Genome-Wide mRNA and Long Non-Coding RNA Analysis of Porcine Trophoblast Cells Infected with Porcine Reproductive and Respiratory Syndrome Virus Associated with Reproductive Failure
Abstract
1. Introduction
2. Results
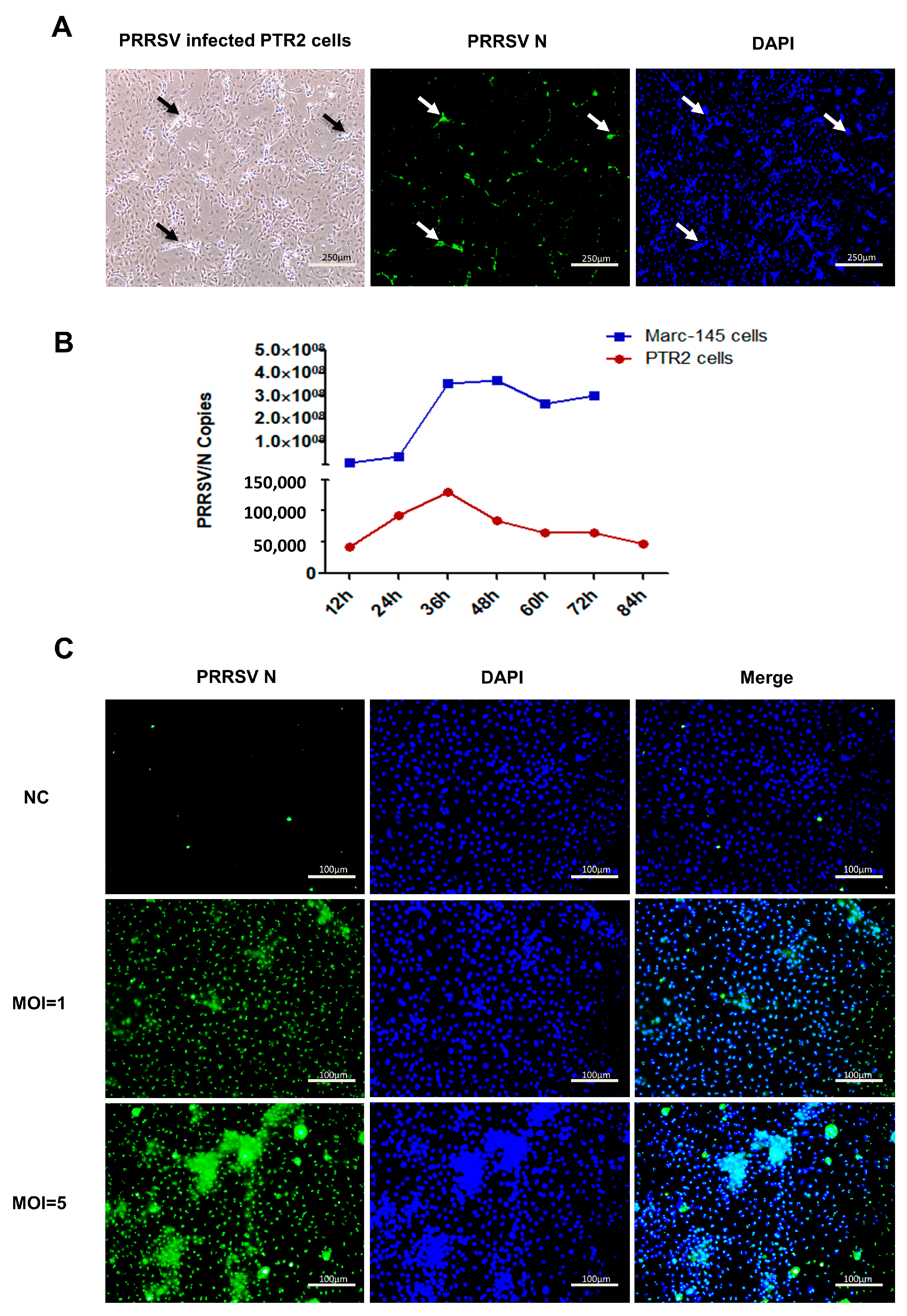
2.1. Cytopathic Effect (CPE) of PTR2 Infected with PRRSV
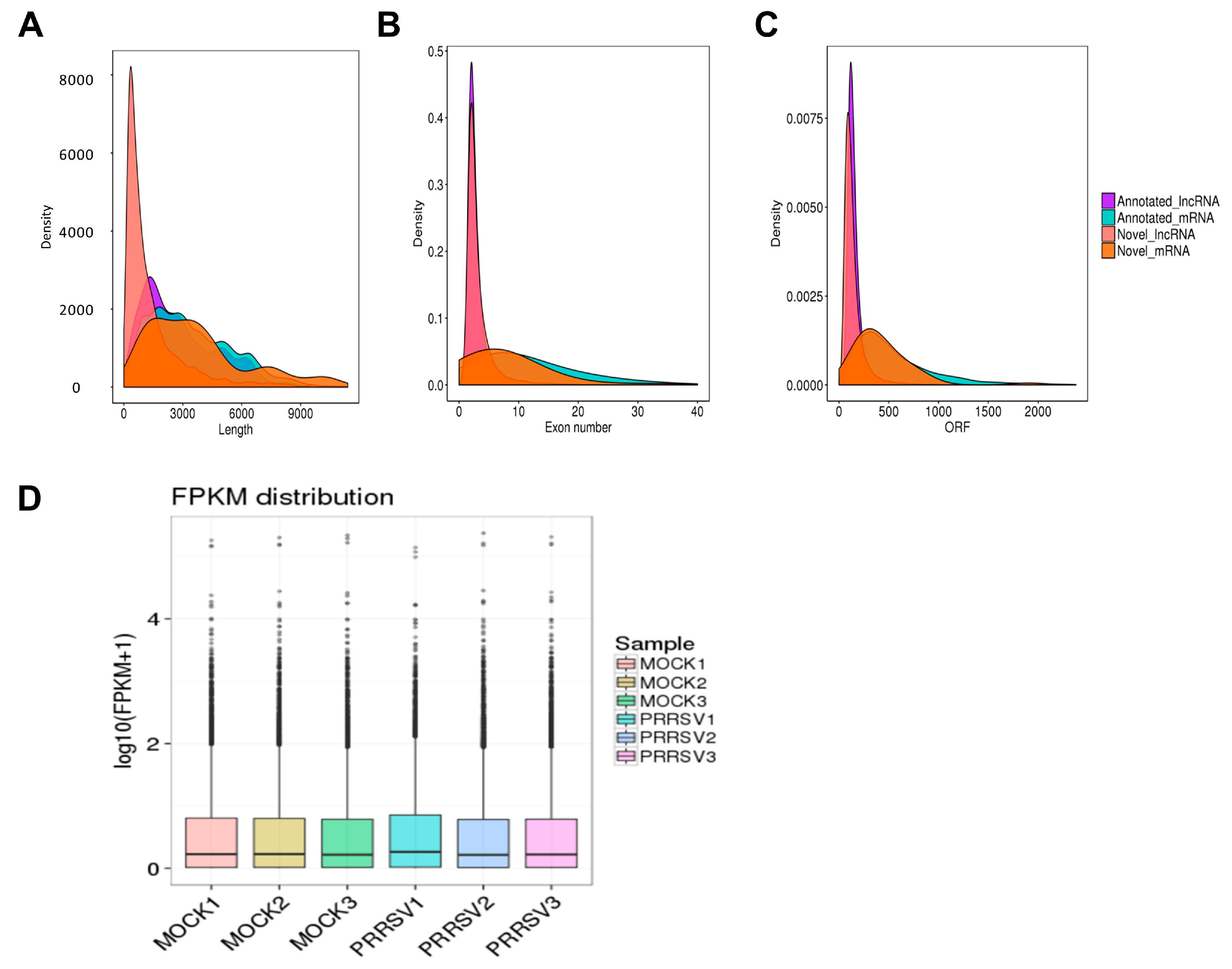
2.2. RNA Sequencing Data Processing and Quality Identification
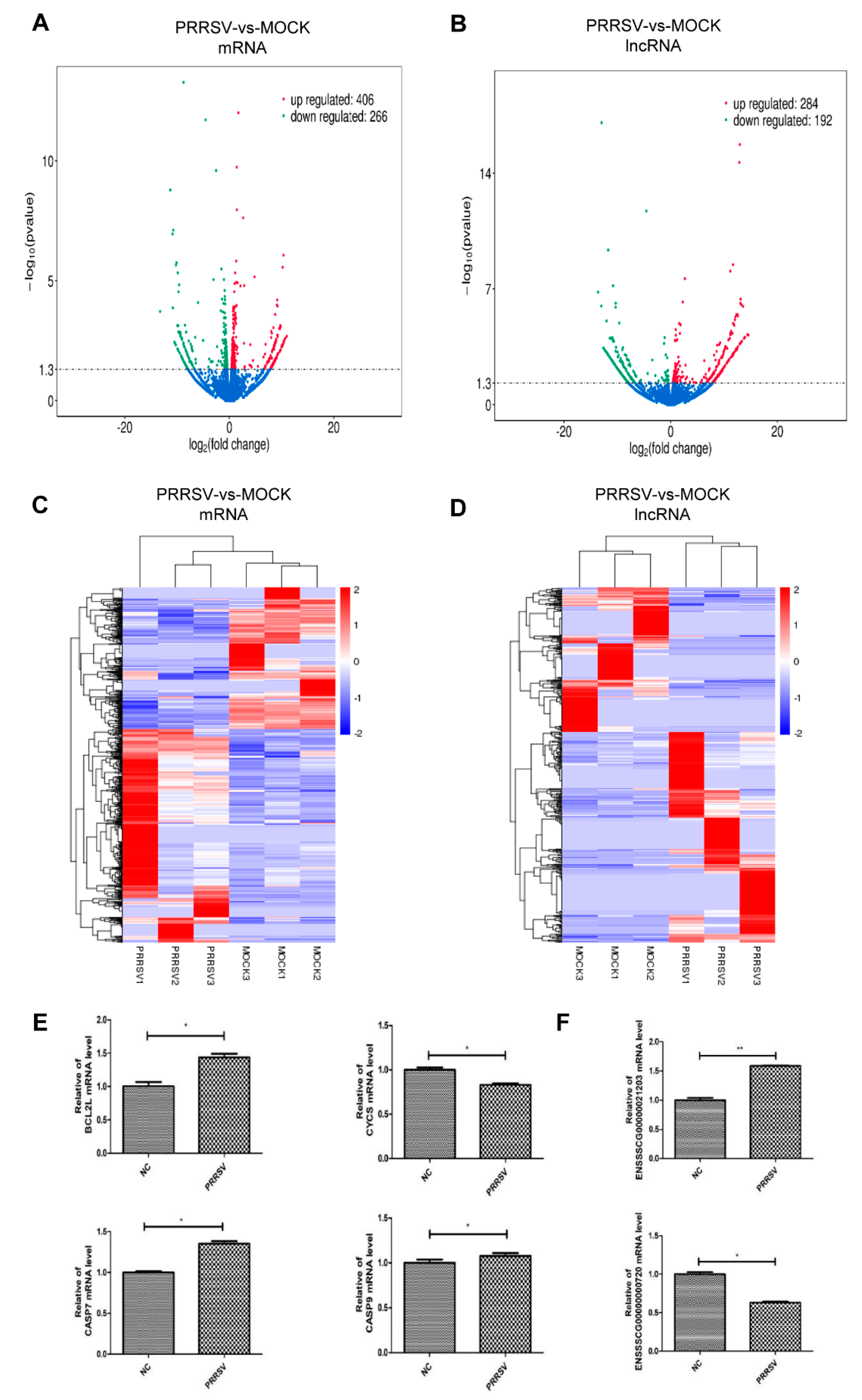
2.3. Expression Profiles of the mRNAs and lncRNAs in the PTR2 Cells with PRRSV Infection
2.4. Pathway Analysis for the Abnormal Expression of mRNAs and lncRNAs in the PTR2 Cells Infected with PRRSV
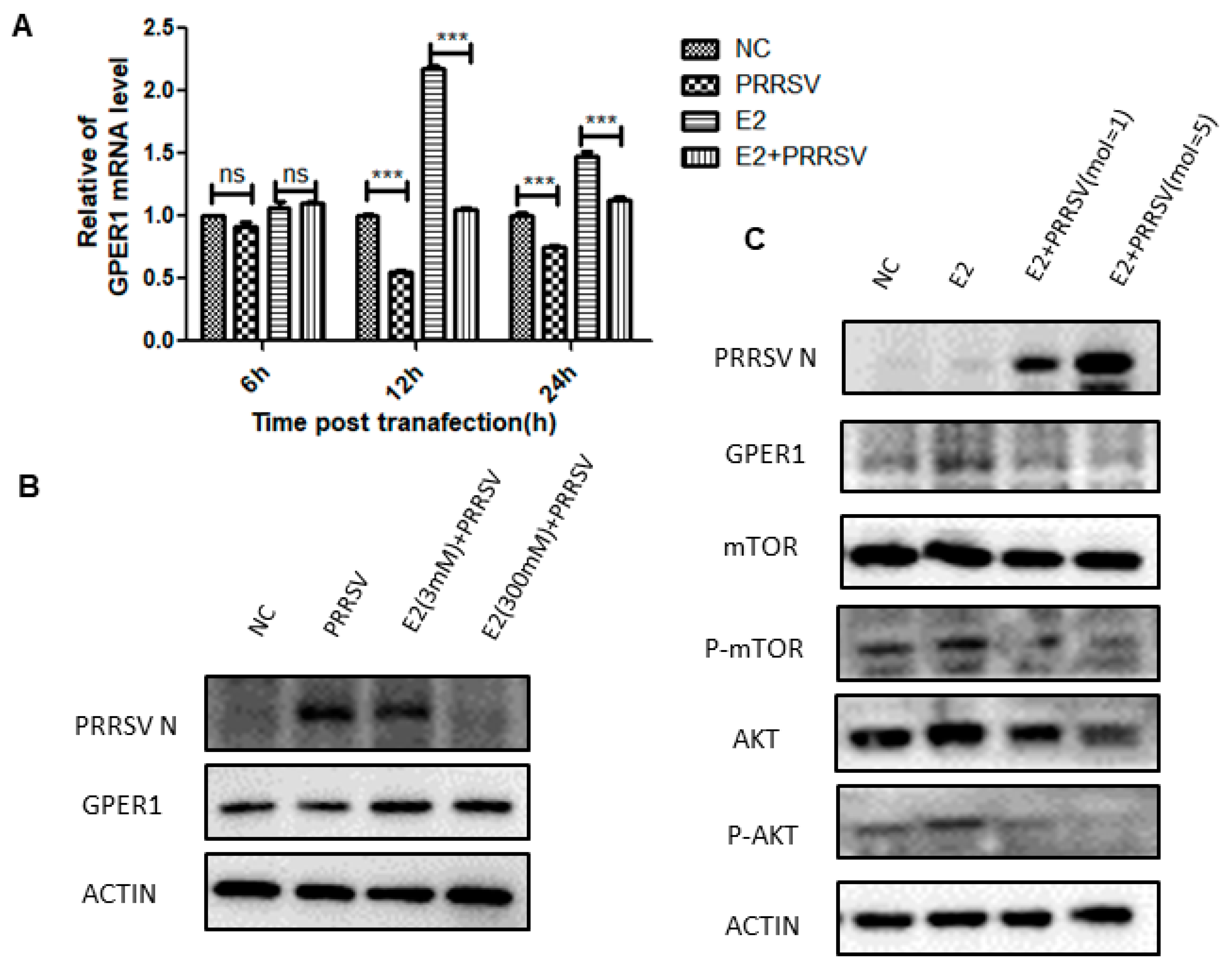
2.5. PRRSV Inhibits the Expression of GPER1 in PTR2 Cells and Downregulates the Activation of the PI3K-AKT-mTOR Signaling Pathway Induced by Estrogen
2.6. PRRSV Induced the Apoptosis of the PTR2 Cells
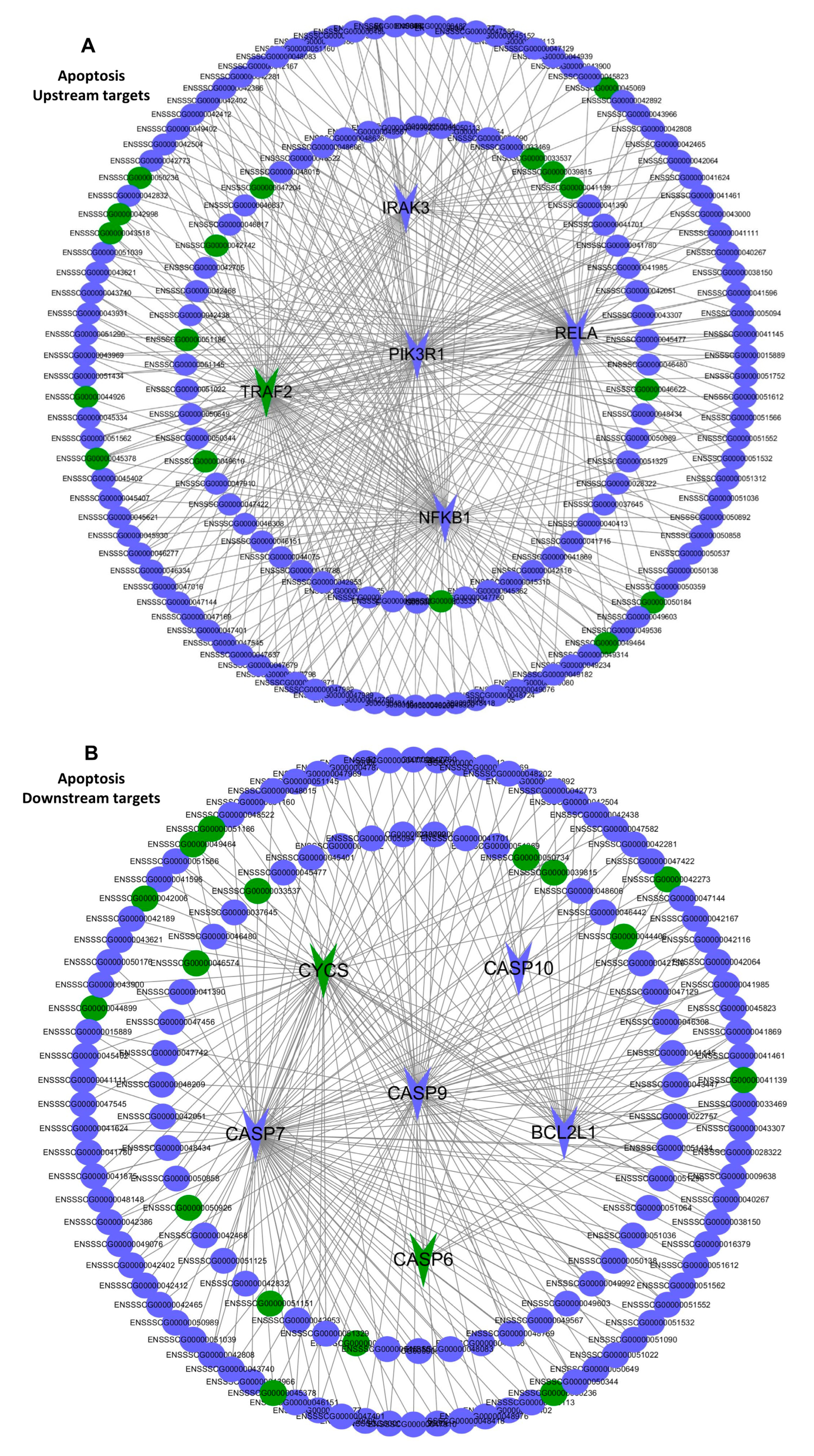
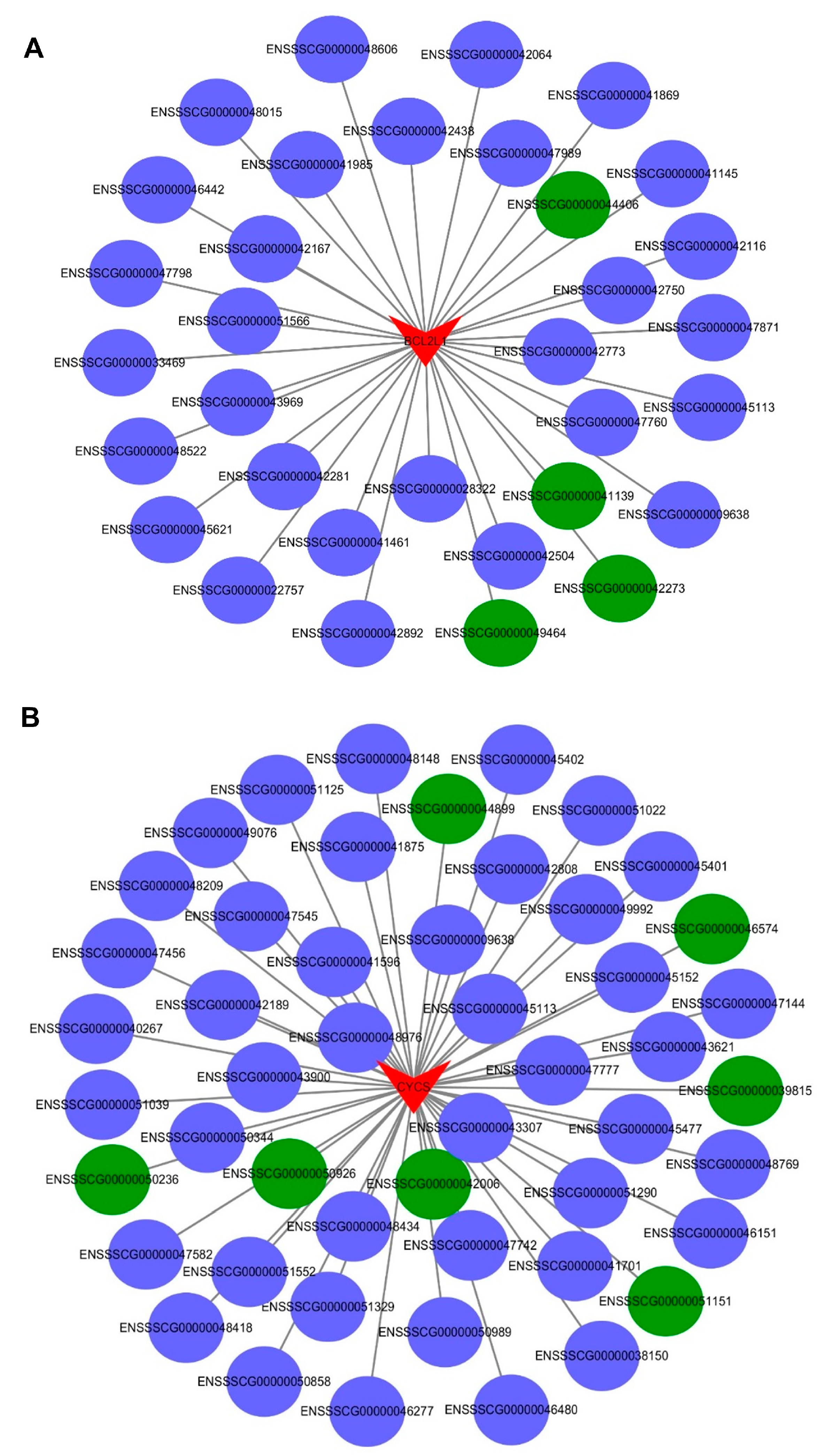
2.7. The Interaction between the Differentially Expressed mRNAs and lncRNAs Associated with Apoptosis in the PTR2 Cells Infected with PRRSV
3. Discussion
4. Material and Methods
4.1. Cell Cultures and Viral Infection
4.2. Immunofluorescence Antibody Assay (IFA)
4.3. RNA Extraction and Qualification
4.4. Preparation of the RNA Library and Transcript Sequencing
4.5. Sequencing Data Analysis
4.6. Differential Expression Analysis and Target Gene Prediction
4.7. Quantitative Real-Time PCR (qPCR) Detection
4.8. GO and KEGG Enrichment Analysis
4.9. Cell Apoptosis Analysis
4.10. Western Blot
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PRRSV | Porcine Reproductive and Respiratory Syndrome Virus |
| PTR2 | Porcine trophoblast cell |
| LncRNA | Long non-coding RNA |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PI3K | Phosphoinositide 3-kinase |
| Akt | The serine/threonine kinase |
| GPER1 | G protein coupled estrogen receptor 1 |
| mTOR | Mechanistic target of rapamycin |
| EGFR | epidermal growth factor receptor |
| MAPK | mitogen-activated protein kinase |
References
- Yuzhakov, A.G.; Raev, S.A.; Shchetinin, A.M.; Gushchin, V.A.; Alekseev, K.P.; Stafford, V.V.; Komina, A.K.; Zaberezhny, A.D.; Gulyukin, A.M.; Aliper, T.I. Full-genome analysis and pathogenicity of a genetically distinct Russian PRRSV-1 Tyu16 strain. Veter-Microbiol. 2020, 247, 108784. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Wang, F.; Liang, W.; Liu, J.; Gao, G.; Wang, Y.; Xu, X.; Su, Q.; Zhang, Q.; Liu, B. Identification of Differentially Expressed Non-coding RNA in Porcine Alveolar Macrophages from Tongcheng and Large White Pigs Responded to PRRSV. Sci. Rep. 2018, 8, 15621. [Google Scholar] [CrossRef]
- An, T.-Q.; Li, J.-N.; Su, C.-M.; Yoo, D. Molecular and Cellular Mechanisms for PRRSV Pathogenesis and Host Response to Infection. Virus Res. 2020, 286, 197980. [Google Scholar] [CrossRef]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Nan, Y.; Xiao, S.; Zhao, Q.; Zhou, E.-M. Antiviral Strategies against PRRSV Infection. Trends Microbiol. 2017, 25, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Friess, A.E.; Sinowatz, F.; Skolek-Winnisch, R.; Träautner, W. The placenta of the pig. I. Fine structural changes of the placental barrier during pregnancy. Anat. Embryol. 1980, 158, 179–191. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, Y.; Wang, L.; Yu, T.; Wang, Z.; Chang, L.; Zhao, X.; Luo, X.; Zhang, L.; Tong, D. Immortalization of porcine placental trophoblast cells through reconstitution of telomerase activity. Theriogenology 2016, 85, 1446–1456. [Google Scholar] [CrossRef]
- Noyola-Martínez, N.; Halhali, A.; Barrera, D. Steroid hormones and pregnancy. Gynecol. Endocrinol. 2019, 35, 376–384. [Google Scholar] [CrossRef]
- Suleman, M.; Malgarin, C.; Detmer, S.; Harding, J.; MacPhee, D. The porcine trophoblast cell line PTr2 is susceptible to porcine reproductive and respiratory syndrome virus-2 infection. Placenta 2019, 88, 44–51. [Google Scholar] [CrossRef]
- Chen, Q.; Men, Y.; Wang, D.; Xu, D.; Liu, S.; Xiao, S.; Fang, L. Porcine reproductive and respiratory syndrome virus infection induces endoplasmic reticulum stress, facilitates virus replication, and contributes to autophagy and apoptosis. Sci. Rep. 2020, 10, 13131. [Google Scholar] [CrossRef]
- Liu, Q.; Qin, Y.; Zhou, L.; Kou, Q.; Guo, X.; Ge, X.; Yang, H.; Hu, H. Autophagy sustains the replication of porcine reproductive and respiratory virus in host cells. Virology 2012, 429, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Peng, X.; Qiao, M.; Zhao, H.; Li, M.; Liu, G.; Mei, S. Genome-wide analysis of long noncoding RNA and mRNA profiles in PRRSV-infected porcine alveolar macrophages. Genomics 2020, 112, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Wang, L.; Wu, H.; Singh, D.; Zhang, X. Integrative analysis of microRNA and mRNA expression profiles in MARC-145 cells infected with PRRSV. Virus Genes 2020, 56, 610–620. [Google Scholar] [CrossRef]
- Zhang, K.; Ge, L.; Dong, S.; Liu, Y.; Wang, D.; Zhou, C.; Ma, C.; Wang, Y.; Su, F.; Jiang, Y. Global miRNA, lncRNA, and mRNA Transcriptome Profiling of Endometrial Epithelial Cells Reveals Genes Related to Porcine Reproductive Failure Caused by Porcine Reproductive and Respiratory Syndrome Virus. Front. Immunol. 2019, 10, 1221. [Google Scholar] [CrossRef]
- Jayakodi, M.; Selvan, S.G.; Natesan, S.; Muthurajan, R.; Duraisamy, R.; Ramineni, J.J.; Rathinasamy, S.A.; Karuppusamy, N.; Lakshmanan, P.; Chokkappan, M. A web accessible resource for investigating cassava phenomics and genomics information: BIOGEN BASE. Bioinformation 2011, 6, 391–392. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fuentes, N.; Silveyra, P. Estrogen receptor signaling mechanisms. Adv. Protein Chem. Struct. Biol. 2019, 116, 135–170. [Google Scholar] [CrossRef]
- Qu, J.; Han, Y.; Zhao, Z.; Wu, Y.; Lu, Y.; Chen, G.; Jiang, J.; Qiu, L.; Gu, A.; Wang, X. Perfluorooctane sulfonate interferes with non-genomic estrogen receptor signaling pathway, inhibits ERK1/2 activation and induces apoptosis in mouse spermatocyte-derived cells. Toxicology 2021, 460, 152871. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Chen, J.; Ai, Y.; Gu, X.; Li, L.; Che, D.; Jiang, Z.; Chen, S.; Huang, H.; Wang, J.; et al. Estrogen-Related Hormones Induce Apoptosis by Stabilizing Schlafen-12 Protein Turnover. Mol. Cell 2019, 75, 1103–1116. [Google Scholar] [CrossRef]
- Geisert, R.D.; Brookbank, J.W.; Roberts, R.M.; Bazer, F.W. Establishment of Pregnancy in the Pig: II. Cellular Remodeling of the Porcine Blastocyst During Elongation on Day 12 of Pregnancy12. Biol. Reprod. 1982, 27, 941–955. [Google Scholar] [CrossRef]
- Ross, J.W.; Ashworth, M.D.; Stein, D.R.; Couture, O.P.; Tuggle, C.K.; Geisert, R.D. Identification of differential gene expression during porcine conceptus rapid trophoblastic elongation and attachment to uterine luminal epithelium. Physiol. Genom. 2009, 36, 140–148. [Google Scholar] [CrossRef]
- Vallet, J.L.; Miles, J.R.; Freking, B.A. Development of the pig placenta. Soc. Reprod. Fertil Suppl. 2009, 66, 265–279. [Google Scholar] [CrossRef]
- Revankar, C.M.; Cimino, D.F.; Sklar, L.A.; Arterburn, J.B.; Prossnitz, E.R. A Transmembrane Intracellular Estrogen Receptor Mediates Rapid Cell Signaling. Science 2005, 307, 1625–1630. [Google Scholar] [CrossRef]
- Li, Z.; Lu, Q.; Ding, B.; Xu, J.; Shen, Y. Bisphenol A promotes the proliferation of leiomyoma cells by GPR30-EGFR signaling pathway. J. Obstet. Gynaecol. Res. 2019, 45, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Filardo, E.J. Epidermal growth factor receptor (EGFR) transactivation by estrogen via the G-protein-coupled receptor, GPR30: A novel signaling pathway with potential significance for breast cancer. J. Steroid Biochem. Mol. Biol. 2001, 80, 231–238. [Google Scholar] [CrossRef]
- Tuo, Y.; Xiang, M. mTOR: A double-edged sword for diabetes. J. Leukoc. Biol. 2018, 106, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, J.; Yang, J.; Xiao, L.; Hua, Q.; Zou, Y. PI3K/AKT/mTOR signaling participates in insulin-mediated regulation of pathological myopia-related factors in retinal pigment epithelial cells. BMC Ophthalmol. 2021, 21, 218. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.L.; Cui, W. Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development 2016, 143, 3050–3060. [Google Scholar] [CrossRef]
- Tong, L.; Li, W.; Zhang, Y.; Zhou, F.; Zhao, Y.; Zhao, L.; Liu, J.; Song, Z.; Yu, M.; Zhou, C.; et al. Tacrolimus inhibits insulin release and promotes apoptosis of Min6 cells through the inhibition of the PI3K/Akt/mTOR pathway. Mol. Med. Rep. 2021, 24, 658. [Google Scholar] [CrossRef]
- Xu, Z.; Han, X.; Ou, D.; Liu, T.; Li, Z.; Jiang, G.; Liu, J.; Zhang, J. Targeting PI3K/AKT/mTOR-mediated autophagy for tumor therapy. Appl. Microbiol. Biotechnol. 2019, 104, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-J.; Zhu, J.-P.; Zhou, T.; Cheng, Q.; Yu, L.-X.; Wang, Y.-X.; Yang, S.; Jiang, Y.-F.; Tong, W.; Gao, F.; et al. Identification of Differentially Expressed Proteins in Porcine Alveolar Macrophages Infected with Virulent/Attenuated Strains of Porcine Reproductive and Respiratory Syndrome Virus. PLoS ONE 2014, 9, e85767. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, M.; Liu, X.; Pan, Y.; Yang, R.; Zhao, Z.; Sun, B. Porcine IFI30 inhibits PRRSV proliferation and host cell apoptosis in vitro. Gene 2018, 649, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-Y.; Li, L.-P. MiR-140-5p targets BCL2L1 to promote cardiomyocyte apoptosis. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1. [Google Scholar] [PubMed]
- Liang, Y.-D.; Zhao, K.; Chen, Y.; Zhang, S.-Q. Role of CYCS in the cytogenesis and apoptosis of male germ cells and its clinical application. Zhonghua Nan Ke Xue 2020, 26, 265–270. [Google Scholar] [PubMed]
- Guo, F.; Li, Z.; Song, L.; Han, T.; Feng, Q.; Guo, Y.; Xu, J.; He, M.; You, C. Increased apoptosis and cysteinyl aspartate specific protease-3 gene expression in human intracranial aneurysm. J. Clin. Neurosci. 2007, 14, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Legate, K.R.; Fässler, R. Mechanisms that regulate adaptor binding to β-integrin cytoplasmic tails. J. Cell Sci. 2009, 122 Pt 2, 187–198. [Google Scholar] [CrossRef]
- Legate, K.R.; Wickström, S.A.; Fässler, R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009, 23, 397–418. [Google Scholar] [CrossRef]
- Vehlow, A.; Cordes, N. Invasion as target for therapy of glioblastoma multiforme. Biochim. Biophys. Acta 2013, 1836, 236–244. [Google Scholar] [CrossRef]
- Reinardy, H.C.; Emerson, C.E.; Manley, J.M.; Bodnar, A.G. Tissue Regeneration and Biomineralization in Sea Urchins: Role of Notch Signaling and Presence of Stem Cell Markers. PLoS ONE 2015, 10, e0133860. [Google Scholar] [CrossRef]
- Wang, H.; Zang, C.; Liu, X.S.; Aster, J.C. The Role of Notch Receptors in Transcriptional Regulation. J. Cell Physiol. 2015, 230, 982–988. [Google Scholar] [CrossRef]
- Li, X.-G.; Wang, X.-Q. Notch Signaling in Mammalian Intestinal Stem Cells: Determining Cell Fate and Maintaining Homeostasis. Curr. Stem Cell Res. Ther. 2019, 14, 583–590. [Google Scholar] [CrossRef]
- Zhu, M.; Iwano, T.; Takeda, S. Estrogen and EGFR Pathways Regulate Notch Signaling in Opposing Directions for Multi-Ciliogenesis in the Fallopian Tube. Cells 2019, 8, 933. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, Y.; Xiang, X.; Sharma, M.; Liu, K.; Wei, J.; Shao, D.; Li, B.; Tong, G.; Olszewski, M.A.; et al. Notch signaling contributes to the expression of inflammatory cytokines induced by highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) infection in porcine alveolar macrophages. Dev. Comp. Immunol. 2020, 108, 103690. [Google Scholar] [CrossRef]
- Han, Y. Analysis of the role of the Hippo pathway in cancer. J. Transl. Med. 2019, 17, 116. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Shin, J.E.; Park, H.W. The Role of Hippo Pathway in Cancer Stem Cell Biology. Mol. Cells 2018, 41, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Moroishi, T.; Guan, K.-L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Zhang, P.; Hung, L.-H.; Lloyd, W.; Yeung, K.Y. Hot-starting software containers for STAR aligner. GigaScience 2018, 7, giy092. [Google Scholar] [CrossRef]



| Sample | Raw Reads | Clean Reads | Q20 (%) | Q30 (%) | GC Content (%) | Total Mapped (%) |
|---|---|---|---|---|---|---|
| Mock 1 | 93772488 | 91548686 | 98.17 | 94.83 | 54.04 | 86282174 (94.25%) |
| Mock 2 | 93224184 | 89721222 | 98.16 | 94.81 | 54.85 | 85032843 (94.77%) |
| Mock3 | 89685878 | 87053146 | 98.16 | 94.80 | 54.50 | 81428341 (93.54%) |
| PRRSV1 | 91868782 | 88302932 | 97.78 | 94.06 | 53.83 | 76908982 (87.1%) |
| PRRSV2 | 96897448 | 94476562 | 98.32 | 95.14 | 52.24 | 81424721 (86.19%) |
| PRRSV3 | 107694804 | 105835468 | 98.31 | 95.14 | 52.59 | 91366566 (86.33%) |
| Average | 95523931 | 92823003 | 98.15 | 94.80 | 53.68 | 83740605 (90.36%) |
| Genes | Forward Sequence (5;-3′) | Reverse Sequence (5;-3′) |
|---|---|---|
| PRRSV | CCAGTCAATCAGCTGTGCCA | GACAGGGTACAADTTCCAGCG |
| BCL2L1 | TTGAACGAACTCTTCCGGGA | GTTCTCCTGGATCCAAGGCT |
| CYCS | GTTCAGAAGTGTGCCCAGTG | CATCAGTGTCTCCTCTCCCC |
| CASP7 | CCGGATGACTCAGACATGGA | TTTCCTGTTCCTCCCCTGAC |
| CASP9 | CCGATTTGGCTTACGTCCTG | CAAAGCCTGGACCATTTGCT |
| ENSSSCG00000021203 | TCCACCAGAGCATGAACCAT | CTCCCACTGACTTGCAACAC |
| ENSSSCG00000000720 | CCCAAGCCAACTAAGGAGGA | AGACACAGCTTCATCACCGA |
| GPER1 | CTTCCTGTCCTGCGTCTACA | GTCGTAGTACTGCTCGTCCA |
| β-actin | CCGAGATCTCACCGACTACC | CTCGTAGCTCTTCTCCAGGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Liu, X.; Peng, J.; Song, S.; Xu, G.; Yang, N.; Wu, S.; Wang, L.; Wang, S.; Zhang, L.; et al. Genome-Wide mRNA and Long Non-Coding RNA Analysis of Porcine Trophoblast Cells Infected with Porcine Reproductive and Respiratory Syndrome Virus Associated with Reproductive Failure. Int. J. Mol. Sci. 2023, 24, 919. https://doi.org/10.3390/ijms24020919
Zhang X, Liu X, Peng J, Song S, Xu G, Yang N, Wu S, Wang L, Wang S, Zhang L, et al. Genome-Wide mRNA and Long Non-Coding RNA Analysis of Porcine Trophoblast Cells Infected with Porcine Reproductive and Respiratory Syndrome Virus Associated with Reproductive Failure. International Journal of Molecular Sciences. 2023; 24(2):919. https://doi.org/10.3390/ijms24020919
Chicago/Turabian StyleZhang, Xinming, Xianhui Liu, Jiawei Peng, Sunyangzi Song, Ge Xu, Ningjia Yang, Shoutang Wu, Lin Wang, Shuangyun Wang, Leyi Zhang, and et al. 2023. "Genome-Wide mRNA and Long Non-Coding RNA Analysis of Porcine Trophoblast Cells Infected with Porcine Reproductive and Respiratory Syndrome Virus Associated with Reproductive Failure" International Journal of Molecular Sciences 24, no. 2: 919. https://doi.org/10.3390/ijms24020919
APA StyleZhang, X., Liu, X., Peng, J., Song, S., Xu, G., Yang, N., Wu, S., Wang, L., Wang, S., Zhang, L., Liu, Y., Liang, P., Hong, L., Xu, Z., & Song, C. (2023). Genome-Wide mRNA and Long Non-Coding RNA Analysis of Porcine Trophoblast Cells Infected with Porcine Reproductive and Respiratory Syndrome Virus Associated with Reproductive Failure. International Journal of Molecular Sciences, 24(2), 919. https://doi.org/10.3390/ijms24020919





