludge as Nutritional Source for Horticultural Soils. Waste Manag. 2006, 26, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Černe, M.; Palčić, I.; Pasković, I.; Major, N.; Romić, M.; Filipović, V.; Igrc, M.D.; Perčin, A.; Goreta Ban, S.; Zorko, B.; et al. The Effect of Stabilization on the Utilization of Municipal Sewage Sludge as a Soil Amendment. Waste Manag. 2019, 94, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Horvatić, V.; Begić, H.B.; Romić, D.; Černe, M.; Goreta Ban, S.; Zovko, M.; Romić, M. Evaluation of Land Potential for Use of Biosolids in the Coastal Mediterranean Karst Region. Land 2021, 10, 1035. [Google Scholar] [CrossRef]
- Mei, X.; Tang, J.; Zhang, Y. Sludge Stabilization: Characteristics of the End-Products and an Alternative Evaluative Methodology. Waste Manag. 2020, 105, 355–363. [Google Scholar] [CrossRef]
- Méndez, A.; Gómez, A.; Paz-Ferreiro, J.; Gascó, G. Effects of Sewage Sludge Biochar on Plant Metal Availability after Application to a Mediterranean Soil. Chemosphere 2012, 89, 1354–1359. [Google Scholar] [CrossRef]
- Chagas, J.K.M.; de Figueiredo, C.C.; da Silva, J.; Paz-Ferreiro, J. The Residual Effect of Sewage Sludge Biochar on Soil Availability and Bioaccumulation of Heavy Metals: Evidence from a Three-Year Field Experiment. J. Environ. Manag. 2021, 279, 111824. [Google Scholar] [CrossRef]
- Lahori, A.H.; Mierzwa-Hersztek, M.; Rashid, M.; Kalhoro, S.A.; Memon, M.; Naheed, Z.; Ahmed, M.; Zhang, Z. Residual Effects of Tobacco Biochar along with Different Fixing Agents on Stabilization of Trace Elements in Multi-Metal Contaminated Soils. J. Environ. Sci. 2020, 87, 299–309. [Google Scholar] [CrossRef]
- Qian, T.; Yang, Q.; Jun, D.C.F.; Dong, F.; Zhou, Y. Transformation of Phosphorus in Sewage Sludge Biochar Mediated by a Phosphate-Solubilizing Microorganism. Chem. Eng. J. 2019, 359, 1573–1580. [Google Scholar] [CrossRef]
- Ai, Y.J.; Li, F.P.; Gu, H.H.; Chi, X.J.; Yuan, X.T.; Han, D.Y. Combined Effects of Green Manure Returning and Addition of Sewage Sludge Compost on Plant Growth and Microorganism Communities in Gold Tailings. Environ. Sci. Pollut. Res. 2020, 27, 31686–31698. [Google Scholar] [CrossRef]
- Bašić, F. The Soils of Croatia. 2013. Available online: https://doi.org/10.1007/978-94-007-5815-5 (accessed on 11 September 2019).
- International Organization for Standardization—ISO 18400-104:2018—Soil Quality—Sampling—Part 104: Strangies, 2018. Available online: http://www.iso.org/standard/65223.html (accessed on 11 September 2019).
- Marschner, H. Mineral Nutrition of Higher Plants, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 1955; p. 889. [Google Scholar]
- Černe, M.; Palčić, I.; Major, N.; Pasković, I.; Perković, J.; Užila, Z.; Filipović, V.; Romić, M.; Goreta Ban, S.; Heath, D.J.; et al. Effect of Sewage Sludge-Derived Amendments on the Nutrient Uptake by Chinese Cabbage from Mediterranean Soils. J. Plant Nutr. 2022, 1–25. [Google Scholar] [CrossRef]
- Canet, R.; Pomares, F.; Cabot, B.; Chaves, C.; Ferrer, E.; Ribó, M.; Albiach, M.R. Composting Olive Mill Pomace and Other Residues from Rural Southeastern Spain. Waste Manag. 2008, 28, 2585–2592. [Google Scholar] [CrossRef] [PubMed]
- Itävaara, M.; Vikman, M.; Liisa, M.; Vuorinen, A. Maturity Tests for Composts—Verification Of a Test Scheme for Assessing Maturity. Compos. Sci. Util. 2013, 18, 174–183. [Google Scholar] [CrossRef]
- Cornelissen, G.; Pandit, N.R.; Taylor, P.; Pandit, B.H.; Sparrevik, M.; Schmidt, H.P. Emissions and Char Quality of Flame-Curtain & quot; Kon Tiki & quot; Kilns for Farmer-Scale Charcoal/Biochar Production. PLoS ONE 2016, 11, e0154617. [Google Scholar] [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit RRNA Sequence Analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal Rna Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-Filtering Vastly Improves Diversity Estimates from Illumina Amplicon Sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S RRNA Sequence Formation and Detection in Sanger and 454-Pyrosequenced PCR Amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S RRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for Comprehensive Statistical, Functional, and Meta-Analysis of Microbiome Data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar] [CrossRef]
- Russel, J.; Thorsen, J.; Brejnrod, A.D.; Bisgaard, H.; Sørensen, S.J.; Burmølle, M. DAtest: A Framework for Choosing Differential Abundance or Expression Method. bioRxiv 2018, 241802. [Google Scholar] [CrossRef]
- Lloret, E.; Pascual, J.A.; Brodie, E.L.; Bouskill, N.J.; Insam, H.; Juárez, M.F.-D.D.; Goberna, M. Sewage Sludge Addition Modifies Soil Microbial Communities and Plant Performance Depending on the Sludge Stabilization Process. Appl. Soil Ecol. 2016, 101, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Bakker, M.G.; Schlatter, D.C.; Otto-Hanson, L.; Kinkel, L.L. Diffuse Symbioses: Roles of Plant–Plant, Plant–Microbe and Microbe–Microbe Interactions in Structuring the Soil Microbiome. Mol. Ecol. 2014, 23, 1571–1583. [Google Scholar] [CrossRef]
- Tayyab, M.; Islam, W.; Lee, C.G.; Pang, Z.; Khalil, F.; Lin, S.; Lin, W.; Zhang, H.; Tayyab, M.; Islam, W.; et al. Short-Term Effects of Different Organic Amendments on Soil Fungal Composition. Sustainability 2019, 11, 198. [Google Scholar] [CrossRef]
- Neumann, G.; Bott, S.; Ohler, M.A.; Mock, H.P.; Lippmann, R.; Grosch, R.; Smalla, K. Root Exudation and Root Development of Lettuce (Lactuca Sativa l. Cv. Tizian) as Affected by Different Soils. Front. Microbiol. 2014, 5, 2. [Google Scholar] [CrossRef]
- Curlango-Rivera, G.; Huskey, D.A.; Mostafa, A.; Kessler, J.O.; Xiong, Z.; Hawes, M.C. Intraspecies Variation in Cotton Border Cell Production: Rhizosphere Microbiome Implications. Am. J. Bot. 2013, 100, 1706–1712. [Google Scholar] [CrossRef]
- Ali, A.; Ghani, M.I.; Elrys, A.S.; Ding, H.; Iqbal, M.; Cheng, Z.; Cai, Z. Different Cropping Systems Regulate the Metabolic Capabilities and Potential Ecological Functions Altered by Soil Microbiome Structure in the Plastic Shed Mono-Cropped Cucumber Rhizosphere. Agric. Ecosyst. Environ. 2021, 318, 107486. [Google Scholar] [CrossRef]
- Stacey, N.E.; Lewis, R.W.; Davenport, J.R.; Sullivan, T.S. Composted Biosolids for Golf Course Turfgrass Management: Impacts on the Soil Microbiome and Nutrient Cycling. Appl. Soil Ecol. 2019, 144, 31–41. [Google Scholar] [CrossRef]
- Bai, Y.; Mei, L.; Zuo, W.; Zhang, Y.; Gu, C.; Shan, Y.; Hu, J.; Dai, Q. Response of Bacterial Communities in Coastal Mudflat Saline Soil to Sewage Sludge Amendment. Appl. Soil Ecol. 2019, 144, 107–111. [Google Scholar] [CrossRef]
- Azeem, M.; Hale, L.; Montgomery, J.; Crowley, D.; McGiffen, M.E. Biochar and Compost Effects on Soil Microbial Communities and Nitrogen Induced Respiration in Turfgrass Soils. PLoS ONE 2020, 15, e0242209. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Major, N.; Schierstaedt, J.; Schikora, A.; Palčić, I.; Černe, M.; Goreta Ban, S.; Pasković, I.; Perković, J.; Užila, Z.; Ban, D. Pyrolyzed or Composted Sewage Sludge Application Induces Short-Term Changes in the Terra Rossa Soil Bacterial and Fungal Communities. Sustainability 2022, 14, 11382. https://doi.org/10.3390/su141811382
Major N, Schierstaedt J, Schikora A, Palčić I, Černe M, Goreta Ban S, Pasković I, Perković J, Užila Z, Ban D. Pyrolyzed or Composted Sewage Sludge Application Induces Short-Term Changes in the Terra Rossa Soil Bacterial and Fungal Communities. Sustainability. 2022; 14(18):11382. https://doi.org/10.3390/su141811382
Chicago/Turabian StyleMajor, Nikola, Jasper Schierstaedt, Adam Schikora, Igor Palčić, Marko Černe, Smiljana Goreta Ban, Igor Pasković, Josipa Perković, Zoran Užila, and Dean Ban. 2022. "Pyrolyzed or Composted Sewage Sludge Application Induces Short-Term Changes in the Terra Rossa Soil Bacterial and Fungal Communities" Sustainability 14, no. 18: 11382. https://doi.org/10.3390/su141811382
APA StyleMajor, N., Schierstaedt, J., Schikora, A., Palčić, I., Černe, M., Goreta Ban, S., Pasković, I., Perković, J., Užila, Z., & Ban, D. (2022). Pyrolyzed or Composted Sewage Sludge Application Induces Short-Term Changes in the Terra Rossa Soil Bacterial and Fungal Communities. Sustainability, 14(18), 11382. https://doi.org/10.3390/su141811382









