Phylogenetic Structure of Soil Bacterial Communities along Age Sequence of Subtropical Cunninghamia Lanceolata Plantations
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Soil Sampling
2.2. DNA Extraction, PCR and High-Throughput Sequencing
2.3. Statistical Data Analysis
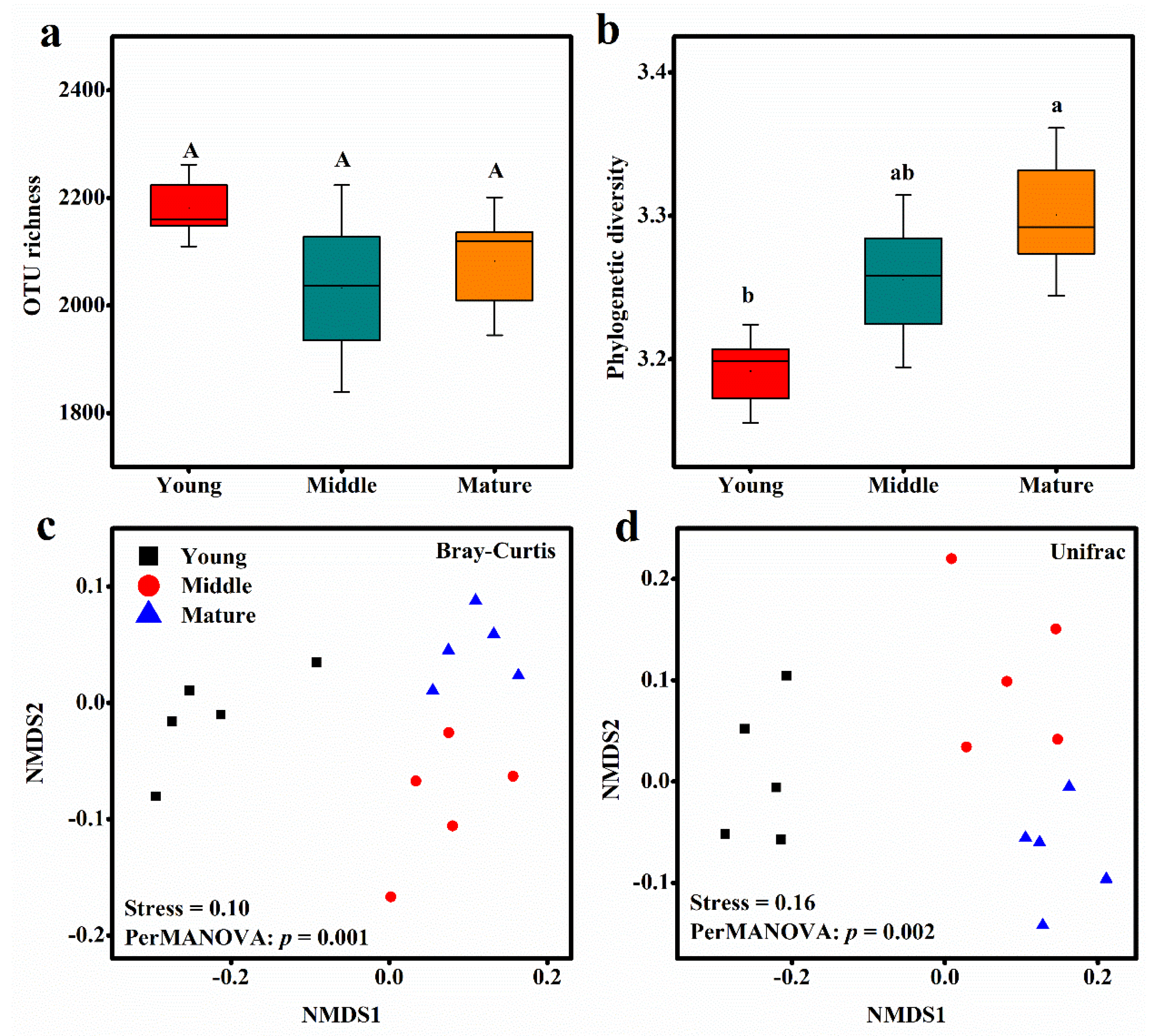
3. Results
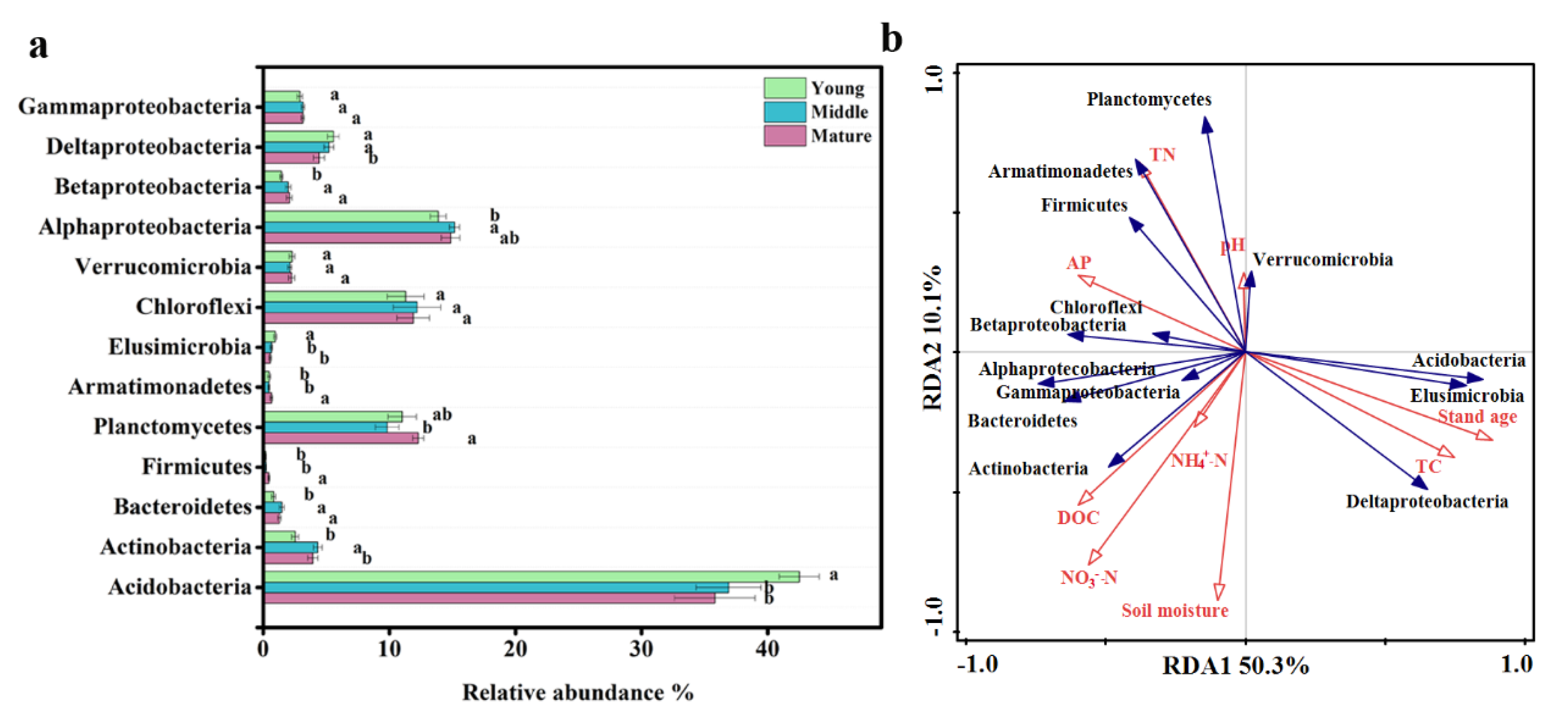
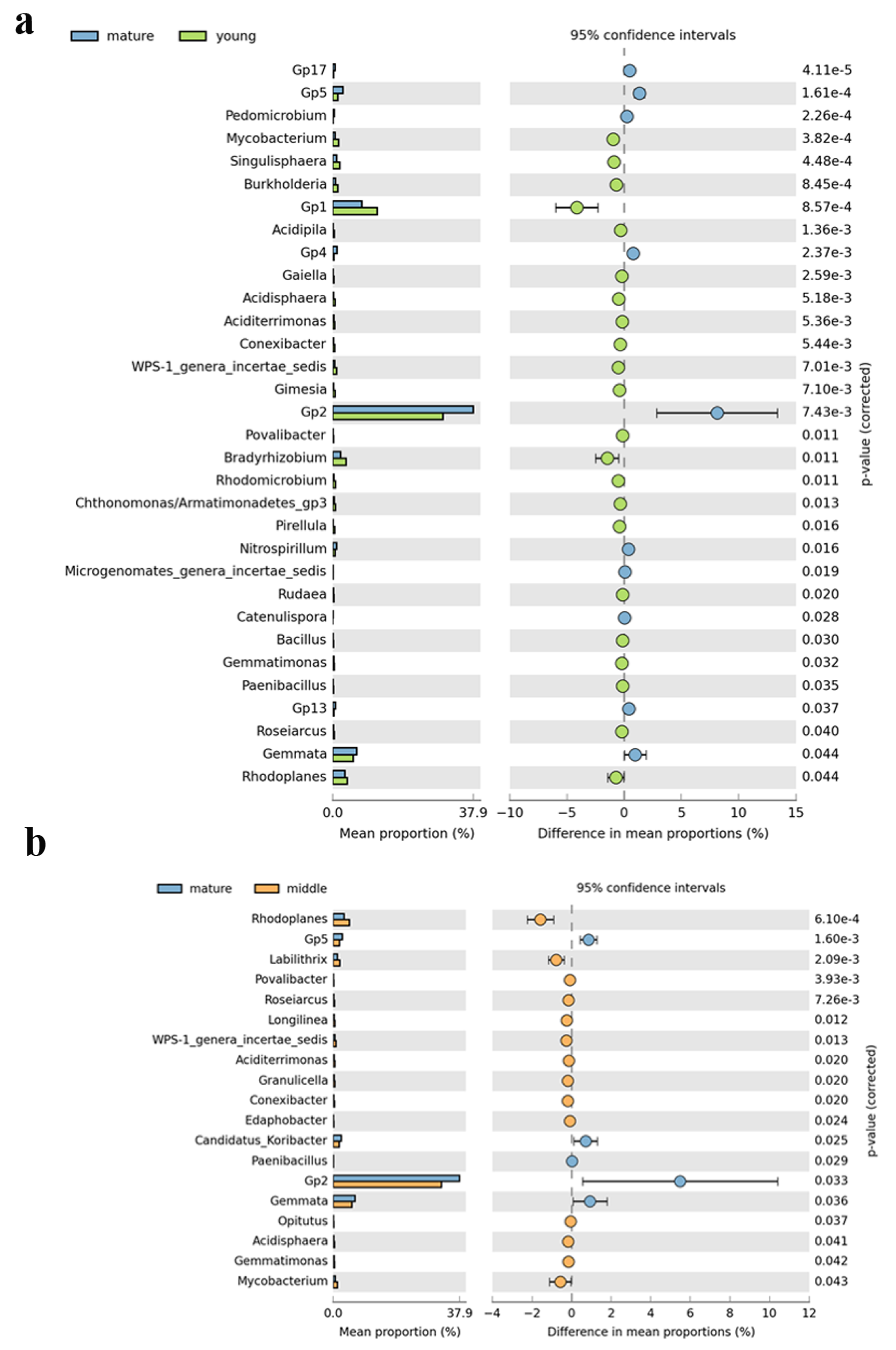
3.1. Distribution and Composition of Soil Bacterial Community
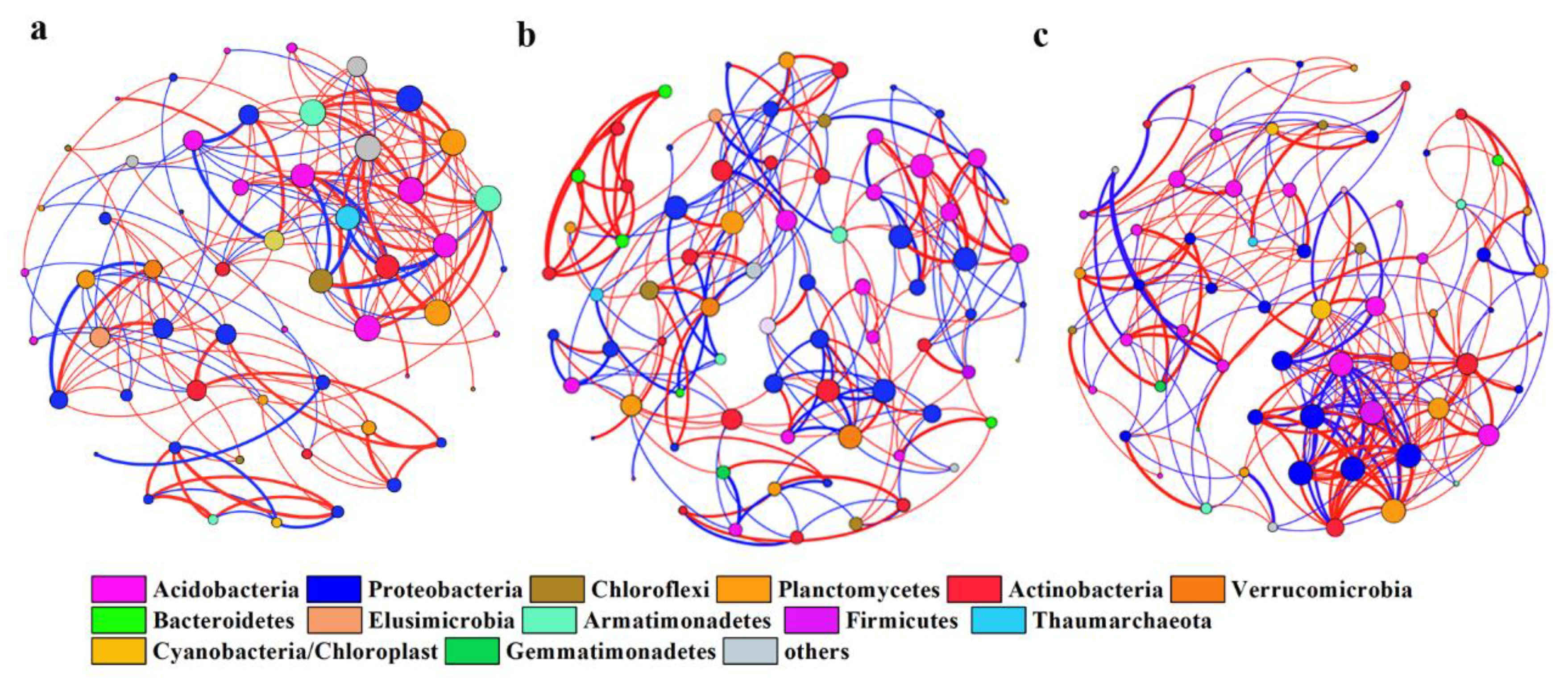
3.2. Network Co-Occurrence of Soil Bacterial Phylotypes
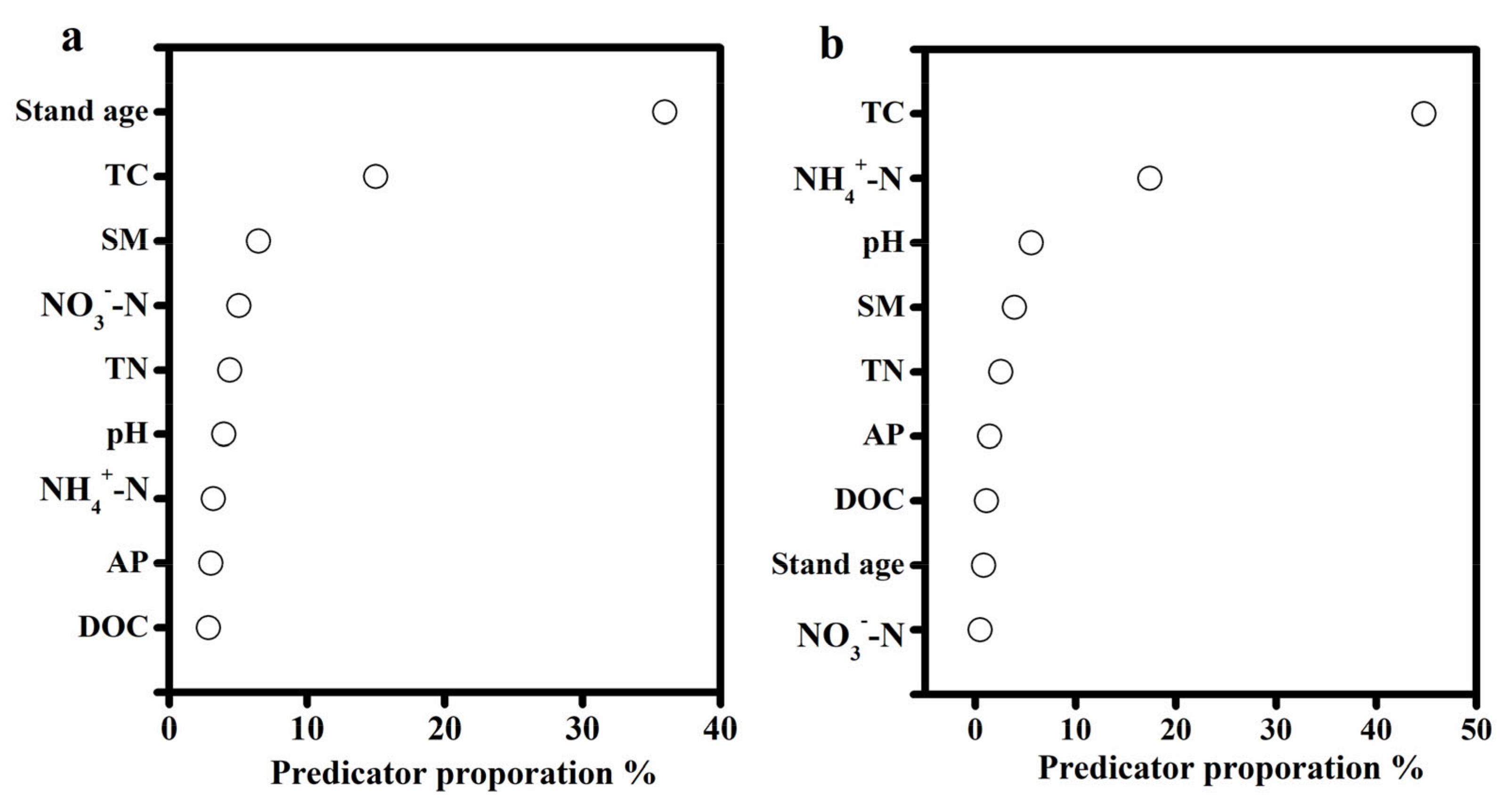
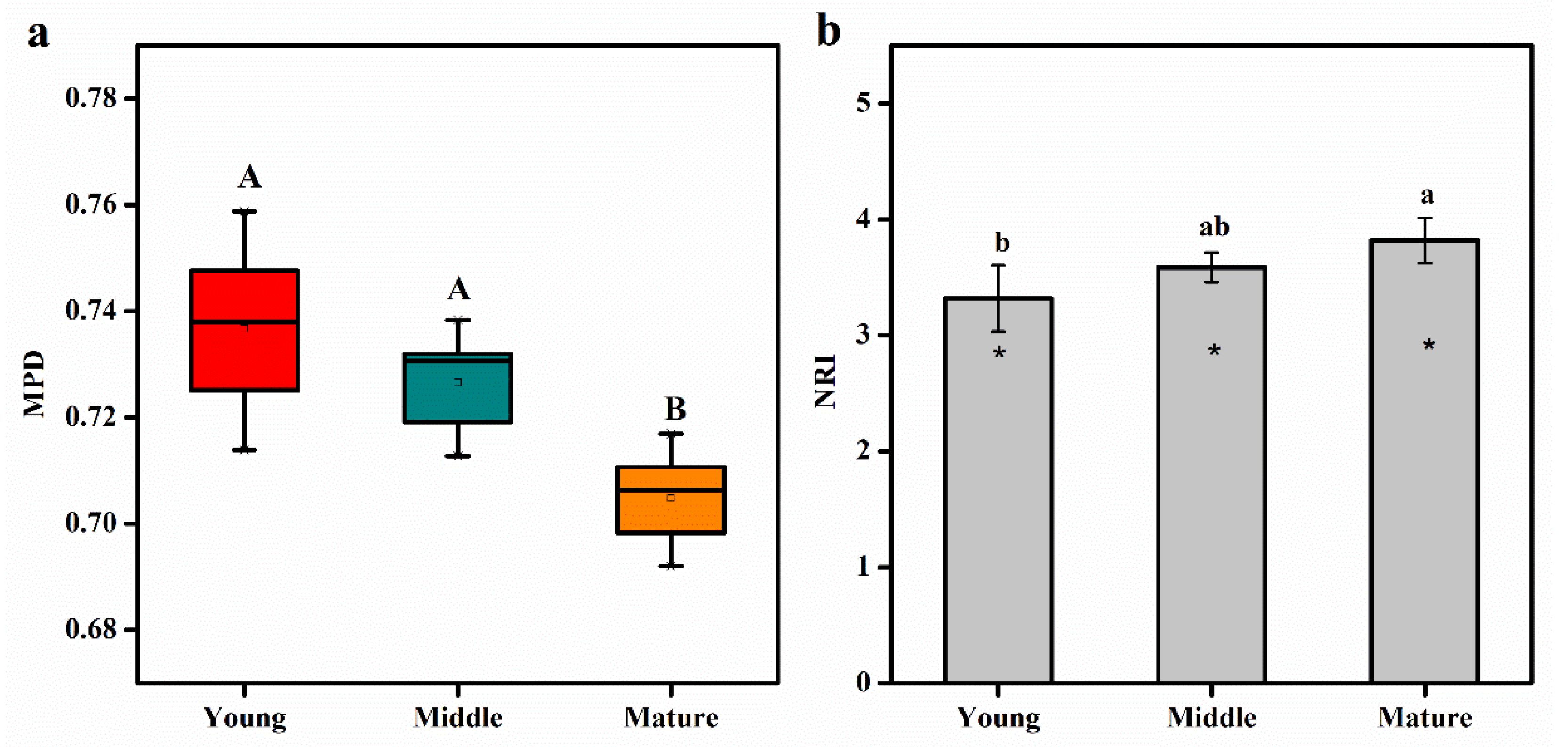
3.3. Phylogenetic Structure of Soil Bacterial Community
4. Discussion
4.1. Changes of Soil Bacterial Community among Different Development Ages of C. lanceolata Plantations
4.2. Phylogenetic Structure of Soil Bacterial Community Assembly under Different Development Ages of C. lanceolata Plantations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Winjum, J.K.; Schroeder, P.E. Forest plantations of the world: Their extent, ecological attributes, and carbon storage. Agric. For. Meteorol. 1997, 84, 153–167. [Google Scholar] [CrossRef]
- Van Dijk, A.I.J.M.; Keenan, R. Planted forests and water in perspective. For. Ecol. Manag. 2007, 251, 1–9. [Google Scholar] [CrossRef]
- FAO. Global Forest Resource Assessment; Food and Agricultural Organization of the United Nations: Rome, Italy, 2010; p. 163. [Google Scholar]
- Ma, X.; Heal, K.V.; Liu, A.; Jarvis, P.G. Nutrient cycling and distribution in different-aged plantations of Chinese fir in southern China. For. Ecol. Manag. 2007, 243, 61–74. [Google Scholar] [CrossRef]
- Lucas-Borja, M.; Hedo, J.; Cerdà, A.; Candel-Pérez, D.; Viñegla, B. Unravelling the importance of forest age stand and forest structure driving microbiological soil properties, enzymatic activities and soil nutrients content in Mediterranean Spanish black pine(Pinus nigra Ar. ssp. salzmannii) Forest. Sci. Total. Environ. 2016, 562, 145–154. [Google Scholar] [CrossRef]
- Carletti, P.; Vendramin, E.; Pizzeghello, D.; Concheri, G.; Zanella, A.; Nardi, S.; Squartini, A. Soil humic compounds and microbial communities in six spruce forests as function of parent material, slope aspect and stand age. Plant Soil 2008, 315, 47–65. [Google Scholar] [CrossRef]
- Waring, B.; Adams, R.; Branco, S.; Powers, J.S. Scale-dependent variation in nitrogen cycling and soil fungal communities along gradients of forest composition and age in regenerating tropical dry forests. New Phytol. 2015, 209, 845–854. [Google Scholar] [CrossRef]
- Liu, J.; Yang, Z.; Dang, P.; Zhu, H.; Gao, Y.; Ha, V.N.; Zhao, Z. Response of soil microbial community dynamics to Robinia pseudoacacia L. afforestation in the loess plateau: A chronosequence approach. Plant Soil 2017, 423, 327–338. [Google Scholar] [CrossRef]
- Barber, N.A.; Chantos-Davidson, K.M.; Peralta, R.A.; Sherwood, J.P.; Swingley, W.D. Soil microbial community composition in tallgrass prairie restorations converge with remnants across a 27-year chronosequence. Environ. Microbiol. 2017, 19, 3118–3131. [Google Scholar] [CrossRef]
- Gao, P.; Zheng, X.; Wang, L.; Liu, B.; Zhang, S. Changes in the Soil Bacterial Community in a Chronosequence of Temperate Walnut-Based Intercropping Systems. Forests 2019, 10, 299. [Google Scholar] [CrossRef]
- Webb, C.O. Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am. Nat. 2000, 156, 145–155. [Google Scholar] [CrossRef]
- Webb, C.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and Community Ecology. Annu. Rev. Ecol. Syst. 2002, 33, 475–505. [Google Scholar] [CrossRef]
- Adler, P.B.; Salguero-Gómez, R.; Compagnoni, A.; Hsu, J.S.; Ray-Mukherjee, J.; Mbeau-Ache, C.; Franco, M. Functional traits explain variation in plant life history strategies. Proc. Natl. Acad. Sci. USA 2013, 111, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Xing, D.; Swenson, N.G.; Weiser, M.D.; Hao, Z. Determinants of species abundance for eastern North American trees. Glob. Ecol. Biogeogr. 2014, 23, 903–911. [Google Scholar] [CrossRef]
- Chase, J.M. Stochastic Community Assembly Causes Higher Biodiversity in More Productive Environments. Science 2010, 328, 1388–1391. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Freedman, Z.; Zak, D. Soil bacterial communities are shaped by temporal and environmental filtering: Evidence from a long-term chronosequence. Environ. Microbiol. 2015, 17, 3208–3218. [Google Scholar] [CrossRef]
- State Forestry Administration of China. Forest Resources Report in China (2009–2013); China Forestry Publishing House: Beijing, China, 2014; p. 86.
- Yu, S.; Wang, D.; Dai, W.; Li, P. Soil carbon budget in different-aged Chinese fir plantations in south China. J. For. Res. 2014, 25, 621–626. [Google Scholar] [CrossRef]
- Chen, G.; Yang, Z.-J.; Gao, R.; Xie, J.; Guo, J.-F.; Huang, Z.; Yang, Y. Carbon storage in a chronosequence of Chinese fir plantations in southern China. For. Ecol. Manag. 2013, 300, 68–76. [Google Scholar] [CrossRef]
- Zhou, L.; He, Z.; Wang, R.; Wu, P.; Ma, X.; Cai, L. Thinning increases understory diversity and biomass, and improves soil properties without decreasing growth of Chinese fir in southern China. Environ. Sci. Pollut. Res. 2016, 23, 24135–24150. [Google Scholar] [CrossRef]
- Zhang, Y.; Tigabu, M.; Yi, Z.; Li, H.; Zhuang, Z.; Yang, Z.; Ma, X. Soil parent material and stand development stage effects on labile soil C and N pools in Chinese fir plantations. Geoderma 2019, 338, 247–258. [Google Scholar] [CrossRef]
- Yu, X.T. The Chinese Fir Silviculture; Fujian Science and Technology Press: Fuzhou, Fujian, China, 1997; pp. 37–44. [Google Scholar]
- Biddle, J.F.; Fitz-Gibbon, S.; Schuster, S.C.; Brenchley, J.E.; House, C.H. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. USA 2008, 105, 10583–10588. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2013, 2, 1–295. [Google Scholar]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef]
- Kembel, S.W. Disentangling niche and neutral influences on community assembly: Assessing the performance of community phylogenetic structure tests. Ecol. Lett. 2009, 12, 949–960. [Google Scholar] [CrossRef]
- Clarke, K.; Gorley, R.N.; Somerfield, P.; Warwick, R. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation; Primer-E Ltd.: Plymouth, UK, 2014; p. 260. [Google Scholar]
- Dini-Andreote, F.; Silva, M.D.C.P.E.; Triadó-Margarit, X.; Casamayor, E.O.; Van Elsas, J.D.; Salles, J.F. Dynamics of bacterial community succession in a salt marsh chronosequence: Evidences for temporal niche partitioning. ISME J. 2014, 8, 1989–2001. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef]
- Sun, H.; Santalahti, M.; Pumpanen, J.; Koster, K.; Berninger, F.; Raffaello, T.; Jumpponen, A.; Asiegbu, F.O.; Heinonsalo, J. Fungal community shifts in structure and function across a boreal forest fFire chronosequence. Appl. Environ. Microb. 2015, 81, 7869–7880. [Google Scholar] [CrossRef]
- Liu, J.; Ha, V.N.; Shen, Z.; Zhu, H.; Zhao, F.; Zhao, Z. Characteristics of bulk and rhizosphere soil microbial community in an ancient Platycladus orientalis forest. Appl. Soil Ecol. 2018, 132, 91–98. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.A.; Lamanna, C.; Morlon, H.; Kerkhoff, A.J.; Enquist, B.J.; Green, J.L. Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. USA 2008, 105, 11505–11511. [Google Scholar] [CrossRef] [PubMed]
- Na, X.; Li, X.; Zhang, Z.; Kardol, P.; Xu, T.T.; Wang, M.; Cao, X.; Ma, F. Bacterial community dynamics in the rhizosphere of a long-lived, leguminous shrub across a 40-year age sequence. J. Soils Sediments 2017, 18, 76–84. [Google Scholar] [CrossRef]
- Nielsen, U.N.; Osler, G.H.R.; Campbell, C.; Burslem, D.F.R.P.; Van Der Wal, R. The influence of vegetation type, soil properties and precipitation on the composition of soil mite and microbial communities at the landscape scale. J. Biogeogr. 2010, 37, 1317–1328. [Google Scholar] [CrossRef]
- Knelman, J.E.; Legg, T.M.; O’Neill, S.P.; Washenberger, C.L.; González, A.; Cleveland, C.C.; Nemergut, D.R. Bacterial community structure and function change in association with colonizer plants during early primary succession in a glacier forefield. Soil Biol. Biochem. 2012, 46, 172–180. [Google Scholar] [CrossRef]
- Landesman, W.; Nelson, D.; Fitzpatrick, M.C. Soil properties and tree species drive ß-diversity of soil bacterial communities. Soil Biol. Biochem. 2014, 76, 201–209. [Google Scholar] [CrossRef]
- Wang, J.; Shen, J.; Wu, Y.; Tu, C.; Soininen, J.; Stegen, J.; He, J.-Z.; Liu, X.; Zhang, L.; Zhang, E. Phylogenetic beta diversity in bacterial assemblages across ecosystems: Deterministic versus stochastic processes. ISME J. 2013, 7, 1310–1321. [Google Scholar] [CrossRef]
- Narwani, A.; Matthews, B.; Fox, J.; Venail, P. Using phylogenetics in community assembly and ecosystem functioning research. Funct. Ecol. 2015, 29, 589–591. [Google Scholar] [CrossRef]
- Goberna, M.; Garcia, C.; Verdú, M. A role for biotic filtering in driving phylogenetic clustering in soil bacterial communities. Glob. Ecol. Biogeogr. 2014, 23, 1346–1355. [Google Scholar] [CrossRef]
- Chase, J.M.; Myers, J.A. Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. B Boil. Sci. 2011, 366, 2351–2363. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Jones, S.E.; Lennon, J.T.; Martiny, A.C. Microbiomes in light of traits: A phylogenetic perspective. Science 2015, 350, aac9323. [Google Scholar] [CrossRef]
- Warwick, R.; Clarke, K.R.; Pienkowski, M.W.; Watkinson, A.R.; Kerby, G. Taxonomic distinctness and environmental assessment. J. Appl. Ecol. 1998, 35, 532–543. [Google Scholar] [CrossRef]
- Letcher, S.G. Phylogenetic structure of angiosperm communities during tropical forest succession. Proc. R. Soc. B Boil. Sci. 2009, 277, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, S.J.; Thrash, J.C.; Temperton, B. Implications of streamlining theory for microbial ecology. ISME J. 2014, 8, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Guo, Z.; Zhong, L.; Zhao, F.; Zhang, J.; Lin, X. Balanced Fertilization Decreases Environmental Filtering on Soil Bacterial Community Assemblage in North China. Front. Microbiol. 2017, 8, 2376. [Google Scholar] [CrossRef]
- Wiens, J.J.; Graham, C. NICHE CONSERVATISM: Integrating Evolution, Ecology, and Conservation Biology. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 519–539. [Google Scholar] [CrossRef]
| Variable | r | p |
|---|---|---|
| Stand age | 0.635 | 0.002 |
| Soil moisture | 0.519 | 0.002 |
| TC | 0.548 | 0.001 |
| TN | 0.528 | 0.001 |
| AP | 0.287 | 0.025 |
| DOC | 0.070 | 0.701 |
| NH4+-N | 0.081 | 0.763 |
| NO3−-N | 0.116 | 0.129 |
| pH | 0.043 | 0.366 |
| Network Metrics | Young | Middle | Mature |
|---|---|---|---|
| Number of nodes | 56 | 76 | 67 |
| Number of edges | 180 | 204 | 236 |
| Average clustering coefficient | 0.222 | 0.259 | 0.282 |
| Modularity | 0.511 | 0.712 | 0.579 |
| Average degree | 3.214 | 2.684 | 3.522 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Zheng, Y.; Yang, Y. Phylogenetic Structure of Soil Bacterial Communities along Age Sequence of Subtropical Cunninghamia Lanceolata Plantations. Sustainability 2020, 12, 1864. https://doi.org/10.3390/su12051864
Cao J, Zheng Y, Yang Y. Phylogenetic Structure of Soil Bacterial Communities along Age Sequence of Subtropical Cunninghamia Lanceolata Plantations. Sustainability. 2020; 12(5):1864. https://doi.org/10.3390/su12051864
Chicago/Turabian StyleCao, Jiling, Yuxiong Zheng, and Yusheng Yang. 2020. "Phylogenetic Structure of Soil Bacterial Communities along Age Sequence of Subtropical Cunninghamia Lanceolata Plantations" Sustainability 12, no. 5: 1864. https://doi.org/10.3390/su12051864
APA StyleCao, J., Zheng, Y., & Yang, Y. (2020). Phylogenetic Structure of Soil Bacterial Communities along Age Sequence of Subtropical Cunninghamia Lanceolata Plantations. Sustainability, 12(5), 1864. https://doi.org/10.3390/su12051864




