Pollutant-Removing Biofilter Strains Associated with High Ammonia and Hydrogen Sulfide Removal Rate in a Livestock Wastewater Treatment Facility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Measurement of Ammonia
2.2. Measurement of Sulfur Compounds
2.3. Sample Collections
2.4. DNA Extraction and Pyrosequencing
2.5. Sequence Processing and Taxonomic Analysis
2.6. Community Analysis Based on Lab-Scale Cultivation
2.7. Statistical Analysis
3. Result and Discussion
3.1. Pollutant Purification Functionality and Community Diversity of Biofilter Facilities
3.2. Taxonomic Composition of the Microbial Community and Pollutant Purification-Related Strains
3.3. The Effectiveness of Illumina MiSeq for Microbial Community Analysis Compared to the Culture-Based Analysis
3.4. Relationship between Pollutant Purification and the Microbial Community
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, B.; Du, Y.; Han, W.; Geng, Y.; Wang, Q.; Duan, Y.; Ren, Y.; Liu, D.; Chang, J.; Ge, Y. Reduce health damage cost of greenhouse gas and ammonia emissions by assembling plant diversity in floating constructed wetlands treating wastewater. J. Clean. Prod. 2020, 244, 118927. [Google Scholar] [CrossRef]
- Hussein, H.M. Environmental impact assessment of aquatic quality index for some private filtration stations in Diwaniyah governorate. Syst. Rev. Pharm. 2021, 12, 286–290. [Google Scholar]
- Zhu, X.; Dao, G.; Tao, Y.; Zhan, X.; Hu, H. A review on control of harmful algal blooms by plant-derived allelochemicals. J. Hazard. Mater. 2020, 123403. [Google Scholar]
- Hashem, S. Investigating the principles of water treatment and industrial wastewater. J. Eng. Indu. Res. 2021, 2, 44–55. [Google Scholar]
- Liu, D.H.; Lipták, B.G. Wastewater Treatment; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Wang, Y.; Li, L.; Xiong, R.; Guo, X.; Liu, J. Effects of aeration on microbes and intestinal bacteria in bioaerosols from the BRT of an indoor wastewater treatment facility. Sci. Total Environ. 2019, 648, 1453–1461. [Google Scholar] [CrossRef]
- Mouratib, R.; Achiou, B.; El Krati, M.; Younssi, S.A.; Tahiri, S. Low-cost ceramic membrane made from alumina-and silica-rich water treatment sludge and its application to wastewater filtration. J. Eur. Ceram. Soc. 2020, 40, 5942–5950. [Google Scholar] [CrossRef]
- Huang, J.L.; Wang, H.H.; Alam, F.; Cui, Y.W. Granulation of halophilic sludge inoculated with estuarine sediments for saline wastewater treatment. Sci. Total Environ. 2019, 682, 532–540. [Google Scholar] [CrossRef]
- Ulu, F.; Kobya, M. Ammonia removal from wastewater by air stripping and recovery struvite and calcium sulphate precipitations from anesthetic gases manufacturing wastewater. J. Water Process. Eng. 2020, 38, 101641. [Google Scholar] [CrossRef]
- Campo, R.; Sguanci, S.; Caffaz, S.; Mazzoli, L.; Ramazzotti, M.; Lubello, C.; Lotti, T. Efficient carbon, nitrogen and phosphorus removal from low C/N real domestic wastewater with aerobic granular sludge. Bioresour. Technol. 2020, 305, 122961. [Google Scholar] [CrossRef]
- Dos Santos, P.; Daniel, L. A review: Organic matter and ammonia removal by biological activated carbon filtration for water and wastewater treatment. Int. J. Environ. Sci. Technol. 2020, 17, 591–606. [Google Scholar] [CrossRef]
- Duesel, B.F., Jr.; Clerkin, C.; Laurent, B.N. Apparatus and Method for Treating Hydrogen Sulfide and Ammonia in Wastewater Streams. U.S. Patent 16/368,317, 3 October 2019. [Google Scholar]
- Watsuntorn, W.; Ruangchainikom, C.; Rene, E.R.; Lens, P.N.; Chulalaksananukul, W. Comparison of sulphide and nitrate removal from synthetic wastewater by pure and mixed cultures of nitrate-reducing, sulphide-oxidizing bacteria. Bioresour. Technol. 2019, 272, 40–47. [Google Scholar] [CrossRef]
- Mondal, M.; Biswas, J.K.; Tsang, Y.F.; Sarkar, B.; Sarkar, D.; Rai, M.; Sarkar, S.K.; Hooda, P.S. A wastewater bacterium Bacillus sp. KUJM2 acts as an agent for remediation of potentially toxic elements and promoter of plant (Lens culinaris) growth. Chemosphere 2019, 232, 439–452. [Google Scholar] [CrossRef]
- Tong, Y.; Wang, M.; Peñuelas, J.; Liu, X.; Paerl, H.W.; Elser, J.J.; Sardans, J.; Couture, R.M.; Larssen, T.; Hu, H. Improvement in municipal wastewater treatment alters lake nitrogen to phosphorus ratios in populated regions. Proc. Natl. Acad. Sci. USA 2020, 117, 11566–11572. [Google Scholar] [CrossRef]
- Al Momani, F.; Örmeci, B. Assessment of algae-based wastewater treatment in hot climate region: Treatment performance and kinetics. Process. Saf. Environ. Prot. 2020, 141, 140–149. [Google Scholar] [CrossRef]
- Sonkar, M.; Kumar, V.; Dutt, D. Use of paper mill sludge and sewage sludge powder as nitrogen and phosphorus sources with bacterial consortium for the treatment of paper industry wastewater. Biocatal. Agric. Biotechnol. 2020, 30, 101843. [Google Scholar] [CrossRef]
- Fan, F.; Xu, R.; Wang, D.; Meng, F. Application of activated sludge for odor control in wastewater treatment plants: Approaches, advances and outlooks. Water Res. 2020, 115915. [Google Scholar] [CrossRef]
- Sookpanya, P.; Suadee, W.; Pharamat, T.; Suwannachot, S.; Ratcha, M. Kinetics of organic and inorganic degradation in biofilter using isolated bacteria from petrochemical wastewater treatment plant. Thai. Environ. Eng. J. 2020, 34, 57–65. [Google Scholar]
- Kim, D.H.; Han, K.I.; Kwon, H.J.; Kim, M.G.; Kim, Y.G.; Choi, D.H.; Lee, K.C.; Suh, M.K.; Kim, H.S.; Lee, J.S. Comamonas flocculans sp. nov., a floc-forming bacterium isolated from livestock wastewater. Curr. Microbiol. 2020, 1–7. [Google Scholar] [CrossRef]
- Blandin, G.; Rosselló, B.; Monsalvo, V.M.; Batlle-Vilanova, P.; Viñas, J.M.; Rogalla, F.; Comas, J. Volatile fatty acids concentration in real wastewater by forward osmosis. J. Membr. Sci. 2019, 575, 60–70. [Google Scholar] [CrossRef]
- Ouyang, E.; Liu, Y.; Ouyang, J.; Wang, X. Effects of different wastewater characteristics and treatment techniques on the bacterial community structure in three pharmaceutical wastewater treatment systems. Environ. Technol. 2019, 40, 329–341. [Google Scholar] [CrossRef]
- Dijkman, H.; Strous, M. Process for the Treatment of Wastewater Containing Organic Material and Ammonia. U.S. Patent 16/317,451, 25 July 2019. [Google Scholar]
- Li, S.; Zhang, Y.; Yin, S.; Wang, X.; Liu, T.; Deng, Z. Analysis of microbial community structure and degradation of ammonia nitrogen in groundwater in cold regions. Environ. Sci. Pollut. Res. 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kalniņš, M.; Bērziņš, A.; Gudrā, D.; Megnis, K.; Fridmanis, D.; Danilko, P.; Muter, O. Selective enrichment of heterotrophic nitrifiers Alcaligenaceae and Alcanivorax spp. from industrial wastewaters. AIMS Microbiol. 2020, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Oceanospirillalesord. nov. In Bergey’s Manual® of Systematic Bacteriology; Springer: Berlin, Germany, 2005; pp. 270–323. [Google Scholar]
- Blázquez, E.; Guisasola, A.; Gabriel, D.; Baeza, J.A. Application of bioelectrochemical systems for the treatment of wastewaters with sulfur species. In Microbial Electrochemical Technology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 641–663. [Google Scholar]
- Kadnikov, V.; Mardanov, A.; Beletsky, A.; Antsiferov, D.; Kovalyova, A.; Karnachuk, O.; Ravin, N. Sulfur-oxidizing bacteria dominate in the water from a flooded coal mine shaft in Kuzbass. Microbiology 2019, 88, 120–123. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Merkel, A.; Muyzer, G. Thioalkalibacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley and Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Tsallagov, S.I.; Sorokin, D.Y.; Tikhonova, T.V.; Popov, V.O.; Muyzer, G. Comparative genomics of Thiohalobacter thiocyanaticus HRh1T and Guyparkeria sp. SCN-R1, halophilic chemolithoautotrophic sulfur-oxidizing gammaproteobacteria capable of using thiocyanate as energy source. Front. Microbiol. 2019, 10, 898. [Google Scholar] [CrossRef] [Green Version]
- Boden, R. Reclassification of Halothiobacillus hydrothermalis and Halothiobacillus halophilus to Guyparkeria gen. nov. in the Thioalkalibacteraceae fam. nov., with emended descriptions of the genus Halothiobacillus and family Halothiobacillaceae. Int. J. Syst. Evol. Microbiol. 2017, 67, 3919–3928. [Google Scholar] [CrossRef]
- Bambauer, A.; Rainey, F.A.; Stackebrandt, E.; Winter, J. Characterization of Aquamicrobium defluvii gen. nov. sp. nov., a thiophene-2-carboxylate-metabolizing bacterium from activated sludge. Arch. Microbiol. 1998, 169, 293–302. [Google Scholar] [CrossRef]
- Fernandez-Gonzalez, N.; Sierra-Alvarez, R.; Field, J.A.; Amils, R.; Sanz, J.L. Adaptation of granular sludge microbial communities to nitrate, sulfide, and/or p-cresol removal. Int. Microbiol. 2019, 22, 305–316. [Google Scholar] [CrossRef]
- Hu, H.Y.; Fujie, K.; Nakagome, H.; Urano, K.; Katayama, A. Quantitative analyses of the change in microbial diversity in a bioreactor for wastewater treatment based on respiratory quinones. Water Res. 1999, 33, 3263–3270. [Google Scholar] [CrossRef]
- Romero, F.; Sabater, S.; Font, C.; Balcázar, J.L.; Acuña, V. Desiccation events change the microbial response to gradients of wastewater effluent pollution. Water Res. 2019, 151, 371–380. [Google Scholar] [CrossRef]
- Wang, X.; Wen, X.; Xia, Y.; Hu, M.; Zhao, F.; Ding, K. Ammonia oxidizing bacteria community dynamics in a pilot-scale wastewater treatment plant. PloS. ONE 2012, 7, e36272. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Tripathi, S.; Chandra, R. Metagenomic analysis for profiling of microbial communities and tolerance in metal-polluted pulp and paper industry wastewater. Bioresour. Technol. 2021, 324, 124681. [Google Scholar] [CrossRef]
- Datta, S.; Rajnish, K.N.; Samuel, M.S.; Pugazlendhi, A.; Selvarajan, E. Metagenomic applications in microbial diversity, bioremediation, pollution monitoring, enzyme and drug discovery. A review. Environ. Chem. Lett. 2020, 18, 1229–1241. [Google Scholar] [CrossRef]
- Korea, S. KOSTAT. Population Trends and Projections of the World and Korea. 2015. Available online: http://kostat.go.kr/portal/eng/pressReleases/8/8/index.board?bmode=read&aSeq=347597&pag (accessed on 8 July 2015).
- Wang, Y.C.; Han, M.F.; Jia, T.P.; Hu, X.R.; Zhu, H.Q.; Tong, Z.; Lin, Y.T.; Wang, C.; Liu, D.Z.; Peng, Y.Z. Emissions, measurement, and control of odor in livestock farms: A review. Sci. Total Environ. 2021, 145735. [Google Scholar] [CrossRef]
- Bolleter, W.; Bushman, C.; Tidwell, P.W. Spectrophotometric determination of ammonia as indophenol. Anal. Chem. 1961, 33, 592–594. [Google Scholar] [CrossRef]
- Leck, C.; Baagander, L.E. Determination of reduced sulfur compounds in aqueous solutions using gas chromatography-flame photometric detection. Anal. Chem. 1988, 60, 1680–1683. [Google Scholar] [CrossRef]
- Hibi, K.; Tsuda, T.; Takeuchi, T.; Nakanishi, T.; Ishii, D. Studies of open-tubular microcapillary liquid chromatography: III. β, β′-oxydipropionitrile and ethylene glycol stationary phases. J. Chromatogr. A 1979, 175, 105–111. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, P.Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PloS ONE 2009, 4, e7401. [Google Scholar] [CrossRef] [Green Version]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.; Moon, T.; Yoon, S.; Weissman, T. DUDE-Seq: Fast, flexible, and robust denoising for targeted amplicon sequencing. PLoS ONE 2017, 12, e0181463. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. CD-HIT: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing Mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Reasoner, D.J.; Geldreich, E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- El Naker, N.A.; Hasan, S.W.; Yousef, A.F. Impact of current density on the function and microbial community structure in electro-bioreactors. J. Hazard. Mater. 2019, 368, 877–884. [Google Scholar] [CrossRef]
- Chen, G.; Huang, J.; Fang, Y.; Zhao, Y.; Tian, X.; Jin, Y.; Zhao, H. Microbial community succession and pollutants removal of a novel carriers enhanced duckweed treatment system for rural wastewater in Dianchi Lake basin. Bioresour. Technol. 2019, 276, 8–17. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Khurana, S.P. Importance of actinobacteria for bioremediation. In Plant Biotechnology: Progress in Genomic Era; Springer: Berlin, Germany, 2019; pp. 277–307. [Google Scholar]
- Zhao, Y.; Shu, X.; Tu, Q.; Yang, Y.; Liu, C.; Fu, D.; Wei, L.; Duan, C. Pollutant removal from agricultural drainage water using a novel double-layer ditch with biofilm carriers. Bioresour. Technol. 2020, 310, 123344. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, H.; Zhang, P.; Wu, Y.; Gou, X.; Song, Y.; Tian, Z.; Zeng, G. Two-stage anoxic/oxic combined membrane bioreactor system for landfill leachate treatment: Pollutant removal performances and microbial community. Bioresour. Technol. 2017, 243, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.J.; Wang, Z.; Huang, T.L.; Zhang, H.; Zhang, H. Simultaneous removal of nitrate, phosphorous and cadmium using a novel multifunctional biomaterial immobilized aerobic strain Proteobacteria Cupriavidus H29. Bioresour. Technol. 2020, 307, 123196. [Google Scholar]
- LaFrentz, S.; Abarca, E.; Mohammed, H.H.; Cuming, R.; Arias, C.R. Characterization of the normal equine conjunctival bacterial community using culture-independent methods. Vet. Ophthalmol. 2020, 23, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Sani, R.K. Molecular techniques to assess microbial community structure, function, and dynamics in the environment. In Microbes and Microbial Technology; Springer: Berlin, Germany, 2011; pp. 29–57. [Google Scholar]
- Suttner, B.; Johnston, E.R.; Orellana, L.H.; Rodriguez, R.L.M.; Hatt, J.K.; Carychao, D.; Carter, M.Q.; Cooley, M.B.; Konstantinidis, K.T. Metagenomics as a public health risk assessment tool in a study of natural creek sediments influenced by agricultural and livestock runoff: Potential and limitations. Appl. Environ. Microbiol. 2020, 86, e02525. [Google Scholar] [CrossRef] [PubMed]
- Efron, B.; Halloran, E.; Holmes, S. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 1996, 93, 13429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pla, L. Bootstrap confidence intervals for the Shannon biodiversity index: A simulation study. J. Agr. Biol. Envir. St. 2004, 9, 42. [Google Scholar] [CrossRef]
- Daubin, V.; Gouy, M.; Perriere, G. A phylogenomic approach to bacterial phylogeny: Evidence of a core of genes sharing a common history. Genome Res. 2002, 12, 1080–1090. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Pandey, A.K.; Udayan, A.; Kumar, S. Role of microbial community and metal-binding proteins in phytoremediation of heavy metals from industrial wastewater. Bioresour. Technol. 2021, 124750. [Google Scholar] [CrossRef]
- Kannan, A.D.; Evans, P.; Parameswaran, P. Long-term microbial community dynamics in a pilot-scale gas sparged anaerobic membrane bioreactor treating municipal wastewater under seasonal variations. Bioresour. Technol. 2020, 310, 123425. [Google Scholar] [CrossRef]
- Ferro, L.; Hu, Y.O.; Gentili, F.G.; Andersson, A.F.; Funk, C. DNA metabarcoding reveals microbial community dynamics in a microalgae-based municipal wastewater treatment open photobioreactor. Algal Res. 2020, 51, 102043. [Google Scholar] [CrossRef]
- Ospina-Betancourth, C.; Acharya, K.; Allen, B.; Entwistle, J.; Head, I.M.; Sanabria, J.; Curtis, T.P. Enrichment of nitrogen-fixing bacteria in a nitrogen-deficient wastewater treatment system. Environ. Sci. Technol. 2020, 54, 3539–3548. [Google Scholar] [CrossRef]
- Chen, J.; Wei, J.; Ma, C.; Yang, Z.; Li, Z.; Yang, X.; Wang, M.; Zhang, H.; Hu, J.; Zhang, C. Photosynthetic bacteria-based technology is a potential alternative to meet sustainable wastewater treatment requirement? Environ. Int. 2020, 137, 105417. [Google Scholar] [CrossRef]
- Pan, Z.; Zhou, J.; Lin, Z.; Wang, Y.; Zhao, P.; Zhou, J.; Liu, S.; He, X. Effects of COD/TN ratio on nitrogen removal efficiency, microbial community for high saline wastewater treatment based on heterotrophic nitrification-aerobic denitrification process. Bioresour. Technol. 2020, 301, 122726. [Google Scholar] [CrossRef]
- Bedoya, K.; Hoyos, O.; Zurek, E.; Cabarcas, F.; Alzate, J.F. Annual microbial community dynamics in a full-scale anaerobic sludge digester from a wastewater treatment plant in Colombia. Sci. Total Environ. 2020, 726, 138479. [Google Scholar] [CrossRef]
- González-Morales, S.I.; Pacheco-Gutiérrez, N.B.; Ramírez-Rodríguez, C.A.; Brito-Bello, A.A.; Estrella-Hernández, P.; Herrera-Estrella, L.; López-Arredondo, D.L. Metabolic engineering of phosphite metabolism in Synechococcus elongatus PCC 7942 as an effective measure to control biological contaminants in outdoor raceway ponds. Biotechnol. Biofuels 2020, 13, 1–19. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Z.; Shen, J.; Kang, J.; Zhang, X.; Li, J.; Zhao, X. Effect of carbon source on pollutant removal and microbial community dynamics in treatment of swine wastewater containing antibiotics by aerobic granular sludge. Chemosphere 2020, 260, 127544. [Google Scholar] [CrossRef]
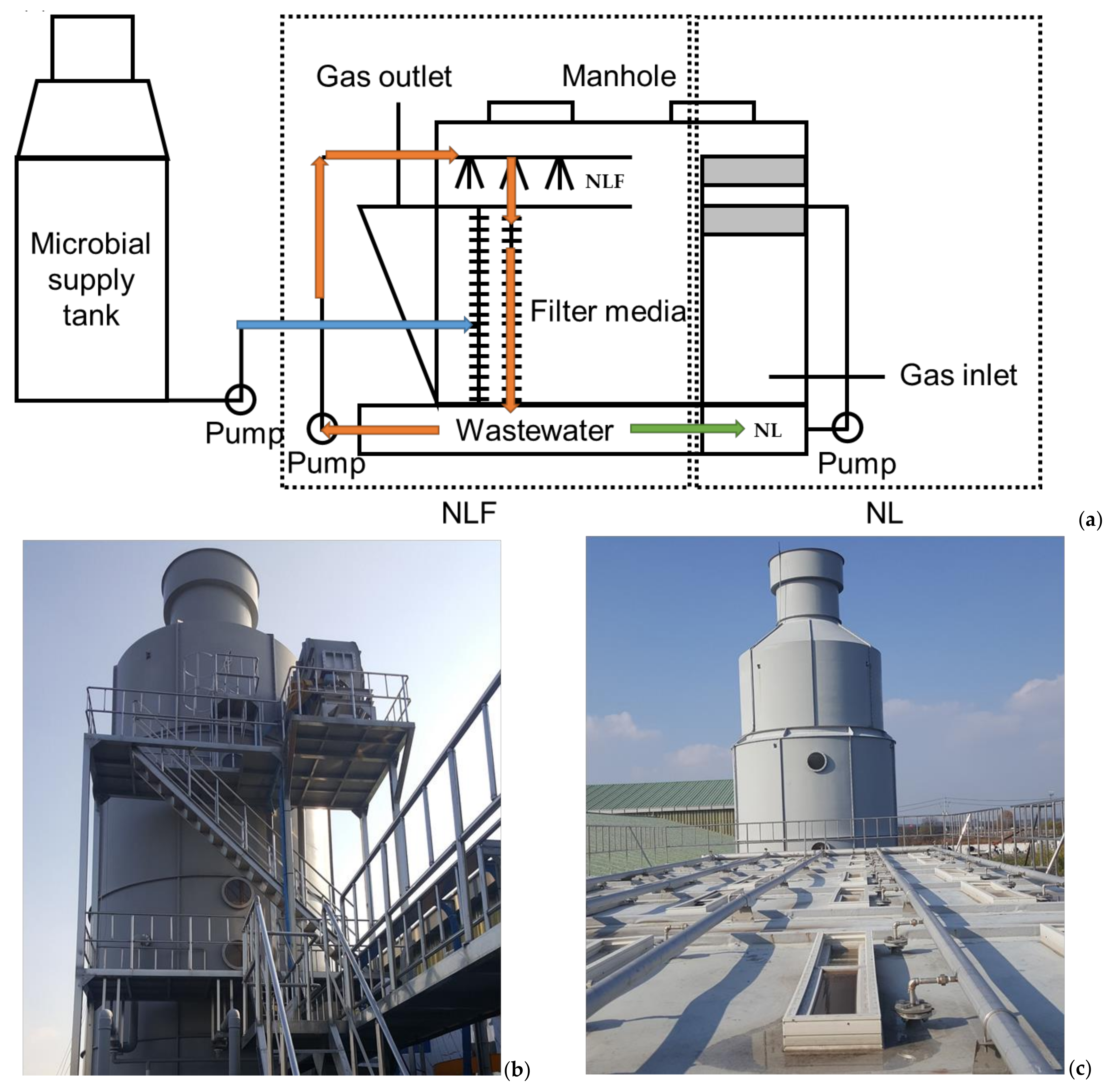
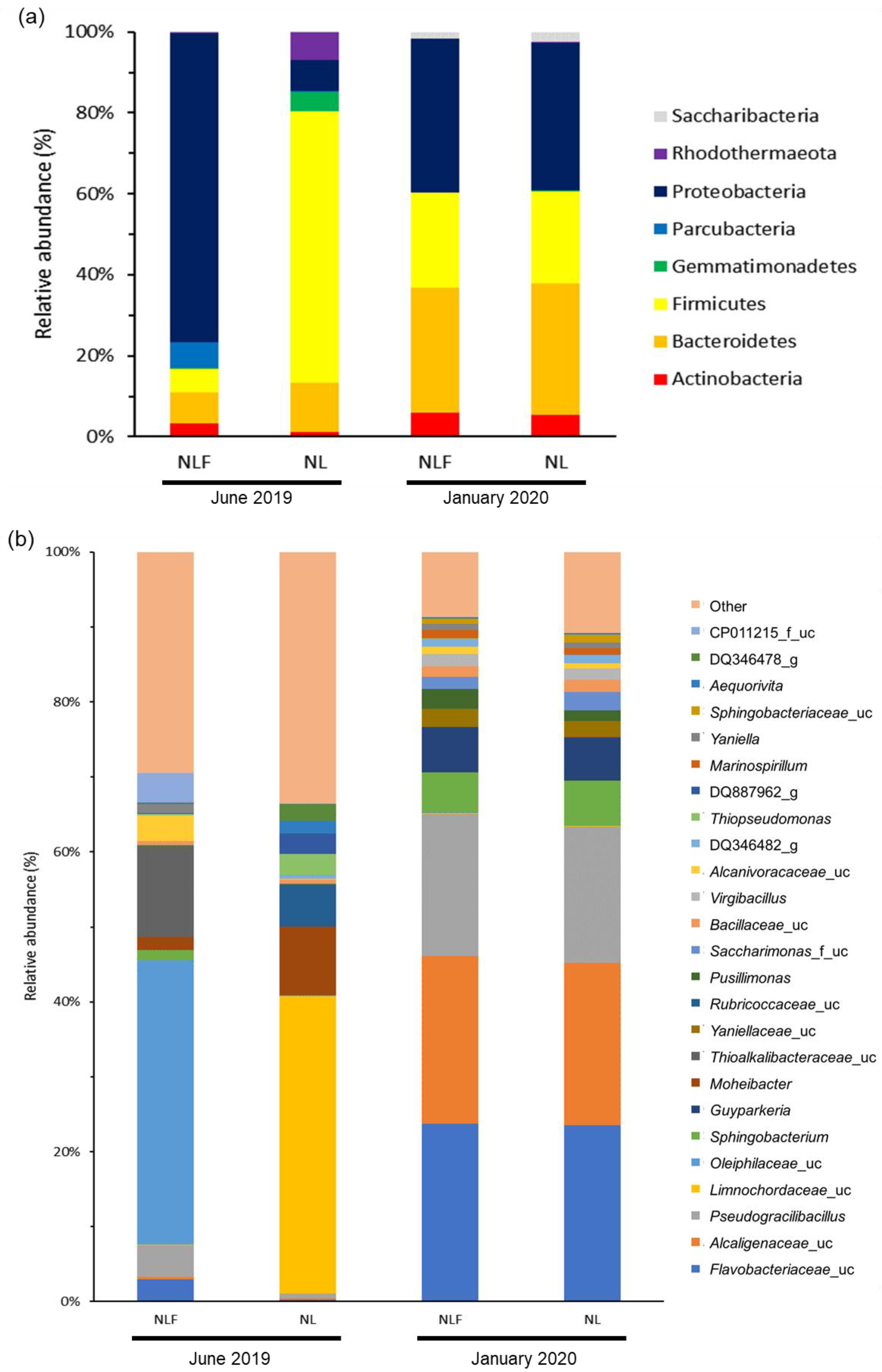
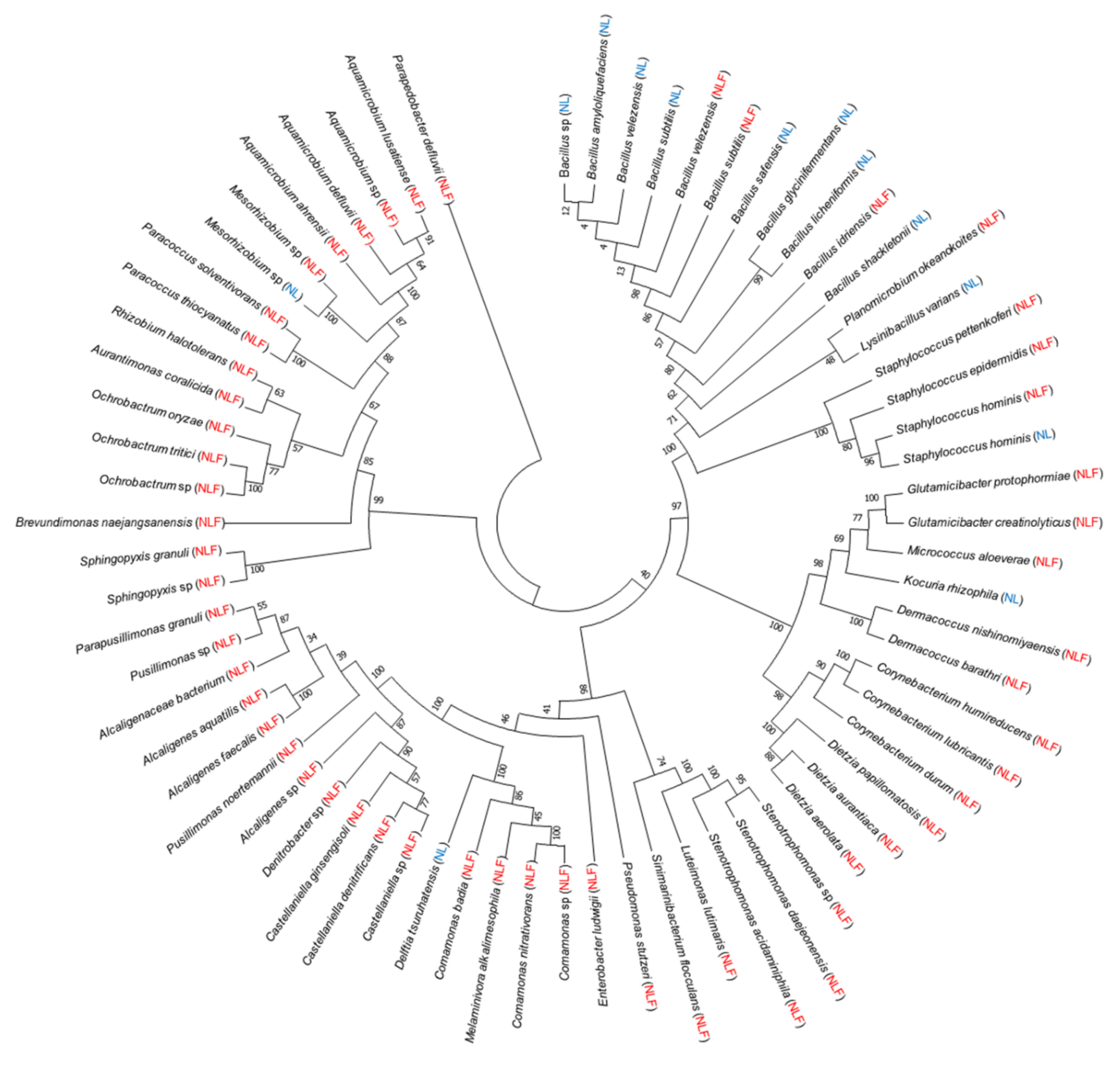
| Evaluation Item | June 2019 | January 2020 | |||
|---|---|---|---|---|---|
| NLF | NL | NLF | NL | ||
| Pollutant concentration | Ammonia (ppm) | 15.30 ± 0.00 | 2.00 ± 0.00 | 15.30 ± 0.14 | 5.50 ± 1.41 |
| Hydrogen sulfide (ppm) | 10.58 ± 0.38 | 0.03 ± 0.01 | 17.17 ± 9.93 | 1.01 ± 0.40 | |
| Sequencing results | Validated reads | 23,958 | 43,833 | 52,922 | 60,290 |
| Number of OTUs a | 740 | 1884 | 852 | 991 | |
| Diversity indicators | Chao1 b | 748.23 | 1891.03 | 857.14 | 997.21 |
| Shannon c | 3.94 | 4.50 | 3.81 | 4.07 | |
| Simpson d | 0.08 | 0.08 | 0.07 | 0.05 | |
| Goods Coverage e | 99.73 | 99.77 | 99.9 | 99.9 | |
| Bacterial Group Characteristic Classification | Taxonomy | June 2019 | January 2020 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NLF | NL | NLF | NL | ||||||||
| Phylum | Genus | Freq a | % b | Freqa | % b | Freq a | % b | Freq a | % b | ||
| Dominant strain | Bacteroidetes | Flavobacteriaceae | 701 | 3.16 | 96 | 0.23 | 12,558 | 23.83 | 14166 | 23.59 | |
| Bacteroidetes | Moheibacter | 421 | 1.89 | 4025 | 9.67 | 6 | 0.01 | 6 | 0.01 | ||
| Bacteroidetes | Sphingobacterium | 319 | 1.44 | 0 | 0.00 | 2865 | 5.44 | 3599 | 5.99 | ||
| Firmicutes | Pseudogracilibacillus | 997 | 4.49 | 284 | 0.68 | 10,019 | 19.01 | 10966 | 18.26 | ||
| Firmicutes | Limnochordaceae | 44 | 0.20 | 17,416 | 41.83 | 41 | 0.08 | 59 | 0.10 | ||
| Functional strain | Strain related to ammonia removal | Proteobacteria | Alcaligenaceae | 89 | 0.40 | 98 | 0.24 | 11,849 | 22.48 | 13,063 | 21.75 |
| Proteobacteria | Oleiphilaceae | 9084 | 40.89 | 7 | 0.02 | 67 | 0.13 | 30 | 0.05 | ||
| Strain related to volatile fatty acid removal | Actinobacteria | Dietzia | 0 | 0.00 | 8 | 0.02 | 24 | 0.05 | 20 | 0.03 | |
| Strain related to hydrogen sulfide removal | Proteobacteria | Comamonas | 159 | 0.72 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Proteobacteria | Thioalkalibacteraceae | 2869 | 12.91 | 0 | 0 | 5 | 0.01 | 21 | 0.04 | ||
| Proteobacteria | Guyparkeria | 0 | 0 | 0 | 0 | 3153 | 5.98 | 3519 | 5.86 | ||
| Proteobacteria | Aquamicrobium | 102 | 0.46 | 5 | 0.01 | 0 | 0 | 6 | 0.01 | ||
| Taxonomic Affinity | Closest Match | GenBank Accession Number | Sequence Similarity (%) | Abundance (%) |
|---|---|---|---|---|
| Alcaligenaceae bacterium | Alcaligenaceae bacterium | MH553029 | 99 | 0.69 |
| Alcaligenes aquatilis | Alcaligenes aquatilis | JX986974 | 100 | 0.69 |
| Alcaligenes faecalis | Alcaligenes faecalis | AUBT01000026 | 99 | 11.71 |
| Alcaligenes sp. | Alcaligenes sp. | DQ421393 | 98 | 0.69 |
| Aquamicrobium ahrensii | Aquamicrobium ahrensii | AM884149 | 100 | 0.69 |
| Aquamicrobium defluvii | Aquamicrobium defluvii | KU163265 | 99 | 1.38 |
| Aquamicrobium lusatiense | Aquamicrobium lusatiense | KM210272 | 99 | 5.52 |
| Aquamicrobium sp. | Aquamicrobium sp. | LN881594 | 99 | 0.69 |
| Aurantimonas coralicida | Aurantimonas coralicida | ATXK01000033 | 100 | 0.69 |
| Bacillus idriensis | Bacillus idriensis | FR682742 | 99 | 3.45 |
| Bacillus subtilis | Bacillus subtilis | AMXN01000021 | 99 | 0.69 |
| Bacillus velezensis | Bacillus velezensis | AY603658 | 99 | 0.69 |
| Brevundimonas naejangsanensis | Brevundimonas naejangsanensis | CP015614 | 99 | 0.69 |
| Castellaniella denitrificans | Castellaniella denitrificans | U82826 | 99 | 0.69 |
| Castellaniella ginsengisoli | Castellaniella ginsengisoli | EU873313 | 99 | 1.38 |
| Castellaniella sp. | Castellaniella sp. | KM210263 | 100 | 1.38 |
| Comamonas badia | Comamonas badia | NR_041011 | 94 | 1.38 |
| Comamonas nitrativorans | Comamonas nitrativorans | AJ251577 | 100 | 3.45 |
| Comamonas sp. | Comamonas sp. | JX271943 | 99 | 2.76 |
| Corynebacterium durum | Corynebacterium durum | Z97069 | 99 | 0.69 |
| Corynebacterium humireducens | Corynebacterium humireducens | CP005286 | 99 | 0.69 |
| Corynebacterium lubricantis | Corynebacterium lubricantis | FM173119 | 96 | 0.69 |
| Denitrobacter sp. | Denitrobacter sp. | EF471227 | 98 | 0.69 |
| Dermacoccus barathri | Dermacoccus barathri | AY894328 | 100 | 0.69 |
| Dermacoccus nishinomiyaensis | Dermacoccus nishinomiyaensis | MF952724 | 100 | 0.69 |
| Dietzia aerolata | Dietzia aerolata | FM995533 | 99 | 8.26 |
| Dietzia aurantiaca | Dietzia aurantiaca | FR821260 | 99 | 2.76 |
| Dietzia papillomatosis | Dietzia papillomatosis | BCSL01000097 | 98 | 0.69 |
| Enterobacter ludwigii | Enterobacter ludwigii | JTLO01000001 | 99 | 0.69 |
| Glutamicibacter creatinolyticus | Glutamicibacter creatinolyticus | CP034412 | 100 | 0.69 |
| Glutamicibacter protophormiae | Glutamicibacter protophormiae | X80745 | 100 | 1.38 |
| Luteimonas lutimaris | Luteimonas lutimaris | NR_117455 | 99 | 0.69 |
| Melaminivora alkalimesophila | Melaminivora alkalimesophila | JQ676982 | 99 | 2.07 |
| Mesorhizobium sp. | Mesorhizobium sp. | KM210274 | 99 | 4.83 |
| Micrococcus aloeverae | Micrococcus aloeverae | KF524364 | 99 | 1.38 |
| Ochrobactrum oryzae | Ochrobactrum oryzae | AM041247 | 100 | 1.38 |
| Ochrobactrum sp. | Ochrobactrum sp. | AM231041 | 100 | 2.07 |
| Ochrobactrum tritici | Ochrobactrum tritici | AJ242584 | 100 | 0.69 |
| Paracoccus solventivorans | Paracoccus solventivorans | NR_042714 | 99 | 1.38 |
| Paracoccus thiocyanatus | Paracoccus thiocyanatus | NR_113663 | 100 | 1.38 |
| Parapedobacter defluvii | Parapedobacter defluvii | KY612414 | 99 | 0.69 |
| Parapusillimonas granuli | Parapusillimonas granuli | DQ466075 | 98 | 0.69 |
| Planomicrobium okeanokoites | Planomicrobium okeanokoites | D55729 | 99 | 0.69 |
| Pseudomonas stutzeri | Pseudomonas stutzeri | CP002881 | 99 | 5.52 |
| Pusillimonas noertemannii | Pusillimonas noertemannii | NR_043129 | 99 | 0.69 |
| Pusillimonas sp. | Pusillimonas sp. | FJ529031 | 99 | 1.38 |
| Sinimarinibacterium flocculans | Sinimarinibacterium flocculans | HQ875491 | 94 | 6.89 |
| Sphingopyxis granuli | Sphingopyxis granuli | BCUA01000059 | 100 | 1.38 |
| Sphingopyxis sp. | Sphingopyxis sp. | CP026381 | 100 | 1.38 |
| Staphylococcus epidermidis | Staphylococcus epidermidis | CP030246 | 99 | 0.69 |
| Staphylococcus hominis | Staphylococcus hominis | X66101 | 99 | 0.69 |
| Staphylococcus pettenkoferi | Staphylococcus pettenkoferi | AF322002 | 99 | 0.69 |
| Stenotrophomonas acidaminiphila | Stenotrophomonas acidaminiphila | LDJO01000053 | 99 | 1.38 |
| Stenotrophomonas daejeonensis | Stenotrophomonas daejeonensis | LDJP01000061 | 99 | 1.38 |
| Stenotrophomonas sp. | Stenotrophomonas sp. | AB180662 | 98 | 1.38 |
| Rhizobium halotolerans | Rhizobium halotolerans | JX307098 | 100 | 0.69 |
| Taxonomic Affinity | Closest Match | GenBank Accession Number | Sequence Similarity (%) | Abundance (%) |
|---|---|---|---|---|
| Bacillus amyloliquefaciens | Bacillus amyloliquefaciens | CP031424 | 100 | 15.62 |
| Bacillus glycinifermentans | Bacillus glycinifermentans | CP023481 | 100 | 3.13 |
| Bacillus licheniformis | Bacillus licheniformis | CP031126 | 100 | 3.13 |
| Bacillus safensis | Bacillus safensis | CP010405 | 100 | 6.24 |
| Bacillus shackletonii | Bacillus shackletonii | NR_025373 | 99 | 3.13 |
| Bacillus sp. | Bacillus sp. | CP030937 | 100 | 12.49 |
| Bacillus subtilis | Bacillus subtilis | CP029052 | 100 | 3.13 |
| Bacillus velezensis | Bacillus velezensis | CP023075 | 100 | 34.37 |
| Delftia tsuruhatensis | Delftia tsuruhatensis | MH478206 | 100 | 3.13 |
| Kocuria rhizophila | Kocuria rhizophila | KP345929 | 100 | 6.24 |
| Lysinibacillus varians | Lysinibacillus varians | AY082370 | 99 | 3.13 |
| Mesorhizobium sp. | Mesorhizobium sp. | KM210274 | 99 | 3.13 |
| Staphylococcus hominis | Staphylococcus hominis | CP014107 | 100 | 3.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.-H.; Yun, H.-S.; Kim, Y.-S.; Kim, J.-G. Pollutant-Removing Biofilter Strains Associated with High Ammonia and Hydrogen Sulfide Removal Rate in a Livestock Wastewater Treatment Facility. Sustainability 2021, 13, 7358. https://doi.org/10.3390/su13137358
Kim D-H, Yun H-S, Kim Y-S, Kim J-G. Pollutant-Removing Biofilter Strains Associated with High Ammonia and Hydrogen Sulfide Removal Rate in a Livestock Wastewater Treatment Facility. Sustainability. 2021; 13(13):7358. https://doi.org/10.3390/su13137358
Chicago/Turabian StyleKim, Dong-Hyun, Hyun-Sik Yun, Young-Saeng Kim, and Jong-Guk Kim. 2021. "Pollutant-Removing Biofilter Strains Associated with High Ammonia and Hydrogen Sulfide Removal Rate in a Livestock Wastewater Treatment Facility" Sustainability 13, no. 13: 7358. https://doi.org/10.3390/su13137358
APA StyleKim, D.-H., Yun, H.-S., Kim, Y.-S., & Kim, J.-G. (2021). Pollutant-Removing Biofilter Strains Associated with High Ammonia and Hydrogen Sulfide Removal Rate in a Livestock Wastewater Treatment Facility. Sustainability, 13(13), 7358. https://doi.org/10.3390/su13137358






