Effects of Rhizophagus intraradices on Plant Growth and the Composition of Microbial Communities in the Roots of Continuous Cropping Soybean at Maturity
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Arbuscular Mycorrhizal Inocula
2.3. Experimental Design
2.4. Plant Growth Conditions
2.5. Plant Harvest
2.6. Mycorrhizal Colonization and Soybean Growth Analysis
2.7. Illumina Sequencing
2.8. Processing of Sequencing Data
2.9. Statistical Analysis
3. Results
3.1. Mycorrhizal Colonization Assessment
3.2. Effects on Soybean Plants Growth
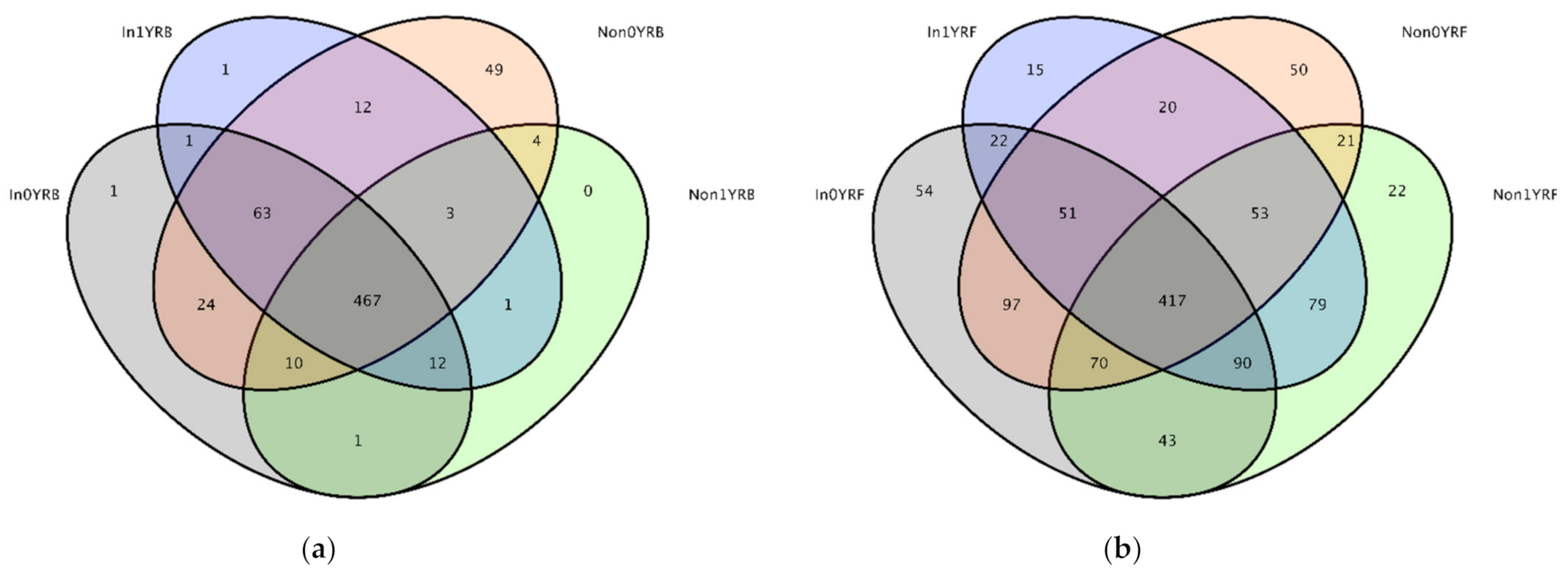
3.3. Diversity and Abundance of Microbes in the Roots of Continuous Cropping Soybean
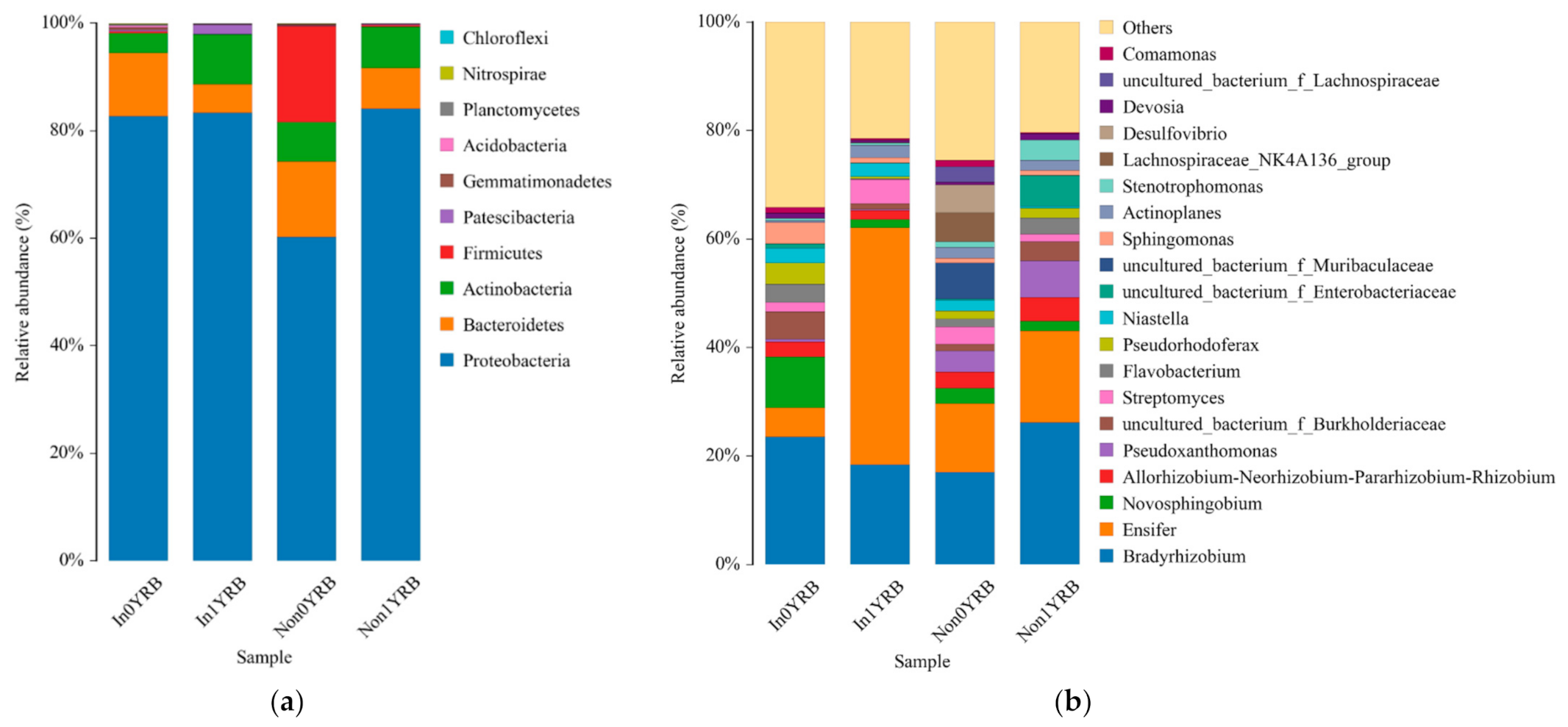
3.4. Effects of R. intraradices and Continuous Cropping Regimes on Bacterial Communities
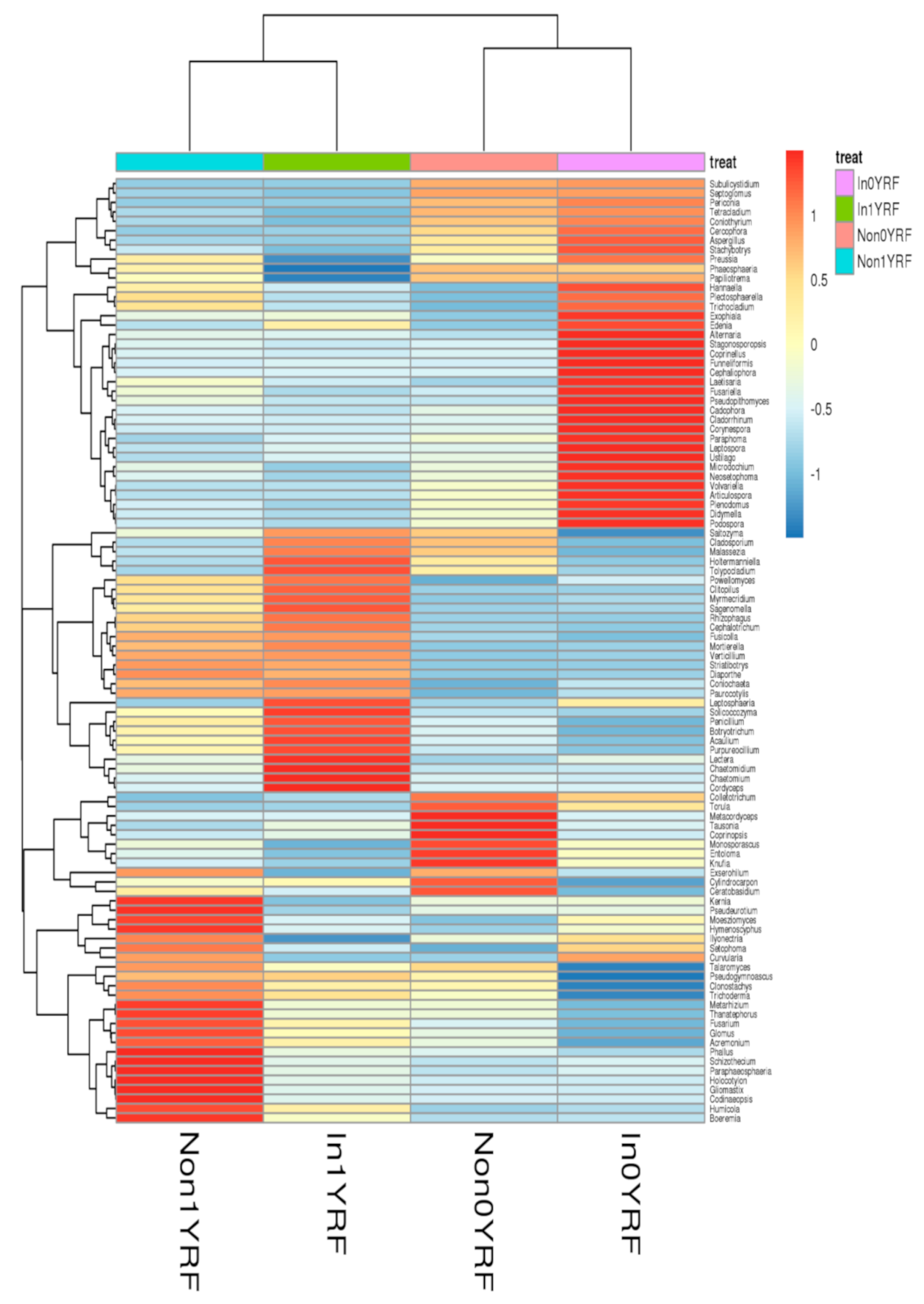
3.5. Effects of R. intraradices and Continuous Cropping Regimes on Fungal Communities
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cui, G.J.; Ai, S.Y.; Chen, K.; Wang, X.R. Arbuscular mycorrhiza augments cadmium tolerance in soybean by altering accumulation and partitioning of nutrient elements, and related gene expression. Ecotox. Environ. Saf. 2019, 171, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Marro, N.; Cofré, N.; Grilli, G.; Alvarez, C.; Labuckas, D.; Maestri, D.; Urcelay, C. Soybean yield, protein content and oil quality in response to interaction of arbuscular mycorrhizal fungi and native microbial populations from mono- and rotation-cropped soils. Appl. Soil Ecol. 2020, 152, 103575. [Google Scholar] [CrossRef]
- Liu, H.; Pan, F.J.; Han, X.Z.; Song, F.B.; Zhang, Z.M.; Yan, J.; Xu, Y.L. A comprehensive analysis of the response of the fungal community structure to long-term continuous cropping in three typical upland crops. J. Integr. Agric. 2020, 19, 866–880. [Google Scholar] [CrossRef]
- Liu, Z.X.; Liu, J.J.; Yu, Z.H.; Yao, Q.; Li, Y.S.; Liang, A.Z.; Zhang, W.; Mi, G.; Jin, J.; Liu, X.B.; et al. Long-term continuous cropping of soybean is comparable to crop rotation in mediating microbial abundance, diversity and community composition. Soil Till. Res. 2020, 197, 104503. [Google Scholar] [CrossRef]
- Tian, L.; Shi, S.H.; Ma, L.N.; Tran, L.S.P.; Tian, X.J. Community structures of the rhizomicrobiomes of cultivated and wild soybeans in their continuous cropping. Microbiol. Res. 2020, 232, 126390. [Google Scholar] [CrossRef]
- Zeng, H.L.; Zhong, W.; Tan, F.X.; Shu, Y.H.; Feng, Y.J.; Wang, J.W. The influence of Bt maize cultivation on communities of arbuscular mycorrhizal fungi revealed by MiSeq sequencing. Front Microbiol. 2019, 9, 3275. [Google Scholar] [CrossRef]
- Rizzo, E.; Sherman, T.; Manosalva, P.; Gomez, S.K. Assessment of local and systemic changes in plant gene expression and aphid responses during potato interactions with arbuscular mycorrhizal fungi and potato aphids. Plants 2020, 9, 82. [Google Scholar] [CrossRef]
- Spagnoletti, F.N.; Leiva, M.; Chiocchio, V.; Lavado, R.S. Phosphorus fertilization reduces the severity of charcoal rot (Macrophomina phaseolina) and the arbuscular mycorrhizal protection in soybean. J. Plant Nutr. Soil Sci. 2018, 181, 855–860. [Google Scholar] [CrossRef]
- Song, Y.Y.; Chen, D.M.; Lu, K.; Sun, Z.X.; Zeng, R.S. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front Plant Sci. 2015, 6, 786. [Google Scholar] [CrossRef] [PubMed]
- Nafady, N.A.; Hashem, M.; Hassan, E.A.; Ahmed, H.A.M.; Alamri, S.A. The combined effect of arbuscular mycorrhizae and plant-growth-promoting yeast improves sunflower defense against Macrophomina phaseolina diseases. Biol. Control 2019, 138, 104049. [Google Scholar] [CrossRef]
- Aseel, D.G.; Rashad, Y.M.; Hammad, S.M. Arbuscular mycorrhizal fungi trigger transcriptional expression of flavonoid and chlorogenic acid biosynthetic pathways genes in tomato against tomato Mosaic virus. Sci. Rep. UK 2019, 9, 9692. [Google Scholar] [CrossRef] [PubMed]
- Dreher, D.; Baldermann, S.; Schreiner, M.; Hause, B. An arbuscular mycorrhizal fungus and a root pathogen induce different volatiles emitted by Medicago truncatula roots. J. Adv. Res. 2019, 19, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Jie, W.G.; Bai, L.; Yu, W.J.; Cai, B.Y. Analysis of interspecific relationships between Funneliformis mosseae and Fusarium oxysporum in the continuous cropping of soybean rhizosphere soil during the branching period. Biocontrol Sci. Technol. 2015, 25, 1036–1051. [Google Scholar] [CrossRef]
- Pawlowski, M.L.; Hartman, G.L. Reduction of sudden death syndrome foliar symptoms and Fusarium virguliforme DNA in roots inoculated with Rhizophagus intraradices. Plant Dis. 2020, 104, 1415–1420. [Google Scholar] [CrossRef]
- Jie, W.G.; Lin, J.X.; Guo, N.; Cai, B.Y.; Yan, X.F. Community composition of rhizosphere fungi as affected by Funneliformis mosseae in soybean continuous cropping soil during seedling period. Chil. J. Agric. Res. 2019, 79, 356–365. [Google Scholar] [CrossRef]
- Jie, W.G.; Lin, J.X.; Guo, N.; Cai, B.Y.; Yan, X.F. Effects of Funneliformis mosseae on mycorrhizal colonization, plant growth and the composition of bacterial community in the rhizosphere of continuous cropping soybean at seedling stage. Int. J. Agric. Biol. 2019, 22, 1173–1180. [Google Scholar]
- Phillips, J.M.; Hayman, D.S. Improved procedures for clearing roots and staining parasitic and cesicula-arbuscular mycorrhizal fungi for rapid, assessment of infection. Trans. Brit. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Long, L.K.; Yang, S.Z.; Yao, Q.; Zhu, H.H. DNA extraction from arbuscular mycorrhizal fungi and analysis by PCR-denaturing gradient gel electrophoresis. Mycosystema 2005, 24, 564–569. [Google Scholar]
- Dorn-In, S.; Bassitta, R.; Schwaiger, K.; Bauer, J.; Holzel, C.S. Specific amplification of bacterial DNA by optimized so-called universal bacterial primers in samples rich of plant DNA. J. Microbiol. Meth. 2015, 113, 50–56. [Google Scholar] [CrossRef]
- Smith, D.P.; Peay, K.G. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS ONE 2014, 9, e90234. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Armougom, F.; Raoult, D. Exploring microbial diversity using 16S rRNA highthroughput methods. J. Comput. Sci. Syst. Biol. 2009, 2, 74–92. [Google Scholar] [CrossRef]
- Hao, Z.P.; Fayolle, L.; van Tuinen, D.; Chatagnier, O.; Li, X.L. Local and systemic mycorrhiza-induced protection against the ectoparasitic nematode Xiphinema index involves priming of defence gene responses in grapevine. J. Exp. Biol. 2012, 63, 3657–3672. [Google Scholar] [CrossRef] [PubMed]
- Aimé, S.; Alabouvette, C.; Steinberg, C.; Olivain, C. The endophytic strain Fusarium oxysporum Fo47: A good candidate for priming the defense responses in tomato roots. Mol. Plant Microbe Interact. 2013, 26, 918–926. [Google Scholar] [CrossRef]
- Slaughter, A.; Daniela, X.; Flors, V.; Luna, E.; Hohn, B.; Brigitte, M.M. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 2012, 158, 835–843. [Google Scholar] [CrossRef]
- Al-Askar, A.A.; Rashad, Y.M. Arbuscular mycorrhizal fungi: A biocontrol agent against common. Plant Pathol. J. 2010, 9, 31–38. [Google Scholar] [CrossRef]
- Sharma, I.P.; Sharma, A.K. Physiological and biochemical changes in tomato cultivar PT-3 with dual inoculation of mycorrhiza and PGPR against root-knot nematode. Symbiosis 2017, 71, 175–183. [Google Scholar] [CrossRef]
- Veresoglou, S.D.; Barto, E.K.; Menexes, G.; Rillig, M.C. Fertilization affects severity of disease caused by fungal plant pathogens. Plant Pathol. 2013, 62, 961–969. [Google Scholar] [CrossRef]
- Nwangburuka, C.C.; Olawuyi, O.J.; Oyekale, K.; Ogunwemimo, K.O.; Denton, O.; Daramola, D.S.; Awotade, D. Effect of Arbuscular mycorrhizal (AM), Poultry Manure (PM), NPK fertilizer and the combination of AM-PM on the Growth and Yield of Okra (Abelmoschus esculentus). Nat. Sci. 2012, 10, 9–16. [Google Scholar]
- Abott, L.K.; Robson, A.D. Infectivity and effectiveness of five Endomycorrhizal fungi: Competition of indigenous fungi in field soils. Aust. J. Agric. Res. 1982, 32, 621–663. [Google Scholar] [CrossRef]
- Verbruggen, E.; Heijden, M.G.; Rillig, M.C.; Kiers, E.T. Mycorrhizal fungal establishment in agricultural soils: Factors determining inoculation success. New Phytol. 2013, 197, 1104–1109. [Google Scholar] [CrossRef]
- Artursson, V.; Finlay, R.D.; Jansson, J.K. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ. Microbiol. 2006, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Caballero, G.; Caravaca, F.; Fernández-González, A.J.; Alguacil, M.M.; Fernández-López, M.; Roldán, A. Arbuscular mycorrhizal fungi inoculation mediated changes in rhizosphere bacterial community structure while promoting revegetation in a semiarid ecosystem. Sci. Total Environ. 2017, 584, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]


| Index | Non0Y | In0Y | Non1Y | In1Y |
|---|---|---|---|---|
| Colonization rate (%) | 18.32 ± 1.34 c | 70.77 ± 1.35 b | 19.25 ± 1.28 c | 73.68 ± 0.94 a |
| Treatments | Plant Height (cm) | Stem Girth (cm) | Root Fresh Weight (g) | Root Dry Weight (g) | Shoot Fresh Weight (g) | Shoot Dry Weight (g) | 100-Seed Weight (g) | Seed-Yield per Plant (g) | Pods per Plant |
|---|---|---|---|---|---|---|---|---|---|
| Non0Y | 82.88 ± 1.17 b | 1.00 ± 0.05 b | 11.10 ± 0.90 b | 3.40 ± 0.39 b | 100.69 ± 1.74 b | 25.30 ± 0.84 b | 24.64 ± 1.21 b | 22.55 ± 0.94 b | 66.85 ± 2.27 b |
| In0Y | 89.50 ± 1.76 a | 1.14 ± 0.06 a | 16.43 ± 0.73 a | 4.39 ± 0.38 a | 113.69 ± 1.66 a | 30.90 ± 1.23 a | 26.94 ± 1.01 a | 25.82 ± 1.48 a | 78.51 ± 1.03 a |
| Non1 Y | 74.01 ± 1.18 c | 0.84 ± 0.06 c | 8.38 ± 0.72 c | 2.28 ± 0.34 c | 81.03 ± 1.24 c | 20.53 ± 0.68 c | 19.48 ± 0.63 d | 17.79 ± 1.01 d | 50.08 ± 2.15 d |
| In1Y | 84.46 ± 1.23 b | 0.99 ± 0.05 b | 11.35 ± 1.01 b | 3.45 ± 0.19 b | 99.36 ± 2.24 b | 25.76 ± 1.02 b | 21.40 ± 1.16 c | 19.77 ± 1.21 c | 59.27 ± 1.60 c |
| Sample ID | OTU | ACE | Chao1 | Simpson | Shannon | Coverage |
|---|---|---|---|---|---|---|
| Non0YRB | 437 ± 89 ab | 503.52 ± 22.88 ab | 505.65 ± 31.89 ab | 0.1033 ± 0.0824 a | 3.5752 ± 0.5533 a | 0.9998 ± 0.0001 a |
| In0YRB | 514 ± 12 a | 539.35 ± 18.87 a | 553.56 ± 30.73 a | 0.0758 ± 0.0287 a | 4.0754 ± 0.1404 a | 0.9994 ± 0.0003 a |
| Non1YRB | 395 ± 50 b | 461.75 ± 14.39 b | 487.28 ± 12.97 b | 0.2637 ± 0.1951 a | 2.8138 ± 1.1539 a | 0.9995 ± 0.0004 a |
| In1YRB | 451 ± 53 ab | 496.19 ± 44.30 ab | 499.93 ± 39.22 ab | 0.1279 ± 0.0966 a | 3.2188 ± 0.8709 a | 0.9998 ± 0.0001 a |
| Non0YRF | 537 ± 10 a | 544.35 ± 12.82 a | 551.15 ± 18.35 a | 0.0271 ± 0.0017 a | 4.6749 ± 0.0890 a | 0.9997 ± 0.0001 a |
| In0YRF | 579 ± 7 a | 500.65 ± 17.55 a | 543.43 ± 55.57 a | 0.0501 ± 0.0079 a | 4.2910 ± 0.0513 a | 0.9997 ± 0.0001 a |
| Non1YRF | 564 ± 86 a | 588.18 ± 5.64 a | 595.86 ± 12.28 a | 0.0225 ± 0.0052 a | 4.7948 ± 0.1975 a | 0.9997 ± 0.0001 a |
| In1YRF | 493 ± 18 a | 571.07 ± 87.10 a | 586.95 ± 74.99 a | 0.0257 ± 0.0052 a | 4.7346 ± 0.1410 a | 0.9997 ± 0.0001 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jie, W.-G.; Yao, Y.-X.; Guo, N.; Zhang, Y.-Z.; Qiao, W. Effects of Rhizophagus intraradices on Plant Growth and the Composition of Microbial Communities in the Roots of Continuous Cropping Soybean at Maturity. Sustainability 2021, 13, 6623. https://doi.org/10.3390/su13126623
Jie W-G, Yao Y-X, Guo N, Zhang Y-Z, Qiao W. Effects of Rhizophagus intraradices on Plant Growth and the Composition of Microbial Communities in the Roots of Continuous Cropping Soybean at Maturity. Sustainability. 2021; 13(12):6623. https://doi.org/10.3390/su13126623
Chicago/Turabian StyleJie, Wei-Guang, Yan-Xuan Yao, Na Guo, Ying-Zhi Zhang, and Wei Qiao. 2021. "Effects of Rhizophagus intraradices on Plant Growth and the Composition of Microbial Communities in the Roots of Continuous Cropping Soybean at Maturity" Sustainability 13, no. 12: 6623. https://doi.org/10.3390/su13126623
APA StyleJie, W.-G., Yao, Y.-X., Guo, N., Zhang, Y.-Z., & Qiao, W. (2021). Effects of Rhizophagus intraradices on Plant Growth and the Composition of Microbial Communities in the Roots of Continuous Cropping Soybean at Maturity. Sustainability, 13(12), 6623. https://doi.org/10.3390/su13126623






