Soil Communities: Who Responds and How Quickly to a Change in Agricultural System?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Agronomic Managements
2.2. Soil Sampling
2.3. Soil Physicochemical Properties
2.4. Soil Living Communities
2.5. Statistical Analysis
3. Results and Discussion
3.1. Soil Physico-Chemical Properties
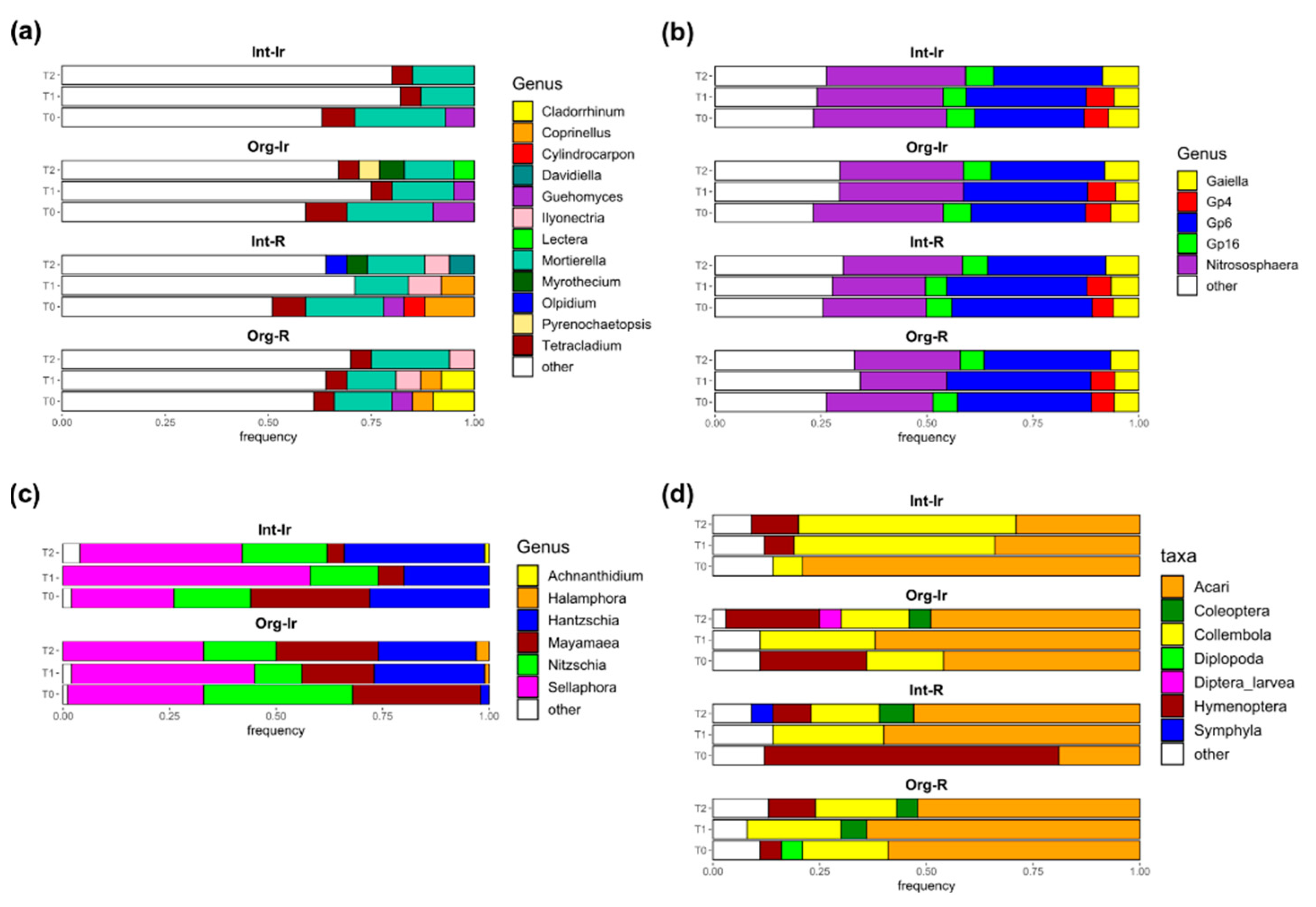
3.2. Effects of Agricultural Practices on Fungal Soil Communities
3.3. Soil Properties Mainly Affected Bacterial Communities
3.4. Diatoms as a Promising Community for Indicating Soil Quality
3.5. Effect of Seasonality on Microarthropods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lavelle, P.; Decaëns, T.; Aubert, M.; Barot, S.; Blouin, M.; Bureau, F.; Margerie, P.; Mora, P.; Rossi, J.P. Soil invertebrates and ecosystem services. Eur. J. Soil Biol. 2006, 42, S3–S15. [Google Scholar] [CrossRef]
- Zimmerer, K.S. Biological diversity in agriculture and global change. Annu. Rev. Environ. Resour. 2010, 35, 137–166. [Google Scholar] [CrossRef] [Green Version]
- Vischetti, C.; Casucci, C.; De Bernardi, A.; Monaci, E.; Tiano, L.; Marcheggiani, F.; Ciani, M.; Comitini, F.; Marini, E.; Taskin, E.; et al. Sub-Lethal Effects of Pesticides on the DNA of Soil Organisms as Early Ecotoxicological Biomarkers. Front. Microbiol. 2020, 11, 1892. [Google Scholar] [CrossRef]
- Yang, T.; Lupwayi, N.; Marc, S.A.; Siddique, K.H.M.; Bainard, L.D. Anthropogenic drivers of soil microbial communities and impacts on soil biological functions in agroecosystems. Glob. Ecol. Conserv. 2021, 27, e01521. [Google Scholar] [CrossRef]
- Ouyang, Y.; Norton, J.M. Short-term nitrogen fertilization affects microbial community composition and nitrogen mineralization functions in an agricultural soil. Appl. Environ. Microbiol. 2020, 86, e02278-19. [Google Scholar] [CrossRef]
- Qaswar, M.; Dongchu, L.; Jing, H.; Tianfu, H.; Ahmed, W.; Abbas, M.; Lu, Z.; Jiangxue, D.; Khan, Z.H.; Ullah, S.; et al. Interaction of liming and long-term fertilization increased crop yield and phosphorus use efficiency (PUE) through mediating exchangeable cations in acidic soil under wheat–maize cropping system. Sci. Rep. 2020, 10, 19828. [Google Scholar] [CrossRef]
- Tully, K.L.; McAskill, C. Promoting soil health in organically managed systems: A review. Org. Agric. 2020, 10, 339–358. [Google Scholar] [CrossRef]
- Garciá-Orenes, F.; Morugań-Coronado, A.; Zornoza, R.; Scow, K. Changes in soil microbial community structure influenced by agricultural management practices in a Mediterranean agro-ecosystem. PLoS ONE 2013, 8, e80522. [Google Scholar] [CrossRef]
- Luo, X.; Wang, M.K.; Hu, G.; Weng, B. Seasonal change in microbial diversity and its relationship with soil chemical properties in an orchard. PLoS ONE 2019, 14, e0215556. [Google Scholar] [CrossRef]
- Coller, E.; Cestaro, A.; Zanzotti, R.; Bertoldi, D.; Pindo, M.; Larger, S.; Albanese, D.; Mescalchin, E.; Donati, C. Microbiome of vineyard soils is shaped by geography and management. Microbiome 2019, 7, 140. [Google Scholar] [CrossRef] [Green Version]
- Whalen, E.D.; Smith, R.G.; Grandy, A.S.; Frey, S.D. Manganese limitation as a mechanism for reduced decomposition in soils under atmospheric nitrogen deposition. Soil Biol. Biochem. 2018, 127, 252–263. [Google Scholar] [CrossRef]
- Liang, B.; Ma, C.; Fan, L.; Wang, Y.; Yuan, Y. Compost amendment alters soil fungal community structure of a replanted apple orchard. Arch. Agron. Soil Sci. 2020, 67, 739–752. [Google Scholar] [CrossRef]
- Schloter, M.; Dilly, O.; Munch, J.C. Indicators for evaluating soil quality. Agric. Ecosyst. Environ. 2003, 98, 255–262. [Google Scholar] [CrossRef]
- Harkes, P.; Suleiman, A.K.A.; van den Elsen, S.J.J.; de Haan, J.J.; Holterman, M.; Kuramae, E.E.; Helder, J. Conventional and organic soil management as divergent drivers of resident and active fractions of major soil food web constituents. Sci. Rep. 2019, 9, 13521. [Google Scholar] [CrossRef] [PubMed]
- Menta, C.; Conti, F.D.; Fondón, C.L.; Staffilani, F.; Remelli, S. Soil arthropod responses in agroecosystem: Implications of different management and cropping systems. Agronomy 2020, 7, 982. [Google Scholar] [CrossRef]
- Mantoni, C.; Pellegrini, M.; Dapporto, L.; Del Gallo, M.; Pace, L.; Silveri, D.; Fattorini, S. Comparison of Soil Biology Quality in Organically and Conventionally Managed Agro-Ecosystems Using Microarthropods. Agriculture 2021, 11, 1022. [Google Scholar] [CrossRef]
- Joimel, S.; Schwartz, C.; Bonfanti, J.; Hedde, M.; Krogh, P.H.; Pérès, G.; Pernin, C.; Rakoto, A.; Salmon, S.; Santorufo, L.; et al. Functional and Taxonomic Diversity of Collembola as Complementary Tools to Assess Land Use Effects on Soils Biodiversity. Front. Ecol. Evol. 2021, 9, 630919. [Google Scholar] [CrossRef]
- Simoni, S.; Caruso, G.; Vignozzi, N.; Gucci, R.; Valboa, G.; Pellegrini, S.; Palai, G.; Goggioli, D.; Gagnarli, E. Effect of long-term soil management practices on tree growth, yield and soil biodiversity in a high-density olive agro-ecosystem. Agronomy 2021, 11, 1036. [Google Scholar] [CrossRef]
- Meyer, S.; Kundel, D.; Birkhofer, K.; Fliessbach, A.; Scheu, S. Soil microarthropods respond differently to simulated drought in organic and conventional farming systems. Ecol. Evol. 2021, 11, 10369–10380. [Google Scholar] [CrossRef]
- Foets, J.; Wetzel, C.E.; Teuling, A.J.; Pfister, L. Temporal and spatial variability of terrestrial diatoms at the catchment scale: Controls on productivity and comparison with other soil algae. PeerJ 2020, 8, e8296. [Google Scholar] [CrossRef] [Green Version]
- Barragán, C.; Wetzel, C.E.; Ector, L. A standard method for the routine sampling of terrestrial diatom communities for soil quality assessment. J. Appl. Phycol. 2018, 30, 1095–1113. [Google Scholar] [CrossRef]
- Bérard, A.; Rimet, F.; Capowiez, Y.; Leboulanger, C. Procedures for Determining the Pesticide Sensitivity of Indigenous Soil Algae: A Possible Bioindicator of Soil Contamination? Arch. Environ. Contam. Toxicol. 2004, 46, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, M.; Wetzel, C.E.; Ector, L.; Teuling, A.J.; Pfister, L. On the potential for terrestrial diatom communities and diatom indices to identify anthropic disturbance in soils. Ecol. Indic. 2017, 75, 73–81. [Google Scholar] [CrossRef]
- Heger, T.J.; Straub, F.; Mitchell, E.A.D. Impact of farming practices on soil diatoms and testate amoebae: A pilot study in the DOK-trial at Therwil, Switzerland. Eur. J. Soil Biol. 2012, 49, 31–36. [Google Scholar] [CrossRef]
- Zhang, Y.; Ouyang, S.; Nie, L.; Chen, X. Soil diatom communities and their relation to environmental factors in three types of soil from four cities in central-west China. Eur. J. Soil Biol. 2020, 98, 103175. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apprill, A.; Mcnally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Walters, W.; Hyde, E.R.; Berg-lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K. Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems 2016, 1, e0009-15. [Google Scholar] [CrossRef] [Green Version]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods Applications; Innis, M.A., Gelfand, D.H., Snisky, J.J., White, T.J., Eds.; Academic Press: New Yourk, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parisi, V.; Menta, C.; Gardi, C.; Jacomini, C.; Mozzanica, E. Microarthropod communities as a tool to assess soil quality and biodiversity: A new approach in Italy. Agric. Ecosyst. Environ. 2005, 105, 323–333. [Google Scholar] [CrossRef]
- Menta, C.; Conti, F.D.; Pinto, S.; Bodini, A. Soil Biological Quality index (QBS-ar): 15 years of application at global scale. Ecol. Indic. 2018, 85, 773–780. [Google Scholar] [CrossRef]
- R Core Development Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Seaton, F.M.; George, P.B.L.; Lebron, I.; Jones, D.L.; Creer, S.; Robinson, D.A. Soil textural heterogeneity impacts bacterial but not fungal diversity. Soil Biol. Biochem. 2020, 144, 107766. [Google Scholar] [CrossRef]
- Cloutier, M.L.; Murrell, E.; Barbercheck, M.; Kaye, J.; Finney, D.; García-González, I.; Bruns, M.A. Fungal community shifts in soils with varied cover crop treatments and edaphic properties. Sci. Rep. 2020, 10, 6198. [Google Scholar] [CrossRef] [PubMed]
- Bach, E.M.; Baer, S.G.; Meyer, C.K.; Six, J. Soil texture affects soil microbial and structural recovery during grassland restoration. Soil Biol. Biochem. 2010, 42, 2182–2191. [Google Scholar] [CrossRef]
- Kepler, R.M.; Epp Schmidt, D.J.; Yarwood, S.A.; Cavigelli, M.A.; Reddy, K.N.; Duke, S.O.; Bradley, C.A.; Williams, M.M.; Maula, J.E. Soil microbial communities in diverse agroecosystems exposed to the herbicide glyphosate. Appl. Environ. Microbiol. 2020, 86, e01744-19. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Duan, Y.; Zhang, R.; Liu, C.; Wang, Y.; Li, M.; Ding, Y.; Awasthi, M.K.; Li, H. Connecting soil dissolved organic matter to soil bacterial community structure in a long-term grass-mulching apple orchard. Ind. Crops Prod. 2020, 149, 112344. [Google Scholar] [CrossRef]
- Liang, B.; Ma, C.; Fan, L.; Wang, Y.; Yuan, Y. Soil amendment alters soil physicochemical properties and bacterial community structure of a replanted apple orchard. Microbiol. Res. 2018, 216, 1–11. [Google Scholar] [CrossRef]
- Girvan, M.S.; Bullimore, J.; Ball, A.S.; Pretty, J.N.; Osborn, A.M. Responses of active bacterial and fungal communities in soils under winter wheat to different fertilizer and pesticide regimens. Appl. Environ. Microbiol. 2004, 70, 2692–2701. [Google Scholar] [CrossRef] [Green Version]
- Smit, E.; Leeflang, P.; Gommans, S.; Van Den Broek, J.; Van Mil, S.; Wernars, K. Diversity and Seasonal Fluctuations of the Dominant Members of the Bacterial Soil Community in a Wheat Field as Determined by Cultivation and Molecular Methods. Appl. Environ. Microbiol. 2001, 67, 2284–2291. [Google Scholar] [CrossRef] [Green Version]
- Whitelaw-Weckert, M.A.; Rahman, L.; Hutton, R.J.; Coombes, N. Permanent swards increase soil microbial counts in two Australian vineyards. Appl. Soil Ecol. 2007, 36, 224–232. [Google Scholar] [CrossRef]
- Guo, A.; Ding, L.; Tang, Z.; Zhao, Z.; Duan, G. Microbial response to CaCO3 application in an acid soil in southern China. J. Environ. Sci. 2019, 79, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Gkisakis, V.D.; Kollaros, D.; Bàrberi, P.; Livieratos, I.C.; Kabourakis, E.M. Soil Arthropod Diversity in Organic, Integrated, and Conventional Olive Orchards and Different Agroecological Zones in Crete, Greece. Agroecol. Sustain. Food Syst. 2015, 39, 276–294. [Google Scholar] [CrossRef]
- Sheikh, A.A.; Rehman, N.; Bhat, T.A.; Sofi, M.A.; Bhat, M.A.; Un Nabi, S.; Aijaz, C.; Sheikh, A.; Lone, G.; Sofi, A. Vertical distribution of soil arthropods in apple ecosystem of Kashmir. J. Entomol. Zool. Stud. JEZS 2017, 843, 843–846. [Google Scholar]
- Mantoni, C.; Di Musciano, M.; Fattorini, S. Use of microarthropods to evaluate the impact of fire on soil biological quality. J. Environ. Manag. 2020, 266, 110624. [Google Scholar] [CrossRef] [PubMed]


| Organism | Index | Int-Ir-T0 | Int-Ir-T1 | Int-Ir-T2 | Org-Ir-T0 | Org-Ir-T1 | Org-Ir-T2 | Int-R-T0 | Int-R-T1 | Int-R-T2 | Org-R-T0 | Org-R-T1 | Org-R-T2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fungi | S | 104 ± 16 | 1046 ± 37 | 1035 ± 20 | 1003 ± 19 | 976 ± 37 | 998 ± 30 | 967 ± 21 | 994 ± 26 | 896 ± 45 | 902 ± 30 (b) | 968 ± 27 (ab) | 977 ± 47 (a) |
| S.Chao1 | 1703 ± 40 | 1755 ± 82 | 1768 ± 47 | 1680 ± 25 | 1612 ± 66 | 1689 ± 60 | 1722 ± 40 (a) | 1696 ± 40 (ab) | 1518 ± 77 (b) | 1572 ± 52 | 1631 ± 43 | 1662 ± 73 | |
| H | 4.7 ± 0.1 | 4.7 ± 0.1 | 4.6 ± 0.1 | 4.6 ± 0.1 | 4.3 ± 0.2 | 4.6 ± 0.1 | 4.5 ± 0.1 | 4.6 ± 0.1 | 4.3 ± 0.2 | 4.3 ± 0.1 (b) | 4.5 ± 0.1 (ab) | 4.6 ± 0.2 (a) | |
| J | 0.67 ± 0.01 | 0.68 ± 0.01 | 0.67 ± 0.01 | 0.66 ± 0.01 | 0.63 ± 0.03 | 0.67 ± 0.01 | 0.66 ± 0.01 | 0.66 ± 0.01 | 0.64 ± 0.02 | 0.64 ± 0.01 (b) | 0.66 ± 0.01 (ab) | 0.67 ± 0.02 (a) | |
| Bacteria | S | 5452 ± 127 | 5508 ± 177 | 5243 ± 119 | 5379 ± 179 | 5499 ± 102 | 5477 ± 108 | 5916 ± 92 (a) | 5802 ± 162 (ab) | 5463 ± 149 (b) | 5991 ± 74 (a) | 5863 ± 105 (ab) | 5686 ± 103 (b) |
| S.Chao1 | 10,945 ± 284 | 10,439 ± 44 | 10,504 ± 343 | 10,824 ± 47 | 10,651 ± 33 | 10,906 ± 30 | 11,784 ± 212 (a) | 11,385 ± 403 (ab) | 10,562 ± 34 (b) | 11,933 ± 180 | 11,244 ± 26 | 11,204 ± 253 | |
| H | 6.99 ± 0.06 (ab) | 7.14 ± 0.06 (a) | 6.92 ± 0.05 (b) | 7.02 ± 0.07 | 7.16 ± 0.06 | 7.10 ± 0.06 | 7.33 ± 0.03 (a) | 7.38 ± 0.04 (a) | 7.16 ± 0.06 (b) | 7.33 ± 0.05 (ab) | 7.42 ± 0.04 (a) | 7.28 ± 0.04 (b) | |
| J | 0.81 ± 0.01 (ab) | 0.83 ± 0.01 (a) | 0.81 ± 0 (b) | 0.82 ± 0.01 | 0.83 ± 0.01 | 0.82 ± 0.01 | 0.84 ± 0.00 (b) | 0.85 ± 0.00 (a) | 0.83 ± 0.01 (b) | 0.84 ± 0.00 (b) | 0.86 ± 0.00 (a) | 0.84 ± 0.00 (b) | |
| Microartrhopods | Abundance | 8146 ± 4454 (ab) | 3261 ± 1183 (b) | 7643 ± 1665 (a) | 4401 ± 740 | 6070 ± 2928 | 4974 ± 1288 | 9776 ± 4563 | 3503 ± 1041 | 3611 ± 415 | 9751 ± 3804 | 6643 ± 1719 | 7305 ± 1197 |
| QBS | 85 ± 7 (a) | 65 ± 11 (b) | 82 ± 7 (ab) | 79 ± 11 | 83 ± 10 | 65 ± 10 | 114 ± 10 (a) | 76 ± 10 (b) | 118 ± 10 (a) | 127 ± 9 (ab) | 116 ± 10 (b) | 148 ± 9 (a) | |
| S | 6.0 ± 0.6 (ab) | 5.4 ± 0.8 (b) | 7.0 ± 0.4 (a) | 7.0 ± 0.6 | 6.0 ± 0.7 | 6.0 ± 0.8 | 8.0 ± 0.7 (a) | 6.0 ± 0.6 (b) | 8.0 ± 0.5 (a) | 8.7 ± 0.7 (ab) | 7.5 ± 0.5 (b) | 9.9 ± 0.5 (a) | |
| S.Chao1 | 9 ± 1 | 6 ± 1 | 8 ± 1 | 8 ± 1 | 7 ± 1 | 7 ± 1 | 11 ± 2 | 7 ± 1 | 10 ± 1 | 10 ± 1 | 11 ± 2 | 11 ± 1 | |
| H | 1.2 ± 0.1 | 1.1 ± 0.1 | 1.2 ± 0.1 | 1.1 ± 0.1 | 1.0 ± 0.1 | 1.1 ± 0.1 | 1.3 ± 0.1 (ab) | 0.9 ± 0.2 (b) | 1.4 ± 0.1 (a) | 1.4 ± 0.1 | 1.1 ± 0.1 | 1.4 ± 0.1 | |
| J | 0.70 ± 0.08 | 0.67 ± 0.06 | 0.63 ± 0.04 | 0.60 ± 0.05 | 0.62 ± 0.05 | 0.59 ± 0.08 | 0.62 ± 0.07 | 0.53 ± 0.06 | 0.65 ± 0.03 | 0.66 ± 0.06 | 0.57 ± 0.05 | 0.63 ± 0.04 | |
| Diatoms | S | 7 ± 2 | 6 ± 1 | 8 ± 2 | 5 ± 1 | 6 ± 1 | 6 ± 1 | ||||||
| S.Chao1 | 7 ± 2 | 6 ± 1 | 8 ± 2 | 5 ± 1 | 7 ± 1 | 7 ± 1 | |||||||
| H | 1.20 ± 0.07 (a) | 0.84 ± 0.05 (b) | 1.12 ± 0.07 (a) | 1.16 ± 0.03 (b) | 1.1 ± 0.1 (ab) | 1.31 ± 0.03 (a) | |||||||
| J | 0.65 ± 0.05 | 0.50 ± 0.04 | 0.58 ± 0.06 | 0.73 ± 0.02 | 0.64 ± 0.07 | 0.71 ± 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coller, E.; Oliveira Longa, C.M.; Morelli, R.; Zanoni, S.; Cersosimo Ippolito, M.C.; Pindo, M.; Cappelletti, C.; Ciutti, F.; Menta, C.; Zanzotti, R.; et al. Soil Communities: Who Responds and How Quickly to a Change in Agricultural System? Sustainability 2022, 14, 383. https://doi.org/10.3390/su14010383
Coller E, Oliveira Longa CM, Morelli R, Zanoni S, Cersosimo Ippolito MC, Pindo M, Cappelletti C, Ciutti F, Menta C, Zanzotti R, et al. Soil Communities: Who Responds and How Quickly to a Change in Agricultural System? Sustainability. 2022; 14(1):383. https://doi.org/10.3390/su14010383
Chicago/Turabian StyleColler, Emanuela, Claudia Maria Oliveira Longa, Raffaella Morelli, Sara Zanoni, Marco Cristiano Cersosimo Ippolito, Massimo Pindo, Cristina Cappelletti, Francesca Ciutti, Cristina Menta, Roberto Zanzotti, and et al. 2022. "Soil Communities: Who Responds and How Quickly to a Change in Agricultural System?" Sustainability 14, no. 1: 383. https://doi.org/10.3390/su14010383
APA StyleColler, E., Oliveira Longa, C. M., Morelli, R., Zanoni, S., Cersosimo Ippolito, M. C., Pindo, M., Cappelletti, C., Ciutti, F., Menta, C., Zanzotti, R., & Ioriatti, C. (2022). Soil Communities: Who Responds and How Quickly to a Change in Agricultural System? Sustainability, 14(1), 383. https://doi.org/10.3390/su14010383








