Response of Soil Bacterial Community to Application of Organic and Inorganic Phosphate Based Fertilizers under Vicia faba L. Cultivation at Two Different Phenological Stages
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Properties
2.2. Fertilizer Characteristics
2.2.1. Organic Fertilizers
2.2.2. Inorganic Fertilizers
2.3. Experimental Design
2.4. Plant Biomass Analysis
2.5. Soil Chemical Analysis
2.6. Soil DNA Extraction and 16S rRNA Gene Sequencing
2.7. Sequencing Data Processing and Data Analysis
3. Results
3.1. Plant Biomass
3.2. Soil Inorganic N Compounds
3.3. Bacterial Alpha Diversity
3.4. Correlation of Alpha Diversity with Soil Edaphic Properties and Plant Biomass
3.5. Changes in Bacterial Community Structure (Beta Diversity) in Response to Time, Plant Presence and Fertilizer Treatment
3.6. Correlation between Bacterial Community Structure and Soil Edaphic Properties and Plant Biomass Data
3.7. Bacterial Community Composition of PP and P Fertilizers
3.8. Influence of Plant Presence, Time, and Fertilizer Treatment on Taxonomic Composition
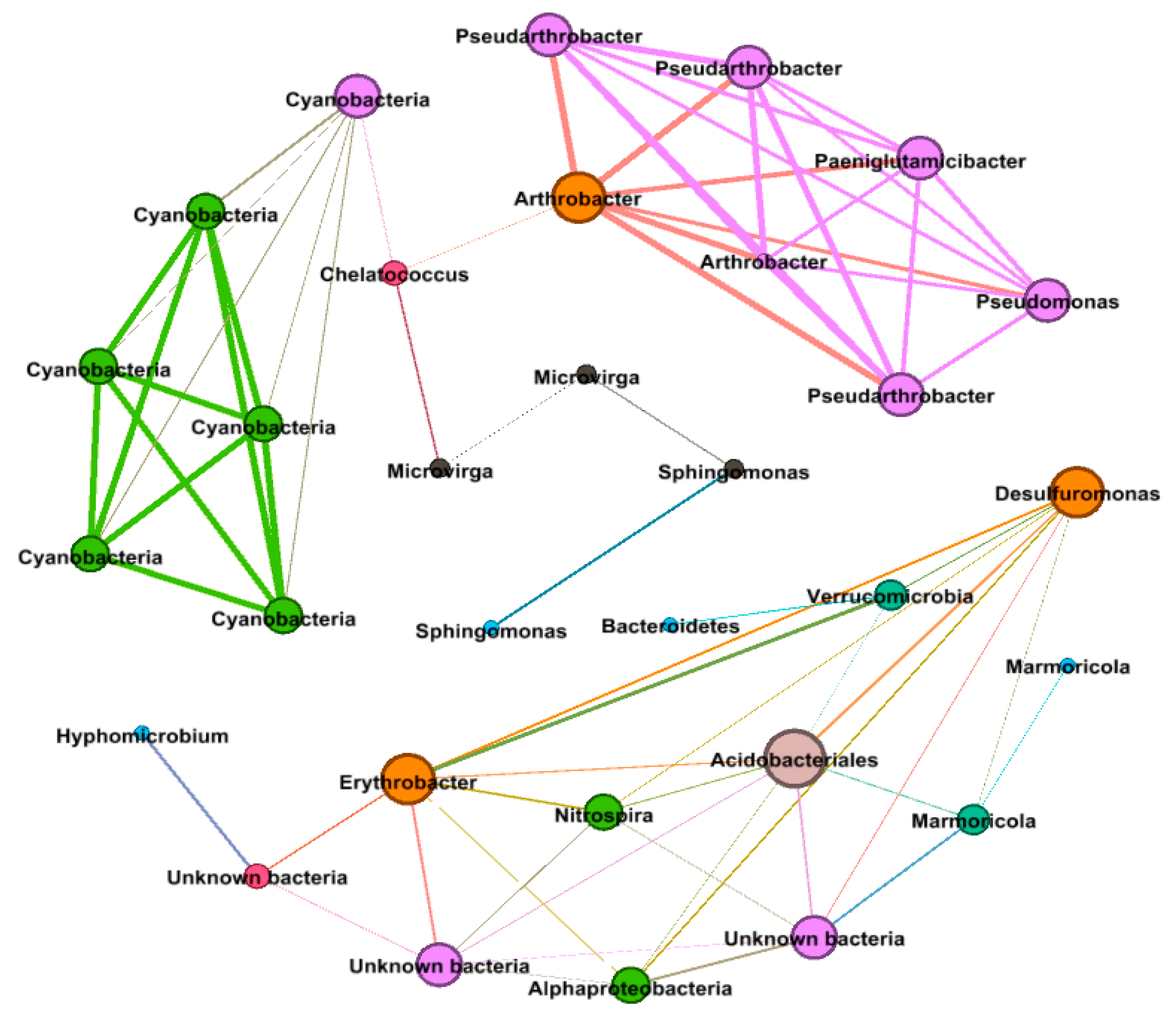
3.9. Bacterial Co-occurrence Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Availability of Data and Material
Conflicts of Interest
References
- Gogoi, N.; Baruah, K.K.; Meena, R.S. Legumes for Soil Health and Sustainable Management. In Legumes for Soil Health and Sustainable Management; Springer: Berlin/Heidelberg, Germany, 2018; pp. 511–539. [Google Scholar]
- Stagnari, F.; Maggio, A.; Galieni, A.; Pisante, M. Multiple benefits of legumes for agriculture sustainability: An overview. Chem. Biol. Technol. Agric. 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Laik, R.; Sharma, S.; Idris, M.; Singh, A.K.; Singh, S.S.; Bhatt, B.P.; Ladha, J.K. Integration of conservation agriculture with best management practices for improving system performance of the rice-wheat rotation in the Eastern Indo-Gangetic Plains of India. Agric. Ecosyst. Environ. 2014, 195, 68–82. [Google Scholar] [CrossRef]
- Gan, Y.; Hamel, C.; O’Donovan, J.T.; Cutforth, H.; Zentner, R.P.; Campbell, C.A.; Poppy, L. Diversifying crop rotations with pulses enhances system productivity. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Meena, R.S.; Lal, R. Legumes and Sustainable Use of soils. In Legumes for Soil Health and Sustainable Management; Meena, R.S., Lal, R., Eds.; Springer: Singapore, 2018; pp. 1–31. [Google Scholar]
- Malezieux, E.; Crozat, Y.; Dupraz, C.; Laurans, M.; Makowski, D.; Ozier-Lafontaine, H.; Rapidel, B.; de Tourdonnet, S.; Valantin-Morison, M. Mixing plant species in cropping systems: Concepts, tools and models. A review. Agron. Sustain. Dev. 2009, 29, 43–62. [Google Scholar] [CrossRef]
- Crépon, K.; Marget, P.; Peyronnet, C.; Carrouée, B.; Arese, P.; Duc, G. Nutritional value of faba bean (Vicia faba L.) seeds for feed and food. Field Crop. Res. 2010, 115, 329–339. [Google Scholar] [CrossRef]
- Etemadi, F.; Hashemi, M.; Barker, A.V.; Zandvakili, O.R.; Liu, X. Agronomy, Nutritional Value, and Medicinal Application of Faba Bean (Vicia faba L.). Hortic. Plant J. 2019, 5, 170–182. [Google Scholar] [CrossRef]
- Galloway, J.N.; Dentener, F.J.; Capone, D.G.; Boyer, E.W.; Howarth, R.W.; Seitzinger, S.P.; Asner, G.P.; Cleveland, C.C.; Green, P.A.; Holland, E.A.; et al. Nitrogen cycles: Past, present, and future. Biogeochemistry 2004, 70, 153–226. [Google Scholar] [CrossRef]
- Etemadi, F.; Hashemi, M.; Zandvakili, O.; Dolatabadian, A.; Sadeghpour, A. Nitrogen Contribution from Winter-Killed Faba Bean Cover Crop to Spring-Sown Sweet Corn in Conventional and No-Till Systems. Agron. J. 2018, 110, 455–462. [Google Scholar] [CrossRef]
- Wei, M.; Takeshima, K.; Yokoyama, T.; Minamisawa, K.; Mitsui, H.; Itakura, M.; Kaneko, T.; Tabata, S.; Saeki, K.; Omori, H.; et al. Temperature-Dependent Expression of Type III Secretion System Genes and Its Regulation in Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 2010, 23, 628–637. [Google Scholar] [CrossRef]
- Cabeza, R.A.; Liese, R.; Lingner, A.; Von Stieglitz, I.; Neumann, J.; Salinas-Riester, G.; Pommerenke, C.; Dittert, K.; Schulze, J. RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N 2 fixation before emerging P deficiency reaches the nodules. J. Exp. Bot. 2014, 65, 6035–6048. [Google Scholar] [CrossRef]
- Míguez-Montero, M.A.; Valentine, A.; Pérez-Fernández, M.A. Regulatory effect of phosphorus and nitrogen on nodulation and plant performance of leguminous shrubs. AoB Plants 2019, 12, 1–11. [Google Scholar] [CrossRef]
- Chen, Y.; Camps-Arbestain, M.; Shen, Q.; Singh, B.; Cayuela, M.L. The long-term role of organic amendments in building soil nutrient fertility: A meta-analysis and review. Nutr. Cycl. Agroecosyst. 2018, 111, 103–125. [Google Scholar] [CrossRef]
- Swanepoel, P.A.; Botha, P.R.; Truter, W.F.; Surridge-Talbot, A.K. No TitleThe effect of soil carbon on symbiotic nitrogen fixation and symbiotic Rhizobium populations in soil with Trifolium repens as host plant. Afr. J. Range Forage Sci. 2011, 28, 121–128. [Google Scholar] [CrossRef]
- Artursson, V.; Finlay, R.D.; Jansson, J.K. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ. Microbiol. 2006, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.J.; Blackwood, C.B. The Ecology of the Soil Biota and their Function. In Soil Microbiology, Ecology and Biochemistry, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 273–309. [Google Scholar]
- Zhou, Y.; Zhu, H.; Fu, S.; Yao, Q. Variation in Soil Microbial Community Structure Associated with Different Legume Species Is Greater than that Associated with Different Grass Species. Front. Microbiol. 2017, 8, 1007. [Google Scholar] [CrossRef] [PubMed]
- Coleman, D.C.; Whitman, W.B. Linking species richness, biodiversity and ecosystem function in soil systems. Pedobiologia 2005, 49, 479–497. [Google Scholar] [CrossRef]
- Sánchez-Cañizares, C.; Jorrín, B.; Poole, P.S.; Tkacz, A. Understanding the holobiont: The interdependence of plants and their microbiome. Curr. Opin. Microbiol. 2017, 38, 188–196. [Google Scholar] [CrossRef]
- McLaren, J.R.; Turkington, R. Plant Identity Influences Decomposition through More Than One Mechanism. PLoS ONE 2011, 6, e23702. [Google Scholar] [CrossRef]
- Mele, P.M.; Crowley, D.E. Application of self-organizing maps for assessing soil biological quality. Agric. Ecosyst. Environ. 2008, 126, 139–152. [Google Scholar] [CrossRef]
- Zhong, W.; Gu, T.; Wang, W.; Zhang, B.; Lin, X.; Huang, Q.; Shen, W. The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant Soil 2010, 326, 511–522. [Google Scholar] [CrossRef]
- Bünemann, E.; Bossio, D.; Smithson, P.; Frossard, E.; Oberson, A. Microbial community composition and substrate use in a highly weathered soil as affected by crop rotation and P fertilization. Soil Biol. Biochem. 2004, 36, 889–901. [Google Scholar] [CrossRef]
- Zhang, N.N.; Sun, Y.M.; Li, L.; Wang, E.T.; Chen, W.X.; Yuan, H. Effects of intercropping and Rhizobium inoculation on yield and rhizosphere bacterial community of faba bean (Vicia faba L.). Biol. Fertil. Soils 2010, 46, 625–639. [Google Scholar] [CrossRef]
- Duchene, O.; Vian, J.-F.; Celette, F. Intercropping with legume for agroecological cropping systems: Complementarity and facilitation processes and the importance of soil microorganisms. A review. Agric. Ecosyst. Environ. 2017, 240, 148–161. [Google Scholar] [CrossRef]
- Chaudhari, D.; Rangappa, K.; Das, A.; Layek, J.; Basavaraj, S.; Kandpal, B.K.; Shouche, Y.; Rahi, P. Pea (Pisum sativum l.) Plant Shapes Its Rhizosphere Microbiome for Nutrient Uptake and Stress Amelioration in Acidic Soils of the North-East Region of India. Front. Microbiol. 2020, 11, 968. [Google Scholar] [CrossRef] [PubMed]
- Latati, M.; Blavet, D.; Alkama, N.; Laoufi, H.; Drevon, J.-J.; Gerard, F.; Pansu, M.; Ounane, S.M. The intercropping cowpea-maize improves soil phosphorus availability and maize yields in an alkaline soil. Plant Soil 2014, 385, 181–191. [Google Scholar] [CrossRef]
- Latati, M.; Bargaz, A.; Belarbi, B.; Lazali, M.; Benlahrech, S.; Tellah, S.; Kaci, G.; Drevon, J.J.; Ounane, S.M. The intercropping common bean with maize improves the rhizobial efficiency, resource use and grain yield under low phosphorus availability. Eur. J. Agron. 2016, 72, 80–90. [Google Scholar] [CrossRef]
- S Pathan, S.I.; Ceccherini, M.T.; Sunseri, F.; Lupini, A. Rhizosphere as Hotspot for Plant-Soil-Microbe Interaction. In Carbon and Nitrogen Cycling in Soil; Springer: Berlin/Heidelberg, Germany, 2019; pp. 17–43. [Google Scholar]
- Bardgett, R.D.; Van Der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nat. Cell Biol. 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Genet. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Dhakal, D.; Islam, M.A. Grass-Legume Mixtures for Improved Soil Health in Cultivated Agroecosystem. Sustainability 2018, 10, 2718. [Google Scholar] [CrossRef]
- Holford, I.C.R. Soil phosphorus: Its measurement, and its uptake by plants. Soil Res. 1997, 35, 227. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Ali, W.; Nadeem, M.; Ashiq, W.; Zaeem, M.; Gilani, S.S.M.; Rajabi-Khamseh, S.; Pham, T.H.; Kavanagh, V.; Thomas, R.; Cheema, M. The effects of organic and inorganic phosphorus amendments on the biochemical attributes and active microbial population of agriculture podzols following silage corn cultivation in boreal climate. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lori, M.; Symnaczik, S.; Mäder, P.; De Deyn, G.; Gattinger, A. Organic farming enhances soil microbial abundance and activity—A meta-analysis and meta-regression. PLoS ONE 2017, e0180442. [Google Scholar] [CrossRef] [PubMed]
- Celestina, C.; Wood, J.L.; Manson, J.B.; Wang, X.; Sale, P.W.G.; Tang, C.; Franks, A.E. Microbial communities in top- and subsoil of repacked soil columns respond differently to amendments but their diversity is negatively correlated with plant productivity. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Schlesinger, H.V. Ferrous nitroso c0mpounds. J. Am. Chem. Soc. 1929, 51, 1323–1331. [Google Scholar] [CrossRef]
- Kustin, K.; Taub, I.A.; Weinstock, E. A Kinetic study of the formation of the Ferrous-Nitric Oxide Complexis. Inorg. Chem. 1966, 5, 1079–1082. [Google Scholar] [CrossRef]
- Irandoust, M.; Shariati-Rad, M.; Haghighi, M. Nitrite determination in water samples based on a modified Griess reaction and central composite design. Anal. Methods 2013, 5, 5977–5982. [Google Scholar] [CrossRef]
- Panico, S.C.; Ceccherini, M.T.; Memoli, V.; Maisto, G.; Pietramellara, G.; Barile, R.; De Marco, A. Effects of different vegetation types on burnt soil properties and microbial communities. Int. J. Wildland Fire 2020, 29, 628. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Větrovský, T.; Baldrian, P. The Variability of the 16S rRNA Gene in Bacterial Genomes and Its Consequences for Bacterial Community Analyses. PLoS ONE 2013, 8, e57923. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naiïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological Statistics software packages for education and data analysis. Palaeontol. Electron. 2001, 4, 4–9. [Google Scholar]
- Anderson, M.; Ter Braak, C. Permutation tests for multi-factorial analysis of variance. J. Stat. Comput. Simul. 2003, 73, 85–113. [Google Scholar] [CrossRef]
- Harrell, J. Hmisc: Harrell Miscellaneous. R Package Version 3.5-2. 2008. Available online: https://cran.r-project.org/web/packages/Hmisc/Hmisc.pdf (accessed on 18 November 2020).
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Vick-Majors, T.J.; Priscu, J.C.; Amaral-Zettler, L.A. Modular community structure suggests metabolic plasticity during the transition to polar night in ice-covered Antarctic lakes. ISME J. 2014, 8, 778–789. [Google Scholar] [CrossRef]
- Lambiotte, R.; Delvenne, J.C.; Barahona, M. Random walks, Markov processes and the multiscale modular organization of complex networks. IEEE Trans Netw. Sci. Eng. 2014, 1, 76–90. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; Van Der Putten, W.H.; Wall, D.H. Ecological Linkages Between Aboveground and Belowground Biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef]
- Siczek, A.; Frąc, M.; Gryta, A.; Kalembasa, S.; Kalembasa, D. Variation in soil microbial population and enzyme activities under faba bean as affected by pentachlorophenol. Appl. Soil Ecol. 2020, 150, 103466. [Google Scholar] [CrossRef]
- Prober, S.M.; Leff, J.W.; Bates, S.T.; Borer, E.T.; Firn, J.; Harpole, W.S.; Lind, E.M.; Seabloom, E.W.; Adler, P.B.; Bakker, J.D.; et al. Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 2015, 18, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Baudoin, E.; Benizri, E.; Guckert, A. Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol. Biochem. 2003, 35, 1183–1192. [Google Scholar] [CrossRef]
- Fischer, S.; Jofré, E.C.; Cordero, P.V.; Mañero, F.J.G.; Mori, G.B. Survival of native Pseudomonas in soil and wheat rhizosphere and antagonist activity against plant pathogenic fungi. Antonie van Leeuwenhoek 2009, 97, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K.; Fatima, T.; Mishra, I.; Verma, M.; Mishra, J.; Mishra, V. Environmental sustainability: Challenges and viable solutions. Environ. Sustain. 2018, 1, 309–340. [Google Scholar] [CrossRef]
- Pankratova, E.M. Functioning of cyanobacteria in soil ecosystems. Eurasian Soil Sci. 2006, 39, S118–S127. [Google Scholar] [CrossRef]
- Jeppesen, E.; Meerhoff, M.; Davidson, T.A.; Trolle, D.; Søndergaard, M.; Lauridsen, T.L.; Beklioglu, M.; Brucet, S.; Volta, P.; González-Bergonzoni, I.; et al. Climate change impacts on lakes: An integrated ecological perspective based on a multi-faceted approach, with special focus on shallow lakes. J. Limnol. 2014, 73, 88–111. [Google Scholar] [CrossRef]
- Singh, J.S.; Kumar, A.; Rai, A.N.; Singh, D.P. Cyanobacteria: A Precious Bio-resource in Agriculture, Ecosystem, and Environmental Sustainability. Front. Microbiol. 2016, 7, 529. [Google Scholar] [CrossRef]
- Savadova, K.; Mazur-Marzec, H.; Karosienė, J.; Kasperovičienė, J.; Vitonytė, I.; Toruńska-Sitarz, A.; Koreivienė, J. Effect of Increased Temperature on Native and Alien Nuisance Cyanobacteria from Temperate Lakes: An Experimental Approach. Toxins 2018, 10, 445. [Google Scholar] [CrossRef]
- Chen, J.; Shen, W.; Xu, H.; Li, Y.; Luo, T. The Composition of Nitrogen-Fixing Microorganisms Correlates With Soil Nitrogen Content During Reforestation: A Comparison Between Legume and Non-legume Plantations. Front. Microbiol. 2019, 10, 508. [Google Scholar] [CrossRef]
- Hijbeek, R.; Van Ittersum, M.; Berge, H.T.; Gort, G.; Spiegel, H.; Whitmore, A. Do organic inputs matter—A meta-analysis of additional yield effects for arable crops in Europe. Plant Soil 2017, 411, 293–303. [Google Scholar] [CrossRef]
- Li, H.-B.; Singh, R.K.; Singh, P.; Song, Q.-Q.; Xing, Y.-X.; Yang, L.-T.; Li, Y.-R. Genetic Diversity of Nitrogen-Fixing and Plant Growth Promoting Pseudomonas Species Isolated from Sugarcane Rhizosphere. Front. Microbiol. 2017, 8, 1268. [Google Scholar] [CrossRef]
- Cozzolino, V.; Di Meo, V.; Monda, H.; Spaccini, R.; Piccolo, A. The molecular characteristics of compost affect plant growth, arbuscular mycorrhizal fungi, and soil microbial community composition. Biol. Fertil. Soils 2015, 52, 15–29. [Google Scholar] [CrossRef]
- Baumann, K.; Marschner, P.; Smernik, R.J.; Baldock, J.A. Residue chemistry and microbial community structure during decomposition of eucalypt, wheat and vetch residues. Soil Biol. Biochem. 2009, 41, 1966–1975. [Google Scholar] [CrossRef]
- Tao, C.; Li, R.; Xiong, W.; Shen, Z.; Liu, S.; Wang, B.; Ruan, Y.; Geisen, S.; Shen, Q.; Kowalchuk, G.A. Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 2020, 8, 1–14. [Google Scholar] [CrossRef]
- Zelezniak, A.; Andrejev, S.; Ponomarova, O.; Mende, D.R.; Bork, P.; Patil, K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. USA 2015, 112, 6449–6454. [Google Scholar] [CrossRef]
- Jiao, S.; Xu, Y.; Zhang, J.; Hao, X.; Lu, Y. Core Microbiota in Agricultural Soils and Their Potential Associations with Nutrient Cycling. mSystems 2019, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Ren, H.; Li, S.; Leng, X.; Yao, X. Soil Bacterial Community Structure and Co-occurrence Pattern during Vegetation Restoration in Karst Rocky Desertification Area. Front. Microbiol. 2017, 8, 2377. [Google Scholar] [CrossRef]
- Liang, W.; Huang, S.; Liu, J.; Zhang, R.; Yan, F. Removal of nitric oxide in a biotrickling filter under thermophilic condition using Chelatococcus daeguensis. J. Air Waste Manag. Assoc. 2012, 62, 509–516. [Google Scholar] [CrossRef]
- Valenzuela-Encinas, C.; Neria-González, I.; Alcántara-Hernández, R.J.; Estrada-Alvarado, I.; De La Serna, F.J.Z.-D.; Dendooven, L.; Marsch, R. Changes in the bacterial populations of the highly alkaline saline soil of the former lake Texcoco (Mexico) following flooding. Extremophiles 2009, 13, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Leys, N.M.E.J.; Ryngaert, A.; Bastiaens, L.; Verstraete, W.; Top, E.M.; Springael, D. Occurrence and Phylogenetic Diversity of Sphingomonas Strains in Soils Contaminated with Polycyclic Aromatic Hydrocarbons. Appl. Environ. Microbiol. 2004, 70, 1944–1955. [Google Scholar] [CrossRef]
- Bending, G.D.; Lincoln, S.D.; Sørensen, S.R.; Morgan, J.A.W.; Aamand, J.; Walker, A. In-Field Spatial Variability in the Degradation of the Phenyl-Urea Herbicide Isoproturon Is the Result of Interactions between Degradative Sphingomonas spp. and Soil pH. Appl. Environ. Microbiol. 2003, 69, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, S.R.; Ronen, Z.; Aamand, J. Isolation from Agricultural Soil and Characterization of a Sphingomonas sp. Able To Mineralize the Phenylurea Herbicide Isoproturon. Appl. Environ. Microbiol. 2001, 67, 5403–5409. [Google Scholar] [CrossRef] [PubMed]
- Ardley, J.; Parker, M.A.; De Meyer, S.E.; Trengove, R.D.; O’Hara, G.W.; Reeve, W.G.; Yates, R.J.; Dilworth, M.J.; Willems, A.; Howieson, J.G. Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov. are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int. J. Syst. Evol. Microbiol. 2012, 62, 2579–2588. [Google Scholar] [CrossRef] [PubMed]
- Pathan, S.I.; Vetrovsky, T.; Giagnoni, L.; Datta, R.; Baldrian, P.; Nannipieri, P.; Renella, G. Microbial expression profiles in the rhizosphere of two maize lines differing in N use efficiency. Plant Soil 2018, 433, 401–413. [Google Scholar] [CrossRef]
- Feeney, D.S.; Crawford, J.W.; Daniell, T.; Hallett, P.D.; Nunan, N.; Ritz, K.; Rivers, M.; Young, I.M. Three-dimensional Microorganization of the Soil–Root–Microbe System. Microb. Ecol. 2006, 52, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Ponisio, L.C.; M’Gonigle, L.K.; Mace, K.C.; Palomino, J.; De Valpine, P.; Kremen, C. Diversification practices reduce organic to conventional yield gap. Proc. R. Soc. B Boil. Sci. 2015, 282, 20141396. [Google Scholar] [CrossRef] [PubMed]
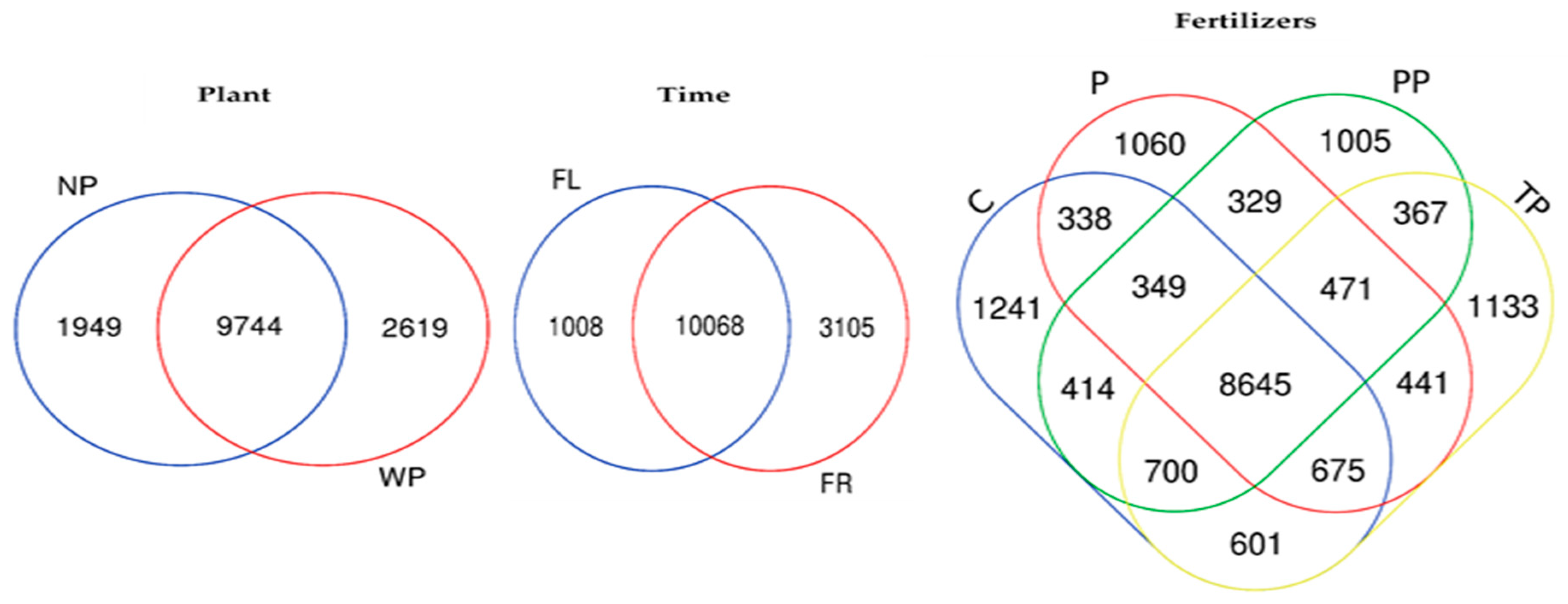


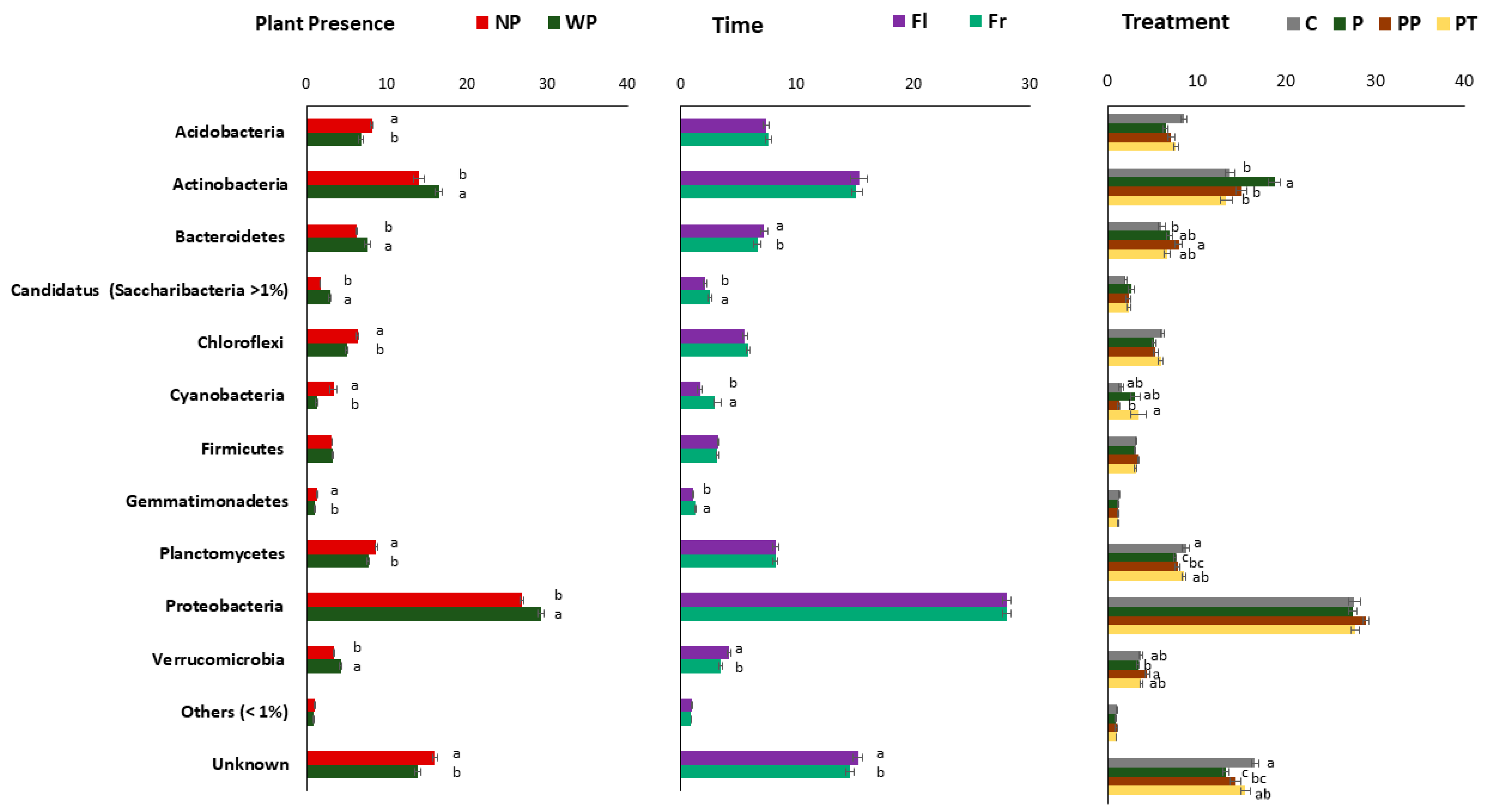
| Sand% | Silt% | Clay% | TotalCaCO3% | pHH2O | OM% | CECcmolc kg−1 | Exchangeable Cations cmolc kg−1 | EC1:2.5mS cm−1 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ca++ | Mg++ | Na+ | K+ | ||||||||
| 88.80 | 7.40 | 3.80 | 0.00 | 6.90 | 0.30 | 3.50 | 1.91 | 0.05 | 0.00 | 0.08 | 0.05 |
| N % | P2O5 % | OC % | SO3 % | |
|---|---|---|---|---|
| VTLW | 3.0 | 11.0 | 17.2 | 12.0 |
| PCM | 3.0 | 2.0 | 71.0 | 0.0 |
| TP | 0.0 | 46.0 | 0.0 | 0.0 |
| U | 46.0 | 0.0 | 0.0 | 0.0 |
| NSRP | 0.0 | 25.0 | 0.0 | 0.0 |
| Treatment | Stems Length (mm) | Stem and Leaves Dry Weight (g) | Roots Collar Diameter (g) | Roots Length (g) | Roots Dry Weight (g) | Seeds (n°) | Seed Dry Weight (g) |
|---|---|---|---|---|---|---|---|
| C | 556.7 ± 23.2 a | 15.7 ± 1.2 a | 9.4 ± 0.8 a | 567.3 ± 40.1 a | 5.5 ± 1.1 ab | 11.4 ± 3.5 ab | 11.9 ± 5.5 ab |
| P | 510 ± 20.4 a | 15.8 ± 6.9 a | 10.4 ± 0.4 a | 413.3 ± 20.5 b | 3.1 ± 1.3 b | 4.6 ± 1.2 b | 5.7 ± 2.4 b |
| PP | 517.6 ± 14.3 a | 18.5 ± 5.3 a | 9.6 ± 0.4 a | 507.9 ± 52.6 ab | 8.9 ± 1.9 a | 15.5 ± 6.5 ab | 13.7 ± 3.3 ab |
| PT | 526.3 ± 13.5 a | 13.3 ± 0.9 a | 10.1 ± 0.4 a | 533.8 ± 39.1 a | 7.2 ± 0.5 a | 17.4 ± 2.6 a | 19.1 ± 3.2 a |
| Factor | df# | Shannon-Wiener Index | Evenness | Chao-1 |
|---|---|---|---|---|
| Fertilizers | 5 | 6.13 ** | 23.86 *** | 0.22 ns |
| Time | 1 | 3.37 * | 25.44 *** | 7.67 ** |
| Plant presence | 1 | 0.07 ns | 0.22 ns | 0.10 ns |
| Fertilizers × Time | 5 | 1.06 ns | 3.79 * | 0.12 ns |
| Fertilizers × Plant presence | 5 | 3.86 * | 3.02 * | 3.93 * |
| Time × Plant presence | 1 | 0.01 ns | 2.27 ns | 0.01 ns |
| Fertilizers × Time × Plant presence | 5 | 2.95 * | 4.06 * | 2.210 ns |
 .
.
 .
.| a | Soil Parameters | Flowering | Fruiting | |||||
| Shannon Index | Evenness | Chao-1 | Shannon Index | Evenness | Chao-1 | |||
| With plant | NO3-N | −0.33 | −0.27 | −0.14 | −0.24 | 0.04 | −0.23 | |
| NO2-N | 0.05 | 0.43 | −0.12 | 0 | 0.02 | −0.19 | ||
| NH4-N | 0.07 | 0.75 | −0.28 | −0.39 | −0.11 | −0.26 | ||
| U-N | 0.22 | −0.11 | 0.3 | −0.14 | 0.22 | −0.41 | ||
| Without plant | NO3-N | −0.47 | −0.59 | −0.18 | 0.25 | 0.47 | −0.3 | |
| NO2-N | 0.38 | 0.78 | 0.08 | 0.33 | 0.07 | 0.37 | ||
| NH4-N | −0.91 | −0.85 | −0.59 | −0.35 | −0.47 | 0.02 | ||
| U-N | −0.54 | −0.79 | −0.2 | −0.64 | −0.75 | −0.06 | ||
| b | Plant parameters | Flowering | Fruiting | |||||
| Shannon Index | Evenness | Chao-1 | Shannon Index | Evenness | Chao-1 | |||
| With plant | Stems length | −0.23 | 0.65 | -0.43 | 0.48 | 0.87 | −0.54 | |
| Stems dry weight | −0.26 | −0.02 | −0.22 | −0.1 | −0.04 | −0.06 | ||
| Roots collar diameter | 0.04 | −0.31 | 0.21 | −0.49 | −0.31 | −0.2 | ||
| Roots length | −0.02 | 0.61 | −0.25 | 0.61 | 0.86 | −0.3 | ||
| Roots dry weight | −0.44 | 0.78 | −0.61 | 0.37 | 0.52 | −0.27 | ||
| Seed n° | 0.2 | 0.45 | −0.05 | 0.26 | 0.36 | −0.04 | ||
| Seed dry weight | −0.04 | 0.43 | −0.14 | 0.38 | 0.28 | 0.14 | ||
| R-Value | p-Value | |
|---|---|---|
| Soil edaphic factors | ||
| NH4-N | 0.13 | 0.028 |
| NO2-N | 0.08 | 0.039 |
| NO3-N | 0.20 | 0.002 |
| Urea-N | 0.11 | 0.032 |
| Plant production data | ||
| Stems length | 0.29 | 0.001 |
| Stems dry weight | 0.07 | 0.231 |
| Roots collar diameter | 0.06 | 0.222 |
| Roots length | 0.36 | 0.0003 |
| Roots dry weight | 0.38 | 0.0002 |
| Seeds | 0.08 | 0.157 |
| Seeds dry weight | 0.08 | 0.138 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pathan, S.I.; Scibetta, S.; Grassi, C.; Pietramellara, G.; Orlandini, S.; Ceccherini, M.T.; Napoli, M. Response of Soil Bacterial Community to Application of Organic and Inorganic Phosphate Based Fertilizers under Vicia faba L. Cultivation at Two Different Phenological Stages. Sustainability 2020, 12, 9706. https://doi.org/10.3390/su12229706
Pathan SI, Scibetta S, Grassi C, Pietramellara G, Orlandini S, Ceccherini MT, Napoli M. Response of Soil Bacterial Community to Application of Organic and Inorganic Phosphate Based Fertilizers under Vicia faba L. Cultivation at Two Different Phenological Stages. Sustainability. 2020; 12(22):9706. https://doi.org/10.3390/su12229706
Chicago/Turabian StylePathan, Shamina Imran, Silvia Scibetta, Chiara Grassi, Giacomo Pietramellara, Simone Orlandini, Maria Teresa Ceccherini, and Marco Napoli. 2020. "Response of Soil Bacterial Community to Application of Organic and Inorganic Phosphate Based Fertilizers under Vicia faba L. Cultivation at Two Different Phenological Stages" Sustainability 12, no. 22: 9706. https://doi.org/10.3390/su12229706
APA StylePathan, S. I., Scibetta, S., Grassi, C., Pietramellara, G., Orlandini, S., Ceccherini, M. T., & Napoli, M. (2020). Response of Soil Bacterial Community to Application of Organic and Inorganic Phosphate Based Fertilizers under Vicia faba L. Cultivation at Two Different Phenological Stages. Sustainability, 12(22), 9706. https://doi.org/10.3390/su12229706








