Evolution of Laboratory Diagnosis of Tuberculosis
Abstract
1. Introduction
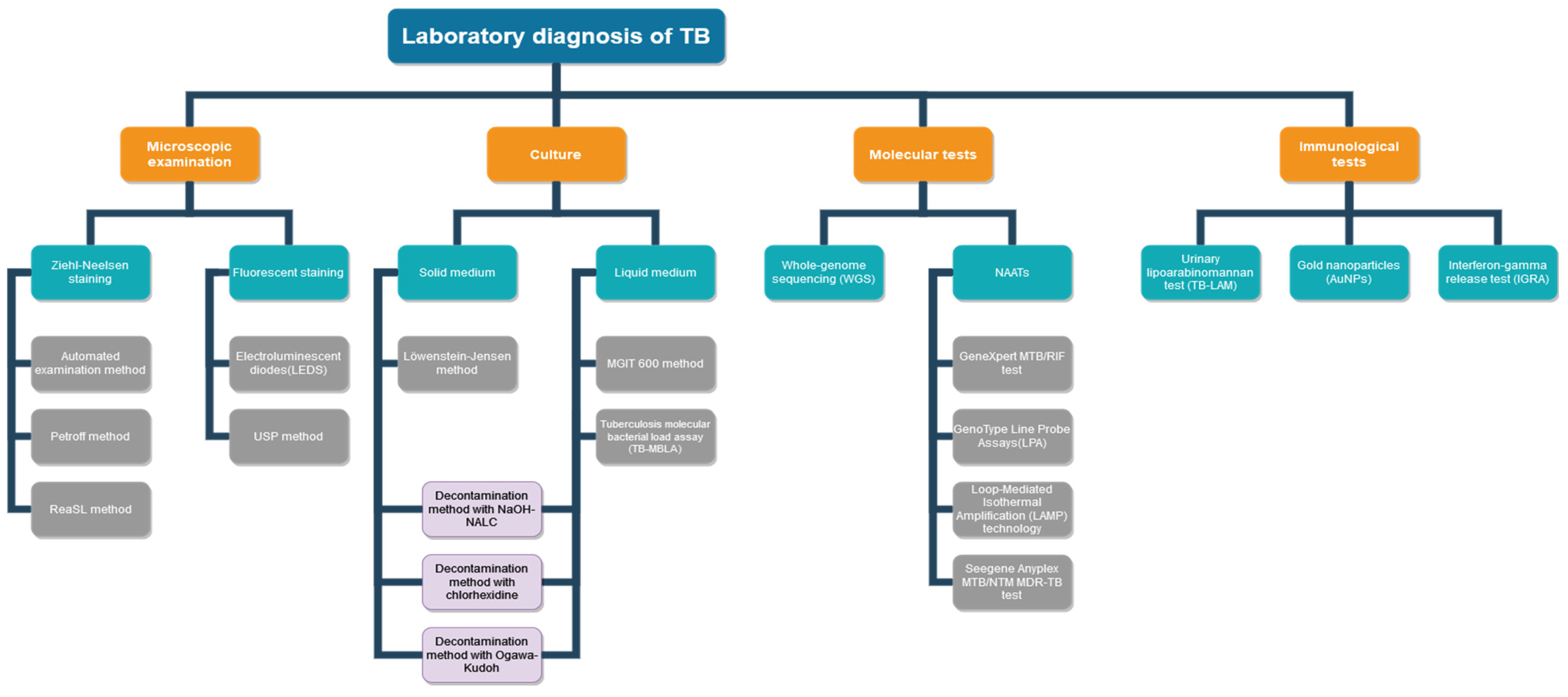
2. Microscopic Diagnosis of TB
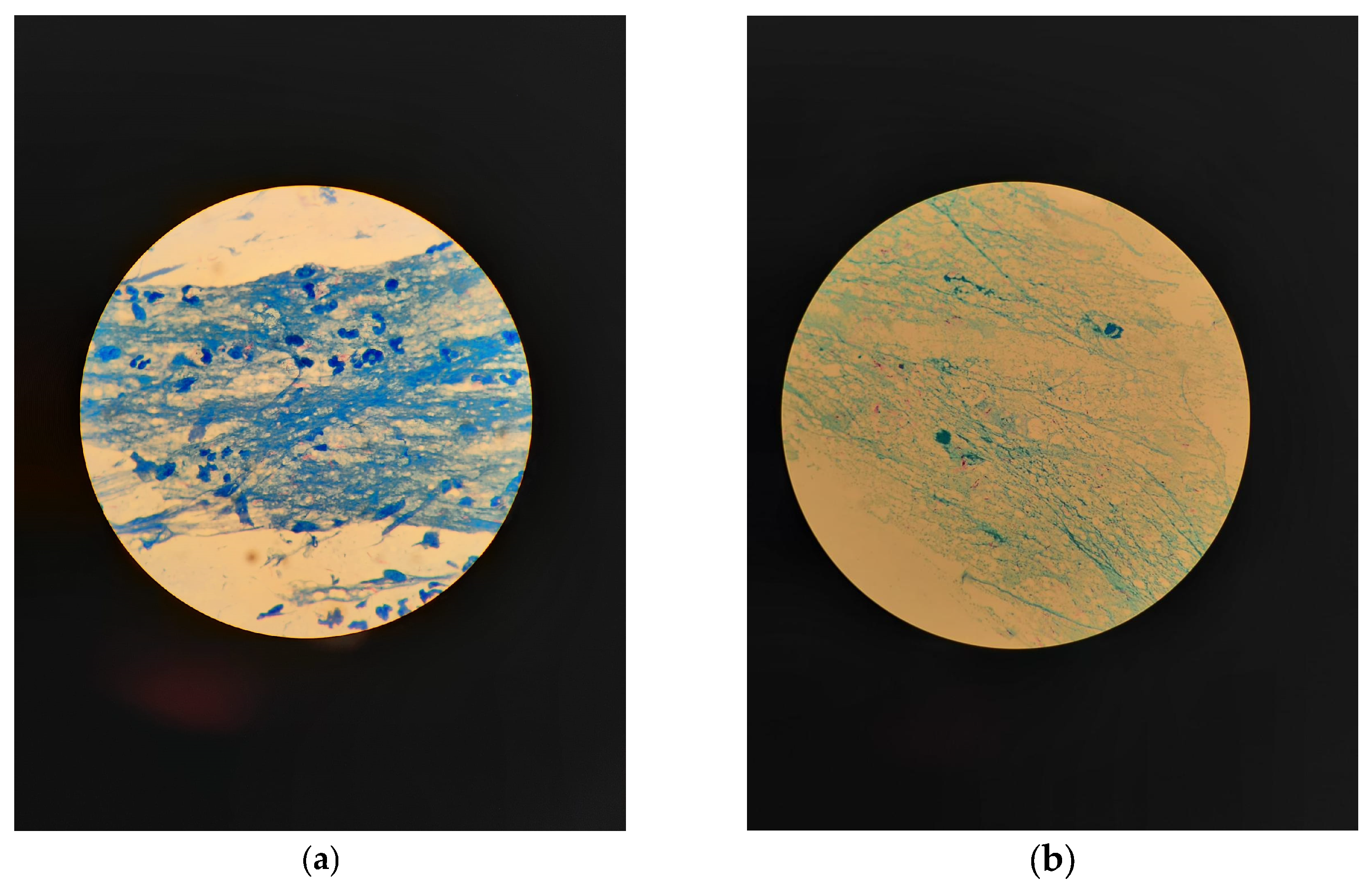
2.1. Microscopic Examination with Ziehl–Neelsen Staining
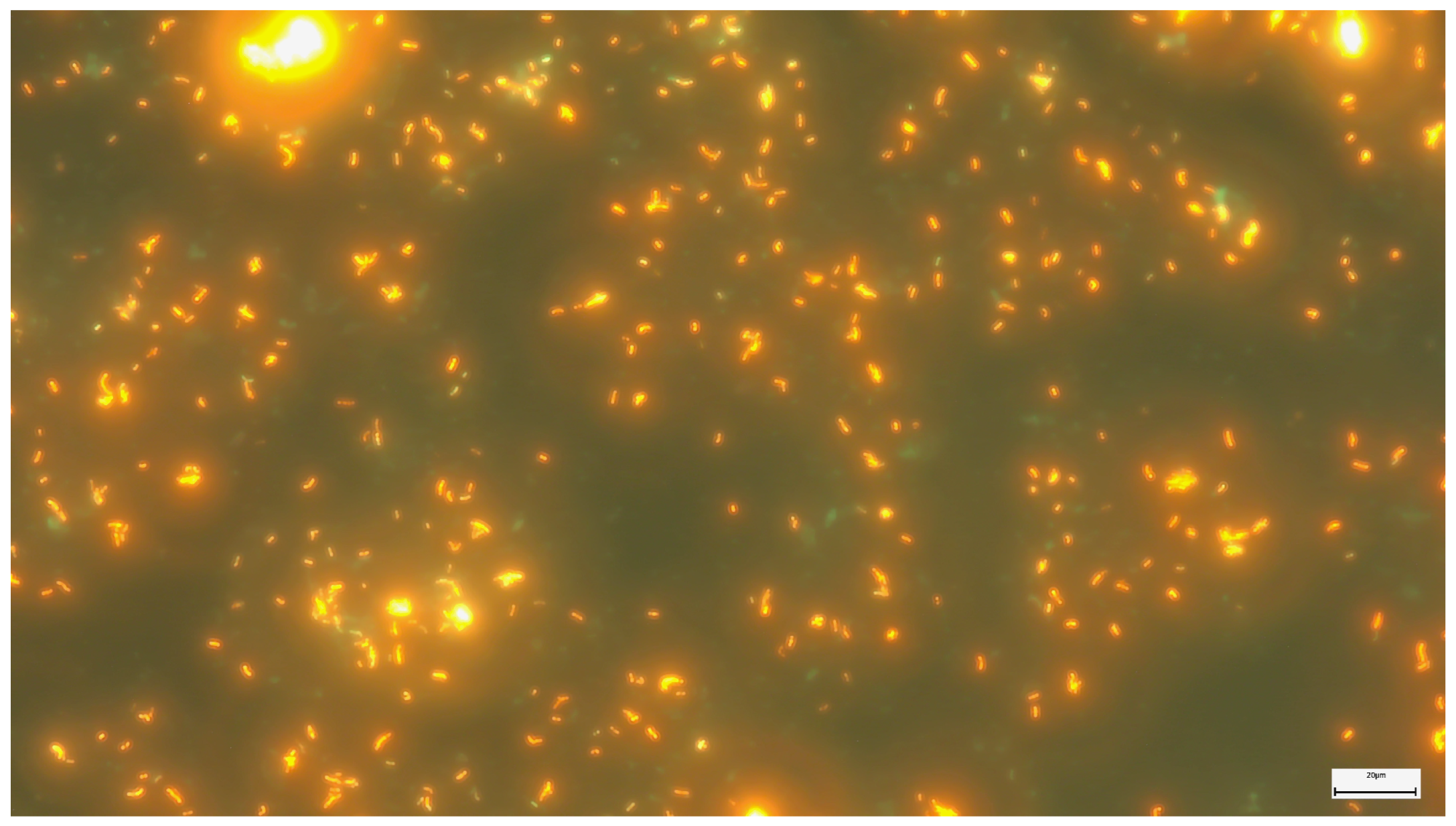
2.2. Microscopic Examination with Fluorescent Staining
2.3. The Fluorescent Microscopy Method with Electroluminescent Diodes (LEDs)
2.4. Automated Microscopic Examination Method
2.5. USP Method (Modified Auramine–Rhodamine Ziehl–Neelsen)
2.6. Petroff Method
2.7. ReaSLR Method
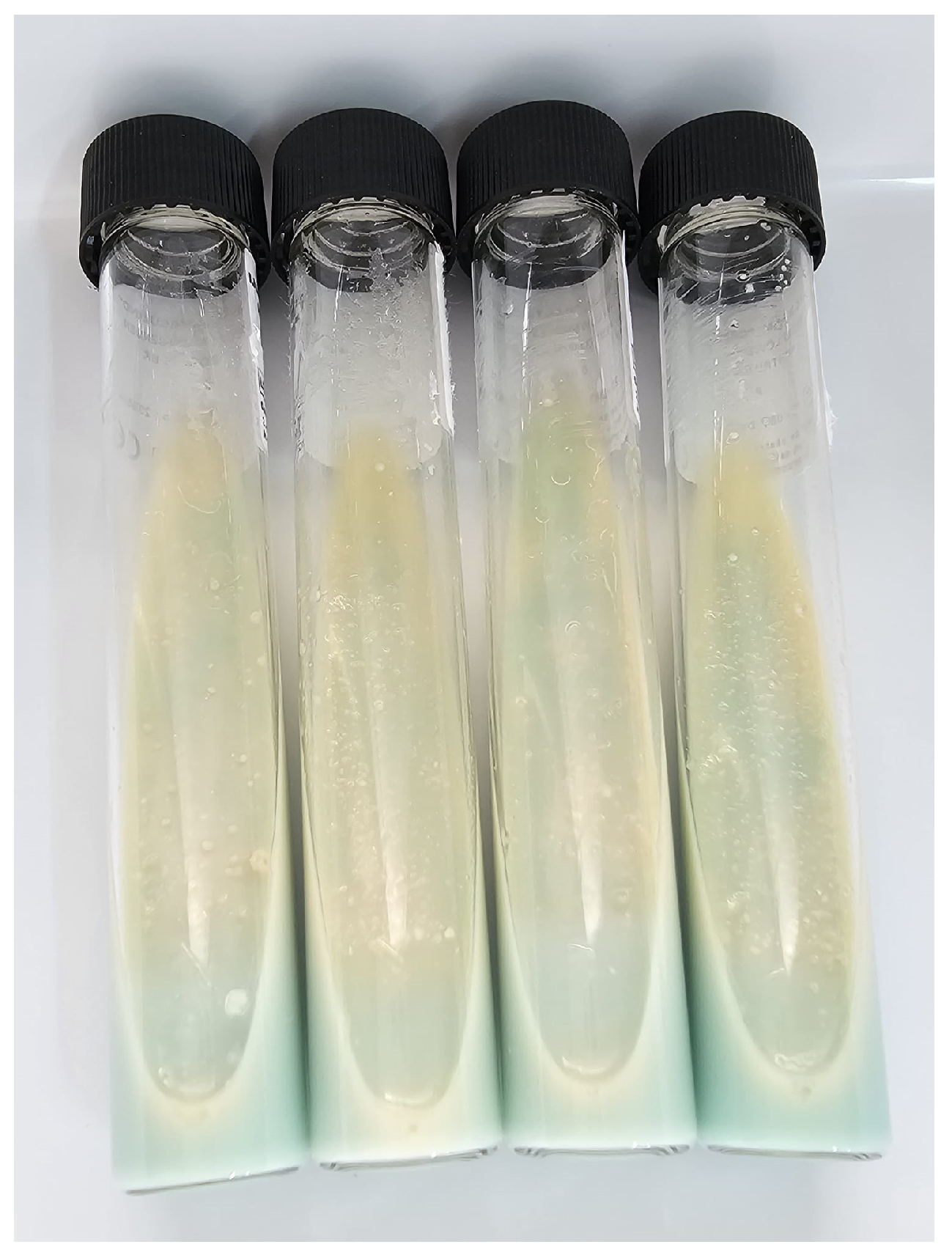
3. Mycobacterial Culture in TB Diagnosis
3.1. Lowenstein–Jensen Method (LJ)
3.2. MGIT 960 Technology
3.3. Decontamination Method with NaOH-NALC
3.4. Tuberculosis Molecular Bacterial Load Assay (TB-MBLA)
3.5. The Method of Decontamination with Chlorhexidine
3.6. Decontamination Method with Ogawa-Kudoh
4. Molecular Methods
4.1. Conventional Nucleic Acid Amplification Tests (NAAT)
4.1.1. GenoType Line Probe Assays (LPA)
4.1.2. Multidrug-Resistant Tuberculosis (MDR-TB)
4.1.3. Extensively Drug-Resistant Tuberculosis (XDR-TB)
4.2. Methods Based on Real-Time Genetic Amplification Technology RT-PCR GeneXpert MTB/RIF
4.3. Loop-Mediated Isothermal Amplification (LAMP) Technology
4.4. PCR Multiplex Seegene Anyplex MTB/NTM MDR-TB
4.5. WGS Sequencing (Whole-Genome Sequencing)
5. Diagnostic immunological tests
5.1. Urinary Lipoarabinomannan Test (TB-LAM)
5.2. Gold Nanoparticles (AuNPs)
5.3. Interferon-Gamma Release Tests (IGRA)
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rakotosamimanana, N.; Rabodoarivelo, M.S.; Palomino, J.C.; Martin, A.; Razanamparany, V.R. Exploring tuberculosis by molecular tests on DNA isolated from smear microscopy slides. Int. J. Infect. Dis. 2017, 56, 248–252. [Google Scholar] [CrossRef][Green Version]
- World Health Organization. Global Tuberculosis Report 2022. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022 (accessed on 10 August 2023).
- Heidary, M.; Shirani, M.; Moradi, M.; Goudarzi, M.; Pouriran, R.; Rezaeian, T.; Khoshnood, S. Tuberculosis challenges: Resistance, co-infection, diagnosis, and treatment. Eur. J. Microbiol. Immunol. 2022, 12, 1–17. [Google Scholar] [CrossRef]
- Aghajani, J.; Farnia, P.; Farnia, P.; Ghanavi, J.; Saif, S.; Marjani, M.; Tabarsi, P.; Moniri, A.; Abtahian, Z.; Hoffner, S.; et al. Effect of COVID-19 Pandemic on Incidence of Mycobacterial Diseases among Suspected Tuberculosis Pulmonary Patients in Tehran, Iran. Int. J. Mycobacteriol. 2022, 11, 415–422. [Google Scholar]
- Can Sarınoğlu, R.; Sili, U.; Eryuksel, E.; Olgun Yildizeli, S.; Cimsit, C.; Karahasan Yagci, A. Tuberculosis and COVID-19: An overlapping situation during pandemic. J. Infect. Dev. Ctries. 2020, 14, 721–725. [Google Scholar] [CrossRef]
- Cole, B.; Nilsen, D.M.; Will, L.; Etkind, S.C.; Burgos, M.; Chorba, T. Essential Components of a Public Health Tuberculosis Prevention, Control, and Elimination Program: Recommendations of the Advisory Council for the Elimination of Tuberculosis and the National Tuberculosis Controllers Association. MMWR Recomm. Rep. 2020, 69, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Lalvani, A.; Pathan, A.A.; Durkan, H.; Wilkinson, K.A.; Whelan, A.; Deeks, J.J.; Reece, W.H.; Latif, M.; Pasvol, G.; Hill, A.V. Enhanced contact tracing and spatial tracking of Mycobacterium tuberculosis infection by enumeration of antigen-specific T cells. Lancet 2001, 357, 2017–2021. [Google Scholar] [CrossRef]
- Abdel, M.; Ms, A.; Ryszewska, K.; Laszlo, A.; Blanc, L. Strategic Approach for the Strengthening of Laboratory Services for Tuberculosis Control. Published Online 2006. Available online: https://www.who.int/publications/i/item/WHO-HTM-TB-2006.364 (accessed on 17 August 2023).
- Procop, G.W. Laboratory Diagnosis and Susceptibility Testing for Mycobacterium tuberculosis. In Tuberculosis and Nontuberculous Mycobacterial Infections; American Society for Microbiology: Washington, DC, USA, 2016; Volume 46. [Google Scholar] [CrossRef]
- Azadi, D.; Motallebirad, T.; Ghaffari, K.; Shojaei, H. Mycobacteriosis and Tuberculosis: Laboratory Diagnosis. Open Microbiol. J. 2018, 12, 41–58. [Google Scholar] [CrossRef]
- Asadi, L.; Croxen, M.; Heffernan, C.; Dhillon, M.; Paulsen, C.; Egedahl, M.L.; Tyrrell, G.; Doro-shenko, A.; Long, R. How much do smear-negative patients really contribute to tuberculosis transmissions? Re-examining an old question with new tools. eClinicalMedicine 2022, 43, 101250. [Google Scholar] [CrossRef]
- Noncommercial Culture and Drug-Susceptibility Testing Methods for Screening Patients at Risk for Multidrug-Resistant Tuberculosis Policy Statement WHO Library Cataloguing-in-Publication Data Noncommercial Culture and Drug-Susceptibility Testing Methods for Screening Patients at Risk for Multidrug-Resistant Tuberculosis: Policy Statement. Published Online 2011. Available online: www.who.int (accessed on 17 August 2023).
- Abebaw, Y.; Kebede, A.; Eshetu, K.; Tesfaye, E.; Tadesse, M.; Sinshaw, W.; Amare, M.; Gamtesa, D.F.; Zerihun, B.; Getu, M.; et al. Quality assurance practices in tuberculosis diagnostic health facilities in Ethiopia. PLoS ONE 2022, 17, e0269601. [Google Scholar] [CrossRef] [PubMed]
- Evelina, L.; Olga, C.; Alina, M.; Adriana, N.; Stela, K.; Alexandru, C. Clinical and paraclinical similarities and differences between pulmonary tuberculosis and community-acquired pneumonia. Public Health Econ. Manag. Med. 2022, 93-S, 258–264. [Google Scholar]
- Shah, M.I.; Mishra, S.; Yadav, V.K.; Chauhan, A.; Sarkar, M.; Sharma, S.K.; Rout, C. Ziehl-Neelsen sputum smear microscopy image database: A resource to facilitate automated bacilli detection for tuberculosis diagnosis. J. Med. Imaging 2017, 4, 027503. [Google Scholar] [CrossRef]
- Caulfield, A.J.; Wengenack, N.L. Diagnosis of active tuberculosis disease: From microscopy to molecular techniques. J. Clin. Tuberc. Other Mycobact. Dis. 2016, 4, 33–43. [Google Scholar] [CrossRef]
- Global Laboratory Initiative Advancing TB Diagnosis a Publication of the Global Laboratory Initiative a Working Group of the Stop TB Partnership Mycobacteriology Laboratory Manual. Available online: https://stoptb.org/wg/gli/gat.asp (accessed on 17 August 2023).
- Selvakumar, N.; Rahman, F.; Rajasekaran, S.; Narayanan, P.R.; Frieden, T.R. Inefficiency of 0.3% carbol fuchsin iniehll-neelsen staining for detecting acid-fast bacilli. J. Clin. Microbiol. 2002, 40, 3041–3043. [Google Scholar] [CrossRef]
- Holani, A.G.; Ganvir, S.M.; Shah, N.N.; Bansode, S.C.; Shende, I.; Jawade, R.; Bijjargi, S.C. Demonstration of mycobacterium tuberculosis in sputum and saliva smears of tuberculosis patients usingiehll neelsen and flurochrome staining—A comparative study. J. Clin. Diagn. Res. 2014, 8, ZC42–ZC45. [Google Scholar]
- Khan, E.A.; Starke, J.R. Diagnosis of tuberculosis in children: Increased need for better methods. Emerg. Infect. Dis. 1995, 1, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Stop TB Partnership. Community System Strengthening and TB. Available online: https://www.stoptb.org/supporting-global-fund/useful-resources-and-links (accessed on 13 August 2023).
- Munyati, S.S.; Dhoba, T.; Makanza, E.D.; Mungofa, S.; Wellington, M.; Mutsvangwa, J.; Gwanzura, L.; Hakim, J.; Nyakabau, M.; Mason, P.R.; et al. Chronic cough in primary health care attendees, Harare, Zimbabwe: Diagnosis and impact of HIV infection. Clin. Infect. Dis. 2005, 40, 1818–1827. [Google Scholar] [CrossRef] [PubMed]
- Zeru, M.A. Prevalence and associated factors of HIV-TB co-infection among HIV patients: A retrospective Study. Afr. Health Sci. 2021, 21, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Romancenco, E. Microbiological Diagnosis of Tuberculosis; Elan Poligraf: Chișinău, Moldova, 2012. [Google Scholar]
- Shen, F.; Sergi, C. Sputum Analysis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK563195/ (accessed on 11 December 2023).
- European Centre for Disease Prevention and Control. European Union Standards for Tuberculosis Care. Available online: https://www.ecdc.europa.eu/en/publications-data/european-union-standards-tuberculosis-care-2017-update (accessed on 13 August 2023).
- Angra, P.; Becx-Bleumink, M.; Gilpin, C.; Joloba, M.; Jost, K.; Kam, K.M.; Kim, S.J.; Lumb, R.; Mitarai, S.; Ramsay, A.; et al. Ziehl-Neelsen staining: Strong red on weak blue, or weak red under strong blue? Int. J. Tuberc. Lung Dis. 2007, 11, 1160–1161. [Google Scholar] [PubMed]
- Tortoli, E.; Bartoloni, A.; Böttger, E.C.; Emler, S.; Garzelli, C.; Magliano, E.; Mantella, A.; Rastogi, N.; Rindi, L.; Scarparo, C.; et al. Burden of unidentifiable mycobacteria in a reference laboratory. J. Clin. Microbiol. 2001, 39, 4058–4065. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Makaen, J.; Maure, T. Bleach processed smear for Acid fast bacilli staining in Papua New Guinea. Lab. Med. 2014, 45, e140–e141. [Google Scholar] [CrossRef][Green Version]
- Olaru, I.D.; Heyckendorf, J.; Grossmann, S.; Lange, C. Time to culture positivity and sputum smear microscopy during tuberculosis therapy. PLoS ONE 2014, 9, e106075. [Google Scholar] [CrossRef]
- Desikan, P. Sputum smear microscopy in tuberculosis: Is it still relevant? Indian J. Med. Res. 2013, 137, 442–444. [Google Scholar]
- Shapiro, H.M.; Hänscheid, T. Fuchsin fluorescence in Mycobacterium tuberculosis: The Ziehl-Neelsen stain in a new light. J. Microbiol. Methods 2008, 74, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Bodal, V.K.; Bal, M.S.; Bhagat, S.; Kishan, J.; Deepika; Brar, R.K. Fluorescent microscopy and Ziehl-Neelsen staining of bronchoalveolar lavage, bronchial washings, bronchoscopic brushing and post bronchoscopic sputum along with cytological examination in cases of suspected tuberculosis. Indian J. Pathol. Microbiol. 2015, 58, 443–447. [Google Scholar] [CrossRef]
- Siddiqi, K.; Lambert, M.L.; Walley, J. Clinical diagnosis of smear-negative pulmonary tuberculosis in low-income countries: The current evidence. Lancet Infect. Dis. 2003, 3, 288–296. [Google Scholar] [CrossRef]
- Abdelaziz, M.M.; Bakr, W.M.; Hussien, S.M.; Amine, A.E. Diagnosis of pulmonary tuberculosis using Ziehl-Neelsen stain or cold staining techniques? J. Egypt Public Health Assoc. 2016, 91, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Steingart, K.R.; Henry, M.; Ng, V.; Hopewell, P.C.; Ramsay, A.; Cunningham, J.; Urbanczik, R.; Perkins, M.; Aziz, M.A.; Pai, M. Fluorescence versus conventional sputum smear microscopy for tuberculosis: A systematic review. Lancet Infect. Dis. 2006, 6, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Oommen, S.; Banaji, N. Laboratory diagnosis of tuberculosis: Advances in technology and drug susceptibility testing. Indian J. Med. Microbiol. 2017, 35, 323–331. [Google Scholar] [CrossRef]
- Dzodanu, E.G.; Afrifa, J.; Acheampong, D.O.; Dadzie, I. Diagnostic Yield of Fluorescence and Ziehl-Neelsen Staining Techniques in the Diagnosis of Pulmonary Tuberculosis: A Comparative Study in a District Health Facility. Tuberc. Res. Treat. 2019, 2019, 4091937. [Google Scholar] [CrossRef]
- World Health Organization. Fluorescent Light-Emitting Diode (LED) Microscopy for Diagnosis of Tuberculosis: Policy Statement; World Health Organization: Geneva, Switzerland, 2011. Available online: https://www.ncbi.nlm.nih.gov/books/NBK131929/ (accessed on 12 August 2023).
- Mnyambwa, N.P.; Ngadaya, E.S.; Kimaro, G.; Kim, D.-J.; Kazwala, R.; Petrucka, P.; Mfinanga, S.G. Assessment of sputum smear-positive but culture-negative results among newly diagnosed pulmonary tuberculosis patients in Tanzania. Int. J. Gen. Med. 2017, 10, 199–205. [Google Scholar] [CrossRef]
- Abbas, A.; Elrayah, E. ILED fluorescent microscope in laboratory diagnosis of tuberculosis. Rawal Med. J. 2017, 42, 396–399. [Google Scholar]
- Gelalcha, A.G.; Kebede, A.; Mamo, H. Light-emitting diode fluorescent microscopy and Xpert MTB/RIF® assay for diagnosis of pulmonary tuberculosis among patients attending Ambo hospital, west-central Ethiopia. BMC Infect. Dis. 2017, 17, 613. [Google Scholar] [CrossRef]
- Zurac, S.; Mogodici, C.; Poncu, T.; Trăscău, M.; Popp, C.; Nichita, L.; Cioplea, M.; Ceachi, B.; Sticlaru, L.; Cioroianu, A.; et al. A New Artificial Intelligence-Based Method for Identifying Mycobacterium Tuberculosis in Ziehl-Neelsen Stain on Tissue. Diagnostics 2022, 12, 1484. [Google Scholar] [CrossRef] [PubMed]
- del Carpio, C.; Dianderas, E.; Zimic, M.; Sheen, P.; Coronel, J.; Lavarello, R.; Kemper, G. An algorithm for detection of Tuberculosis bacilli in Ziehl-Neelsen sputum smear images. Int. J. Electr. Comput. Eng. 2019, 9, 2968–2981. [Google Scholar] [CrossRef]
- Xiong, Y.; Ba, X.; Hou, A.; Zhang, K.; Chen, L.; Li, T. Automatic detection of mycobacterium tuberculosis using artificial intelligence. J. Thorac. Dis. 2018, 10, 1936–1940. [Google Scholar] [CrossRef] [PubMed]
- Zingue, D.; Weber, P.; Soltani, F.; Raoult, D.; Drancourt, M. Automatic microscopic detection of mycobacteria in sputum: A proof-of-concept. Nat. India Sci. Rep. 2018, 8, 11308. [Google Scholar] [CrossRef] [PubMed]
- Mittal, V.; Haider, F.; Singhal, S.; Jamal, S. Is universal sample processing methodology better than conventional techniques for detection of tuberculosis? Indian J. Med. Microbiol. 2014, 32, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Annam, V.; Kulkarni, M.H.; Puranik, R.B. Comparison of the modified fluorescent method and conventional Ziehl-Neelsen method in the detection of acidfast bacilli in lymphnode aspirates. Cytojournal 2009, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, S.; Dudeja, M.; Hanif, M.; Tyagi, J.S. Utility of universal sample processing methodology, combining smear microscopy, culture, and PCR, for diagnosis of pulmonary tuberculosis. J. Clin. Microbiol. 2005, 43, 2703–2708. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cattamanchi, A.; Davis, J.L.; Worodria, W.; Yoo, S.; Matovu, J.; Kiidha, J.; Nankya, F.; Kyeyune, R.; Andama, A.; Joloba, M.; et al. Poor performance of universal sample processing method for diagnosis of pulmonary tuberculosis by smear microscopy and culture in Uganda. J. Clin. Microbiol. 2008, 46, 3325–3329. [Google Scholar] [CrossRef][Green Version]
- Verma, S.; Dhole, T.N.; Kumar, M.; Kashyap, S. Novel approach for improving sensitivity of microscopic detection of acid-fast bacilli (AFB) by use of the ReaSLR method. J. Clin. Microbiol. 2013, 51, 3597–3601. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues da Costa, R.; Silva, M.R.; Augusto, C.J.; Gonçalves Leite, I.C. Fast, simple and cheap: Method modified from conventional cultivation for tuberculosis diagnosis allows seeding on Löwenstein-Jensen of any swab-embedded pulmonary samples decontaminated with sodium hydroxide. Trans. R. Soc. Trop. Med. Hyg. 2022, 116, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Moreira Ada, S.; Huf, G.; Vieira, M.A.; da Costa, P.A.; Aguiar, F.; Marsico, A.G.; de Souza Fonseca, L.; Ricks, M.; Oliveira, M.M.; Detjen, A.; et al. Liquid vs Solid Culture Medium to Evaluate Proportion and Time to Change in Management of Suspects of Tuberculosis—A Pragmatic Randomized Trial in Secondary and Tertiary Health Care Units in Brazil. PLoS ONE 2015, 10, e0127588. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.G.; Lindsey, P.H.; Harvey, C.F.; Bradley, K.K. Recognizing laboratory cross-contamination: Two false-positive cultures of Mycobacterium tuberculosi—Oklahoma, 2011. Chest 2013, 144, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Mohammed Adam, M.A.; Ebraheem, R.S.M.; Bedri, S.A. Statistical Investigation of High Culture Contamination Rates in Mycobacteriology Laboratory. Front. Microbiol. 2022, 13, 789725. [Google Scholar] [CrossRef] [PubMed]
- Battaglioli, T.; Rintiswati, N.; Martin, A.; Palupi, K.R.; Bernaerts, G.; Dwihardiani, B.; Ahmad, R.A.; Matthys, F.; Mahendradhata, Y.; Van der Stuyft, P. Comparative performance of Thin Layer Agar and Löwenstein-Jensen culture for diagnosis of tuberculosis. Clin. Microbiol. Infect. 2013, 19, E502–E508. [Google Scholar] [CrossRef][Green Version]
- Srivastava, S.; Chapagain, M.; Gumbo, T. Effect of specimen processing, growth supplement, and different metabolic population on Mycobacterium tuberculosis laboratory diagnosis. PLoS ONE 2020, 15, e0230927. [Google Scholar] [CrossRef]
- Hasan, M.; Munshi, S.K.; Banu Momi, M.S.; Rahman, F.; Noor, R. Evaluation of the effectiveness of BACTEC MGIT 960 for the detection of mycobacteria in Bangladesh. Int. J. Mycobacteriol. 2013, 2, 214–219. [Google Scholar] [CrossRef]
- Ryu, Y.J. Diagnosis of pulmonary tuberculosis: Recent advances and diagnostic algorithms. Tuberc. Respir. Dis. 2015, 78, 64–71. [Google Scholar] [CrossRef]
- Zabaleta-Vanegas, A.P.; Llerena-Polo, C.; Orjuela-Gamboa, D.L.; Valbuena-Arias, Y.A.; García-González, L.M.; Mejía-Restrepo, G.; Bueno, J.; Garzón-Torres, M.C. Evaluation of BACTEC™ MGIT™ 960 and the nitrate reductase assay in the National Laboratory Network of Colombia. Int. J. Tuberc. Lung Dis. 2013, 17, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Yegian, D.; Budd, V. Toxic effect of sodium hydroxide on tubercle bacilli. Am. J. Clin. Pathol. 1952, 22, 456–460. [Google Scholar] [CrossRef]
- Wallace, E.; Hendrickson, D.; Tolli, N.; Mehaffy, C.; Peña, M.; Nick, J.A.; Knabenbaur, P.; Watkins, J.; Simpson, A.; Amin, A.G.; et al. Culturing Mycobacteria. Methods Mol. Biol. 2021, 2314, 1–58. [Google Scholar] [PubMed]
- Thornton, C.G.; MacLellan, K.M.; Brink, T.L., Jr.; Passen, S. In vitro comparison of NALC-NaOH, tween 80, and C18-carboxypropylbetaine for processing of specimens for recovery of mycobacteria. J. Clin. Microbiol. 1998, 36, 3558–3566. [Google Scholar] [CrossRef] [PubMed]
- Asmar, S.; Drancourt, M. Chlorhexidine decontamination of sputum for culturing Mycobacterium tuberculosis. BMC Microbiol. 2015, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.R.D.; Silva, S.F.D.; Fochat, R.C.; Macedo, R.L.; Pereira, T.V.; Silva, M.R.; Pinto, C.P.G.; Leite, I.C.G. Comparison between Ogawa-Kudoh and modified Petroff techniques for mycobacteria cultivation in the diagnosis of pulmonary tuberculosis. Einstein 2018, 16, eAO4214. [Google Scholar] [CrossRef] [PubMed]
- Mtafya, B.; Sabiiti, W.; Sabi, I.; John, J.; Sichone, E.; Ntinginya, N.E.; Gillespie, S.H. Molecular Bacterial Load Assay Concurs with Culture on NaOH-Induced Loss of Mycobacterium tuberculosis Viability. J. Clin. Microbiol. 2019, 57, e01992-18. [Google Scholar] [CrossRef] [PubMed]
- Gressens, S.B.; Billard-Pomares, T.; Leboité, H.; Cruaud, P.; Bouchaud, O.; Carbonnelle, E.; Méchaï, F. Pulmonary tuberculosis: Evaluation of current diagnostic strategy. Infect. Dis. Now 2021, 51, 273–278. [Google Scholar] [CrossRef] [PubMed]
- OMS. WHO Consolidated Guidelines on Tuberculosis. Module 3: Diagnosis-Rapid Diagnostics for Tuberculosis Detection, 2021 Update. World Health Organization. 2021, pp. 1–164. Available online: https://www.who.int/publications/i/item/9789240029415 (accessed on 5 November 2023).
- Bhirud, P.; Joshi, A.; Hirani, N.; Chowdhary, A. Rapid laboratory diagnosis of pulmonary tuberculosis. Int. J. Mycobacteriol. 2017, 6, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Desikan, P.; Panwalkar, N.; Mirza, S.B.; Chaturvedi, A.; Ansari, K.; Varathe, R.; Chourey, M.; Kumar, P.; Pandey, M. Line probe assay for detection of Mycobacterium tuberculosis complex: An experience from Central India. Indian J. Med. Res. 2017, 145, 70–73. [Google Scholar] [CrossRef]
- Ebonyi, A.O.; Oguche, S.; Abok, I.I.; Isa, Y.O.; Ani, C.C.; Akhiwu, H.O.; Ihekaike, M.M.; Yiltok, E.S.; Ochoga, M.O.; Sagay, A.S. Improving the diagnosis of pulmonary tuberculosis using line probe assay and determining the factors associated with the disease in children in Jos, Nigeria. Germs 2020, 10, 328–337. [Google Scholar] [CrossRef]
- Dicks, K.V.; Stout, J.E. Molecular Diagnostics for Mycobacterium tuberculosis Infection. Annu. Rev. Med. 2019, 70, 77–90. [Google Scholar] [CrossRef]
- Nathavitharana, R.R.; Hillemann, D.; Schumacher, S.G.; Schlueter, B.; Ismail, N.; Omar, S.V.; Sikhondze, W.; Havumaki, J.; Valli, E.; Boehme, C.; et al. Multicenter Noninferiority Evaluation of Hain GenoType MTBDRplus Version 2 and Nipro NTM+MDRTB Line Probe Assays for Detection of Rifampin and Isoniazid Resistance. J. Clin. Microbiol. 2016, 54, 1624–1630. [Google Scholar] [CrossRef][Green Version]
- Singhal, R.; Myneedu, V.P.; Arora, J.; Singh, N.; Bhalla, M.; Verma, A.; Sarin, R. Early detection of multi-drug resistance and common mutations in Mycobacterium tuberculosis isolates from Delhi using GenoType MTBDRplus assay. Indian J. Med. Microbiol. 2015, 33 (Suppl. S1), 46–52. [Google Scholar] [CrossRef]
- Yadav, R.N.; Singh, B.K.; Sharma, S.K.; Sharma, R.; Soneja, M.; Sreenivas, V.; Myneedu, V.P.; Hanif, M.; Kumar, A.; Sachdeva, K.S.; et al. Comparative evaluation of GenoType MTBDRplus line probe assay with solid culture method in early diagnosis of multidrug resistant tuberculosis (MDR-TB) at a tertiary care centre in India. PLoS ONE 2013, 8, e72036. [Google Scholar] [CrossRef]
- Meaza, A.; Kebede, A.; Yaregal, Z.; Dagne, Z.; Moga, S.; Yenew, B.; Diriba, G.; Molalign, H.; Tadesse, M.; Adisse, D.; et al. Evaluation of genotype MTBDRplus VER 2.0 line probe assay for the detection of MDR-TB in smear positive and negative sputum samples. BMC Infect. Dis. 2017, 17, 280. [Google Scholar] [CrossRef]
- Shibabaw, A.; Gelaw, B.; Gebreyes, W.; Robinson, R.; Wang, S.H.; Tessema, B. The burden of pre-extensively and extensively drug-resistant tuberculosis among MDR-TB patients in the Amhara region, Ethiopia. PLoS ONE 2020, 15, e0229040. [Google Scholar] [CrossRef]
- Momen, G.; Aainouss, A.; Lamaammal, A.; Chettioui, F.; Blaghen, M.; Messoudi, M.; Belghmi, K.; Mouslim, J.; El Mzibri, M.; El Messaoudi, M.D.; et al. Molecular characterization of mutations associated with resistance to second line drugs in Mycobacterium tuberculosis patients from Casablanca, Morocco. Rev. Inst. Med. Trop. Sao Paulo 2021, 63, e19. [Google Scholar] [CrossRef] [PubMed]
- Theron, G.; Peter, J.; Richardson, M.; Barnard, M.; Donegan, S.; Warren, R.; Steingart, K.R.; Dheda, K. The diagnostic accuracy of the GenoType(®) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst. Rev. 2014, 10, CD010705. [Google Scholar] [CrossRef]
- Yadav, R.; Saini, A.; Mankotia, J.; Khaneja, R.; Agarwal, P.; Sethi, S. Genetic Characterization of Second-Line Drug-Resistant and Extensively Drug-Resistant Mycobacterium tuberculosis from the Northern Region of India. J. Epidemiol. Glob. Health 2018, 8, 220–224. [Google Scholar] [CrossRef]
- Deggim, V.; Somoskovi, A.; Voit, A.; Böttger, E.C.; Bloemberg, G.V. Integrating the Xpert MTB/RIF assay into a diagnostic workflow for rapid detection of Mycobacterium tuberculosis in a low-prevalence area. J. Clin. Microbiol. 2013, 51, 2396–2399. [Google Scholar] [CrossRef] [PubMed]
- Weyer, K.; Mirzayev, F.; Migliori, G.B.; Van Gemert, W.; D’Ambrosio, L.; Zignol, M.; Floyd, K.; Centis, R.; Cirillo, D.M.; Tortoli, E.; et al. Rapid molecular TB diagnosis: Evidence, policy making and global implementation of Xpert MTB/RIF. Eur. Respir. J. 2013, 42, 252–271. [Google Scholar] [CrossRef] [PubMed]
- Harries, A.D.; Kumar, A.M.V. Challenges and Progress with Diagnosing Pulmonary Tuberculosis in Low- and Middle-Income Countries. Diagnostics 2018, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Shinnick, T.M.; Starks, A.M.; Alexander, H.L.; Castro, K.G. Evaluation of the Cepheid Xpert MTB/RIF assay. Expert Rev. Mol. Diagn. 2015, 15, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Ajantha, G.S.; Shetty, P.C.; Kulkarni, R.D.; Biradar, U. PCR as a diagnostic tool for extra-pulmonary tuberculosis. J. Clin. Diagn. Res. 2013, 7, 1012–1015. [Google Scholar] [CrossRef] [PubMed]
- Rafael, L.L.; Raquel, M.S.; Rogelio, F.A.; Miroslava, F.P.; Alejandra-Isabel, J.G.; Paola, R.S. Discordant results between genotypic and phenotypic assays (Xpert MTB/RIF vs. BACTEC MGIT 960 system) for detection of RIF-resistant Mycobacterium tuberculosis isolates in a high burden region. Infect. Genet. Evol. 2021, 96, 105142. [Google Scholar] [CrossRef]
- Mechal, Y.; Benaissa, E.; El Mrimar, N.; Benlahlou, Y.; Bssaibis, F.; Zegmout, A.; Chadli, M.; Malik, Y.S.; Touil, N.; Abid, A.; et al. Evaluation of GeneXpert MTB/RIF system performances in the diagnosis of extrapulmonary tuberculosis. BMC Infect. Dis. 2019, 19, 1069. [Google Scholar] [CrossRef] [PubMed]
- Daum, L.T.; Fourie, P.B.; Peters, R.P.; Rodriguez, J.D.; Worthy, S.A.; Khubbar, M.; Bhattacharyya, S.; Gradus, M.S.; Mboneni, T.; Marubini, E.E.; et al. Xpert(®) MTB/RIF detection of Mycobacterium tuberculosis from sputum collected in molecular transport medium. Int. J. Tuberc. Lung Dis. 2016, 20, 1118–1124. [Google Scholar] [CrossRef]
- Ssebambulidde, K.; Gakuru, J.; Ellis, J.; Cresswell, F.V.; Bahr, N.C. Improving Technology to Diagnose Tuberculous Meningitis: Are We There Yet? Front. Neurol. 2022, 13, 892224. [Google Scholar] [CrossRef]
- de Almeida, S.M.; Kussen, G.M.B.; Cogo, L.; Carvalho, J.H.; Nogueira, K. Diagnostic characteristics of Xpert MTB/RIF assay for the diagnosis of tuberculous meningitis and rifampicin resistance in Southern Brazil. Arq. Neuropsiquiatr. 2020, 78, 700–707. [Google Scholar] [CrossRef]
- Kim, C.H.; Woo, H.; Hyun, I.G.; Kim, C.; Choi, J.H.; Jang, S.H.; Park, S.M.; Kim, D.G.; Lee, M.G.; Jung, K.S.; et al. A comparison between the efficiency of the Xpert MTB/RIF assay and nested PCR in identifying Mycobacterium tuberculosis during routine clinical practice. J. Thorac. Dis. 2014, 6, 625–631. [Google Scholar]
- Kabir, S.; Parash, M.T.H.; Emran, N.A.; Hossain, A.B.M.T.; Shimmi, S.C. Diagnostic challenges and Gene-Xpert utility in detecting Mycobacterium tuberculosis among suspected cases of Pulmonary tuberculosis. PLoS ONE 2021, 16, e0251858. [Google Scholar] [CrossRef]
- Mekkaoui, L.; Hallin, M.; Mouchet, F.; Payen, M.C.; Maillart, E.; Clevenbergh, P.; Georgala, A.; Van den Wijngaert, S. Performance of Xpert MTB/RIF Ultra for diagnosis of pulmonary and extra-pulmonary tuberculosis, one year of use in a multi-centric hospital laboratory in Brussels, Belgium. PLoS ONE 2021, 16, e0249734. [Google Scholar] [CrossRef]
- Stevenson, D.R.; Biswas, J.R.; Lievesley, A.; Quinn, G.; Buckley, J.; John, L.; Davidson, R.N. False-positive Xpert(®) MTB/RIF more than seven years after cure. Int. J. Tuberc. Lung Dis. 2015, 19, 1264–1265. [Google Scholar] [CrossRef]
- Kay, A.W.; Ness, T.; Verkuijl, S.E.; Viney, K.; Brands, A.; Masini, T.; González Fernández, L.; Eisenhut, M.; Detjen, A.K.; Mandalakas, A.M.; et al. Xpert MTB/RIF Ultra assay for tuberculosis disease and rifampicin resistance in children. Cochrane Database Syst. Rev. 2022, 9, CD013359. [Google Scholar]
- Ozkutuk, N.; Surucüoglu, S. Orta prevalanslı bölgede akciğer ve akciğer dışı tüberküloz tanısında Xpert MTB/RIF testinin değerlendirilmesi [Evaluation of the Xpert MTB/RIF assay for the diagnosis of pulmonary and extrapulmonary tuberculosis in an intermediate-prevalence setting]. Mikrobiyol. Bul. 2014, 48, 223–232. [Google Scholar] [CrossRef]
- Jafari, C.; Olaru, I.D.; Daduna, F.; Lange, C.; Kalsdorf, B. Rapid Diagnosis of Recurrent Paucibacillary Tuberculosis. Pathog. Immun. 2023, 7, 189–202. [Google Scholar] [CrossRef]
- Danchuk, S.N.; McIntosh, F.; Jamieson, F.B.; May, K.; Behr, M.A. Bacillus Calmette-Guérin strains with defined resistance mutations: A new tool for tuberculosis laboratory quality control. Clin. Microbiol. Infect. 2020, 26, 384.e5–384.e8. [Google Scholar] [CrossRef] [PubMed]
- Daum, L.T.; Choi, Y.; Worthy, S.A.; Rodriguez, J.D.; Chambers, J.P.; Fischer, G.W. A molecular transport medium for collection, inactivation, transport, and detection of Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 2014, 18, 847–849. [Google Scholar] [CrossRef] [PubMed]
- Millman, A.J.; Dowdy, D.W.; Miller, C.R.; Brownell, R.; Metcalfe, J.Z.; Cattamanchi, A.; Davis, J.L. Rapid molecular testing for TB to guide respiratory isolation in the U.S.: A cost-benefit analysis. PLoS ONE 2013, 8, e79669. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.; Albert, H.; Gilpin, C.; Alexander, H.; DeGruy, K.; Stevens, W. Multicenter feasibility study to assess external quality assessment panels for Xpert MTB/RIF assay in South Africa. J. Clin. Microbiol. 2014, 52, 2493–2499. [Google Scholar] [CrossRef] [PubMed]
- Pai, M.; Kalantri, S.; Dheda, K. New tools and emerging technologies for the diagnosis of tuberculosis: Part II. Active tuberculosis and drug resistance. Expert Rev. Mol. Diagn. 2006, 6, 423–432. [Google Scholar] [CrossRef]
- Jekloh, N.; Keawliam, P.; Mukem, D.; Rudeeaneksin, J.; Srisungngam, S.; Bunchoo, S.; Klayut, W.; Bhakdeenaun, P.; Phetsuksiri, B. Evaluation of an in-house loop-mediated isothermal amplification for Mycobacterium tuberculosis detection in a remote reference laboratory, Thailand. Rev. Inst. Med. Trop. Sao Paulo 2022, 64, e57. [Google Scholar] [CrossRef]
- Manage, D.P.; Chui, L.; Pilarski, L.M. Sub-microliter scale in-gel loop-mediated isothermal amplification (LAMP) for detection of Mycobacterium tuberculosis. Microfluid. Nanofluid. 2013, 14, 731–741. [Google Scholar] [CrossRef]
- Bojang, A.L.; Mendy, F.S.; Tientcheu, L.D.; Otu, J.; Antonio, M.; Kampmann, B.; Agbla, S.; Sutherland, J.S. Comparison of TB-LAMP, GeneXpert MTB/RIF and culture for diagnosis of pulmonary tuberculosis in The Gambia. J. Infect. 2016, 72, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Li, D.; Zhang, S.; Yao, L.; Hao, X.; Gu, J.; Li, H.; Niu, J.; Zhang, Z.; Zhu, C. Parallel Tests Using Culture, Xpert MTB/RIF, and SAT-TB in Sputum Plus Bronchial Alveolar Lavage Fluid Significantly Increase Diagnostic Performance of Smear-Negative Pulmonary Tuberculosis. Front. Microbiol. 2018, 9, 1107. [Google Scholar] [CrossRef]
- Liu, C.F.; Song, Y.M.; He, P.; Liu, D.X.; He, W.C.; Li, Y.M.; Zhao, Y.L. Evaluation of Multidrug Resistant Loop-mediated Isothermal Amplification Assay for Detecting the Drug Resistance of Mycobacterium tuberculosis. Biomed. Environ. Sci. 2021, 34, 616–622. [Google Scholar]
- Sali, M.; De Maio, F.; Caccuri, F.; Campilongo, F.; Sanguinetti, M.; Fiorentini, S.; Delogu, G.; Giagulli, C. Multicenter Evaluation of Anyplex Plus MTB/NTM MDR-TB Assay for Rapid Detection of Mycobacterium tuberculosis Complex and Multidrug-Resistant Isolates in Pulmonary and Extrapulmonary Specimens. J. Clin. Microbiol. 2016, 54, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.J.; Morlock, G.P.; Sikes, R.D.; Dalton, T.L.; Metchock, B.; Starks, A.M.; Hooks, D.P.; Cowan, L.S.; Plikaytis, B.B.; Posey, J.E. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2011, 55, 2032–2041. [Google Scholar] [CrossRef]
- Safi, H.; Lingaraju, S.; Amin, A.; Kim, S.; Jones, M.; Holmes, M.; McNeil, M.; Peterson, S.N.; Chatterjee, D.; Fleischmann, R.; et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat. Genet. 2013, 45, 1190–1197. [Google Scholar] [CrossRef]
- Clark, T.G.; Mallard, K.; Coll, F.; Preston, M.; Assefa, S.; Harris, D.; Ogwang, S.; Mumbowa, F.; Kirenga, B.; O’Sullivan, D.M.; et al. Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing. PLoS ONE 2013, 8, e83012. [Google Scholar] [CrossRef]
- Murray, P.R.; Rosenthal, K.S.; Pfaller, M.A. Medical Microbiology E-Book, 7th ed.; Elsevier: Philadelphia, PA, USA, 2013; p. 1023. [Google Scholar]
- Bjerrum, S.; Schiller, I.; Dendukuri, N.; Kohli, M.; Nathavitharana, R.R.; Zwerling, A.A.; Denkinger, C.M.; Steingart, K.R.; Shah, M. Lateral flow urine lipoarabinomannan assay for detecting active tuberculosis in people living with HIV. Cochrane Database Syst. Rev. 2019, 10, CD011420. [Google Scholar] [CrossRef]
- Doublier, S.; Zennaro, C.; Spatola, T.; Lupia, E.; Bottelli, A.; Deregibus, M.C.; Carraro, M.; Conaldi, P.G.; Camussi, G. HIV-1 Tat reduces nephrin in human podocytes: A potential mechanism for enhanced glomerular permeability in HIV-associated nephropathy. AIDS 2007, 21, 423–432. [Google Scholar] [CrossRef]
- Correia-Neves, M.; Fröberg, G.; Korshun, L.; Viegas, S.; Vaz, P.; Ramanlal, N.; Bruchfeld, J.; Hamasur, B.; Brennan, P.; Källenius, G. Biomarkers for tuberculosis: The case for lipoarabinomannan. ERJ Open Res. 2019, 5, 00115–2018. [Google Scholar] [CrossRef]
- Huerga, H.; Cossa, L.; Manhiça, I.; Bastard, M.; Telnov, A.; Molfino, L.; Sanchez-Padilla, E. Systematic, Point-of-Care Urine Lipoarabinomannan (Alere TB-LAM) Assay for Diagnosing Tuberculosis in Severely Immunocompromised HIV-Positive Ambulatory Patients. Am. J. Trop. Med. Hyg. 2020, 102, 562–566. [Google Scholar] [CrossRef]
- Drain, P.K.; Heichman, K.A.; Wilson, D. A new point-of-care test to diagnose tuberculosis. Lancet Infect. Dis. 2019, 19, 794–796. [Google Scholar] [CrossRef] [PubMed]
- Simieneh, A.; Tadesse, M.; Kebede, W.; Gashaw, M.; Abebe, G. Combination of Xpert® MTB/RIF and DetermineTM TB-LAM Ag improves the diagnosis of extrapulmonary tuberculosis at Jimma University Medical Center, Oromia, Ethiopia. PLoS ONE 2022, 17, e0263172. [Google Scholar] [CrossRef]
- Logan, N.; Lou-Franco, J.; Elliott, C.; Cao, C. Catalytic gold nanostars for SERS-based detection of mercury ions (Hg2+) with inverse sensitivity. Environ. Sci. Nano 2021, 8, 2718–2730. [Google Scholar] [CrossRef]
- Gilbride, B.; Schmidt Garcia Moreira, G.M.; Hust, M.; Cao, C.; Stewart, L. Catalytic ferromagnetic gold nanoparticle immunoassay for the detection and differentiation of Mycobacterium tuberculosis and Mycobacterium bovis. Anal. Chim. Acta 2021, 1184, 339037. [Google Scholar] [CrossRef] [PubMed]
- Salmanzadeh, S.; Abbasissifar, H.; Alavi, S.M. Comparison study of QuantiFERON test with tuberculin skin testing to diagnose latent tuberculosis infection among nurses working in teaching hospitals of Ahvaz, Iran. Casp. J. Intern. Med. 2016, 7, 82–87. [Google Scholar]
- Nafae, R.M.; Mohammad, M.A.; El-Gammal, M.S.; Abdullah, M.A.M. Use of enzyme linked immunospot assay (ELISpot) for monitoring treatment response of pulmonary tuberculosis patients. Egypt. J. Chest Dis. Tuberc. 2013, 62, 409–417. [Google Scholar] [CrossRef]
- Yun, J.W.; Chung, H.S.; Koh, W.J.; Chung, D.R.; Kim, Y.J.; Kang, E.S. Significant reduction in rate of indeterminate results of the QuantiFERON-TB Gold In-Tube test by shortening incubation delay. J. Clin. Microbiol. 2014, 52, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Kobashi, Y. Current status and future landscape of diagnosing tuberculosis infection. Respir. Investig. 2023, 61, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.S.; Kim, Y.H.; Jeon, K.; Jeong, B.H.; Ryu, Y.J.; Choi, J.C.; Kim, H.C.; Koh, W.J. Factors that Predict Negative Results of QuantiFERON-TB Gold In-Tube Test in Patients with Culture-Confirmed Tuberculosis: A Multicenter Retrospective Cohort Study. PLoS ONE 2015, 10, e0129792. [Google Scholar] [CrossRef]
- Takasaki, J.; Manabe, T.; Morino, E.; Muto, Y.; Hashimoto, M.; Iikura, M.; Izumi, S.; Sugiyama, H.; Kudo, K. Sensitivity and specificity of QuantiFERON-TB Gold Plus compared with QuantiFERON-TB Gold In-Tube and T-SPOT.TB on active tuberculosis in Japan. J. Infect. Chemother. 2018, 24, 188–192. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaporojan, N.; Negrean, R.A.; Hodișan, R.; Zaporojan, C.; Csep, A.; Zaha, D.C. Evolution of Laboratory Diagnosis of Tuberculosis. Clin. Pract. 2024, 14, 388-416. https://doi.org/10.3390/clinpract14020030
Zaporojan N, Negrean RA, Hodișan R, Zaporojan C, Csep A, Zaha DC. Evolution of Laboratory Diagnosis of Tuberculosis. Clinics and Practice. 2024; 14(2):388-416. https://doi.org/10.3390/clinpract14020030
Chicago/Turabian StyleZaporojan, Natalia, Rodica Anamaria Negrean, Ramona Hodișan, Claudiu Zaporojan, Andrei Csep, and Dana Carmen Zaha. 2024. "Evolution of Laboratory Diagnosis of Tuberculosis" Clinics and Practice 14, no. 2: 388-416. https://doi.org/10.3390/clinpract14020030
APA StyleZaporojan, N., Negrean, R. A., Hodișan, R., Zaporojan, C., Csep, A., & Zaha, D. C. (2024). Evolution of Laboratory Diagnosis of Tuberculosis. Clinics and Practice, 14(2), 388-416. https://doi.org/10.3390/clinpract14020030






