Cataloging Actionable Pharmacogenomic Variants for Indian Clinical Practice: A Scoping Review
Abstract
1. Background
2. Objectives
2.1. To Catalog the PGx of WHO Essential Medicines
2.2. To Catalog the PGx of NICU Medicines
2.3. To Enlist the Critical Genes That Require Initial Testing for Drug Adverse Effects for Personalized Medicine
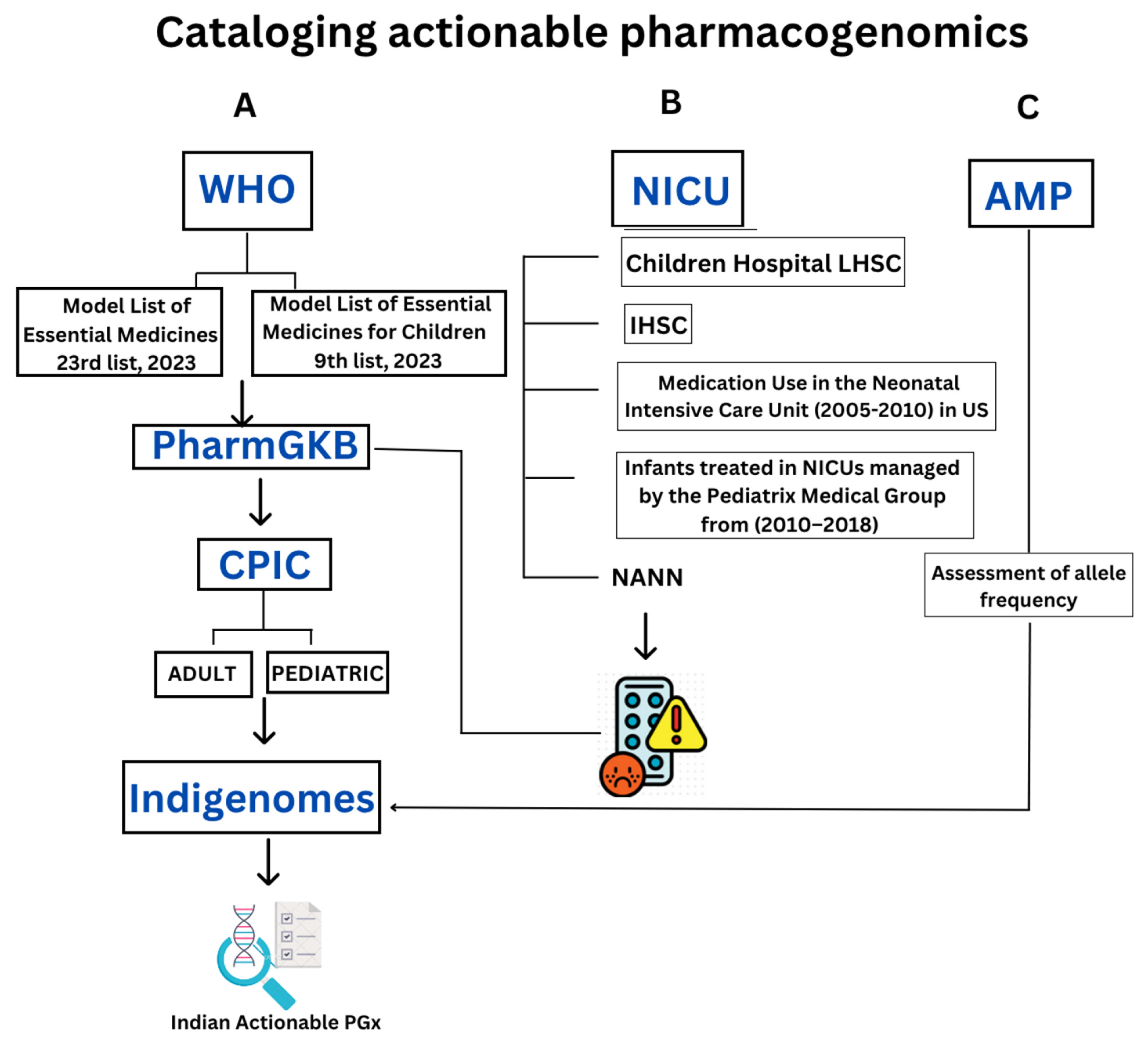
3. Methods
3.1. Essential Drugs from WHO
3.1.1. Obtaining Drug Data from the WHO Website and WHO List of Essential Drugs for Children
3.1.2. Identifying Gene Pairs, Star Alleles, and Reference Single Nucleotide Polymorphism ID Number (rsIDs)
3.1.3. Checking PharmGKB Score and CPIC Level
3.1.4. Extracting Minor Allele Frequency (MAF) Data from IndiGenomes
3.1.5. Data Analysis and Interpretation
3.2. PGx of NICU Drugs
3.2.1. Obtaining Drug Data from Different NICU Medication Databases
3.2.2. Identification of Drug Names and Prescriptions
3.2.3. Assessment of Adverse Drug Reaction (ADR)
3.3. Minimum Set of Alleles for PGx Testing Based on AMP Guidelines
3.3.1. Compile the Gene List Recommended by AMP and Enlist the Drugs Affected by Allele Polymorphisms to Have a Varied Response
3.3.2. Assessment of Allele Frequency
4. Results
4.1. Essential Drugs from WHO
4.1.1. WHO Model List of Essential Medicines—23rd List, 2023
4.1.2. WHO Model List of Essential Medicines for Children—9th List (2023)
4.2. PGx of NICU Drugs
4.3. Minimum Set of Alleles for PGx Testing Based on AMP Guidelines
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PGx | Pharmacogenomics |
| PharmGKB | Pharmacogenomics Knowledge Base |
| PGRN | Pharmacogenomics Research Network |
| UPGx | Ubiquitous Pharmacogenomics Consortium |
| ISPG | International Society of Pharmacogenomics |
| FDA | Food and Drug Administration |
| CPIC | Clinical Pharmacogenetics Implementation Consortium |
| DPWG | Dutch Pharmacogenetics Working Group |
| MAF | Minor allele frequency |
| WHO | World Health Organization |
| NICU | Neonatal Intensive Care Unit |
| LHSC | London Health Sciences Centre |
| AMP | Association for Molecular Pathology |
| SNP | Single Nucleotide Polymorphism |
| rsID | Reference SNP identifiers |
| PPI | Proton Pump Inhibitors |
| ADRs | Adverse drug reactions |
| AST | Aspartate Transaminase |
| ALT | Alanine Transaminase |
| HIV | Human immunodeficiency viruses |
| UNICEF | United Nations International Children’s Emergency Fund |
References
- Rollinson, V.; Turner, R.; Pirmohamed, M. Pharmacogenomics for Primary Care: An Overview. Genes 2020, 11, 1337. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Matilla, M.; Blanco-Verea, A.; Santori, M.; Ansede-Bermejo, J.; Ramos-Luis, E.; Gil, R.; Bermejo, A.; Lotufo-Neto, F.; Hirata, M.; Brisighelli, F.; et al. Genetic susceptibility in pharmacodynamic and pharmacokinetic pathways underlying drug-induced arrhythmia and sudden unexplained deaths. Forensic Sci. Int. Genet. 2019, 42, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Micaglio, E.; Locati, E.T.; Monasky, M.M.; Romani, F.; Heilbron, F.; Pappone, C. Role of Pharmacogenetics in Adverse Drug Reactions: An Update towards Personalized Medicine. Front. Pharmacol. 2021, 12, 651720. [Google Scholar] [CrossRef] [PubMed]
- Lewis, T.; Stone, W.L. Biochemistry, Proteins Enzymes. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Susa, S.T.; Hussain, A.; Preuss, C.V. Drug Metabolism. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Daly, A.K. Pharmacogenetics: A general review on progress to date. Br. Med. Bull. 2017, 124, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Kane, M. CYP2D6 Overview: Allele and Phenotype Frequencies. In Medical Genetics Summaries [Internet]; Pratt, V.M., Scott, S.A., Pirmohamed, M., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2021. [Google Scholar]
- Dean, L.; Kane, M. Clopidogrel Therapy and CYP2C19 Genotype. In Medical Genetics Summaries [Internet]; Pratt, V.M., Scott, S.A., Pirmohamed, M., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2012. [Google Scholar]
- Hahn, M.; Roll, S.C. The Influence of Pharmacogenetics on the Clinical Relevance of Pharmacokinetic Drug-Drug Interactions: Drug-Gene, Drug-Gene-Gene and Drug-Drug-Gene Interactions. Pharmaceuticals 2021, 14, 487. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.J.; Thomas, C.D.; Barbarino, J.; Desta, Z.; Van Driest, S.L.; El Rouby, N.; Johnson, J.A.; Cavallari, L.H.; Shakhnovich, V.; Thacker, D.L.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing. Clin. Pharmacol. Ther. 2021, 109, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Barbarino, J.M.; Whirl-Carrillo, M.; Altman, R.B.; Klein, T.E. PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, 10, e1417. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bousman, C.A.; Maruf, A.A.; Marques, D.F.; Brown, L.C.; Müller, D.J. The emergence, implementation, and future growth of pharmacogenomics in psychiatry: A narrative review. Psychol. Med. 2023, 53, 7983–7993. [Google Scholar] [CrossRef] [PubMed]
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; A Mensah, G.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M. Is pharmacogenomics a reality? Challenges and oppurtunities for India. Indian. J. Hum. Genet. 2011, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Kabbani, D.; Akika, R.; Wahid, A.; Daly, A.K.; Cascorbi, I.; Zgheib, N.K. Pharmacogenomics in practice: A review and implementation guide. Front. Pharmacol. 2023, 14, 1189976. [Google Scholar] [CrossRef] [PubMed]
- Skokou, M.; Karamperis, K.; Koufaki, M.-I.; Tsermpini, E.-E.; Pandi, M.-T.; Siamoglou, S.; Ferentinos, P.; Bartsakoulia, M.; Katsila, T.; Mitropoulou, C.; et al. Clinical implementation of preemptive pharmacogenomics in psychiatry. EBioMedicine 2024, 101, 105009. [Google Scholar] [CrossRef] [PubMed]
- eEML—Electronic Essential Medicines List. list.essentialmeds.org. Available online: https://list.essentialmeds.org/ (accessed on 18 January 2024).
- WHO Model List of Essential Medicines for Children—9th list, 2023 [Internet]. www.who.int. Available online: https://www.who.int/publications/i/item/WHO-MHP-HPS-EML-2023.03 (accessed on 16 March 2024).
- Thorn, C.F.; Klein, T.E.; Altman, R.B. PharmGKB: The pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol. 2005, 311, 179–191. [Google Scholar] [CrossRef] [PubMed]
- PharmGKB. DPWG: Dutch Pharmacogenetics Working Group. Available online: https://www.pharmgkb.org/page/dpwg (accessed on 19 January 2024).
- Clinical Pharmacogenetics Implementation Consortium (CPIC). Cpicpgx.org. 2009. Available online: https://cpicpgx.org/ (accessed on 19 January 2024).
- Table of Pharmacogenomic Biomarkers in Drug Labeling. Available online: https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling (accessed on 21 January 2024).
- Jain, A.; Bhoyar, R.C.; Pandhare, K.; Mishra, A.; Sharma, D.; Imran, M.; Senthivel, V.; Divakar, M.K.; Rophina, M.; Jolly, B.; et al. IndiGenomes: A comprehensive resource of genetic variants from over 1000 Indian genomes. Nucleic Acids Res. 2021, 49, D1225–D1232. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Neonatal Intensive Care Unit (NICU). Medication Manual|LHSC. www.lhsc.on.ca. Available online: https://www.lhsc.on.ca/nicu/neonatal-intensive-care-unit-nicu-medication-manual (accessed on 7 January 2024).
- Pharmacology: NICU Handbook. University of Iowa Stead Family Children’s Hospital. Available online: https://uihc.org/childrens/educational-resources/pharmacology-nicu-handbook (accessed on 10 March 2024).
- Hsieh, E.; Hornik, C.; Clark, R.; Laughon, M.; Benjamin, D.; Smith, P. Medication Use in the Neonatal Intensive Care Unit. Am. J. Perinatol. 2013, 31, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.; Smith, P.B.; Hornik, C.P.; Zimmerman, K.O.; Hornik, C.D.; Pradeep, S.; Clark, R.H.; Benjamin, D.K.; Laughon, M.; Greenberg, R.G. Medication Use in the Neonatal Intensive Care Unit and Changes from 2010 to 2018. J. Pediatr. 2021, 240, 66–71.e4. [Google Scholar] [CrossRef] [PubMed]
- Medication Safety in the NICU. Available online: https://nann.org/wp-content/uploads/2025/04/FINAL-2021_Medication-Safety-in-the-NICU-9ea.pdf (accessed on 7 January 2024).
- PharmGKB. AMP’s Minimum Sets of Alleles for PGx Testing. Available online: https://www.pharmgkb.org/ampAllelesToTest (accessed on 12 March 2024).
- UNICEF. Neonatal Mortality. UNICEF DATA. 2024. Available online: https://data.unicef.org/topic/child-survival/neonatal-mortality/#:~:text=The%20first%2028%20days%20of,1%2C000%20live%20births%20in%201990 (accessed on 5 May 2025).
- Newborn Mortality. www.who.int. 2024. Available online: https://www.who.int/news-room/fact-sheets/detail/newborn-mortality#:~:text=Among%20neonates%2C%20the%20leading%20causes,under%205%20years%20of%20age (accessed on 5 May 2025).
- Sahana, S.; Bhoyar, R.C.; Sivadas, A.; Jain, A.; Imran, M.; Rophina, M.; Senthivel, V.; Diwakar, M.K.; Sharma, D.; Mishra, A.; et al. Pharmacogenomic landscape of Indian population using whole genomes. Clin. Transl. Sci. 2022, 15, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Sivadas, A.; Rathore, S.; Sahana, S.; Jolly, B.; Bhoyar, R.C.; Jain, A.; Sharma, D.; Imran, M.; Senthilvel, V.; Divakar, M.K.; et al. The genomic landscape of CYP2D6 variation in the Indian population. Pharmacogenomics 2024, 25, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Sivadas, A.; Sahana, S.; Jolly, B.; Bhoyar, R.C.; Jain, A.; Sharma, D.; Imran, M.; Senthivel, V.; Divakar, M.K.; Mishra, A.; et al. Landscape of pharmacogenetic variants associated with non-insulin antidiabetic drugs in the Indian population. BMJ Open Diab Res. Care 2024, 12, e003769. [Google Scholar] [CrossRef] [PubMed]
- Genome India. genomeindia.in. Available online: https://genomeindia.in/ (accessed on 15 July 2024).


| ADULTS | |||
|---|---|---|---|
| CONDITION | DRUG | GENE | STAR ALLEL/rsID |
| HIV | Atazanavir | UGT1A1 | *1, *6, *80 |
| Efavirenz | CYP2B6 | *4, *6, *7, *8, *9, *12, *13*18, *19, *20, *26, *34, *36, *37, *38, *39, *40, *41, *42, *43, | |
| Abacavir | HLA-B | *57:01 | |
| Hypertension | Metoprolol | CYP2D6 | *4, *10,*31,*6,,*9,*29,*3, 5, *161, *156, *144, *143, *129, *124, *120, *114, *101, *100, *99, *96, *92, *81, *69, *62, *60, *56, *51, *47, *44, *42, *38, *36, *21, *20, *19, *18, *15, *12 |
| Malignancy | Capecitabine Fluorouracil | DPYD | rs75017182, rs3918290 |
| Irinotecan | UGT1A1 | *6, *80 | |
| Mercaptopurine | NUDT15 | *1, *2, *3, | |
| TPMT | *1, *3A, *3B, *3C, *41 | ||
| Tamoxifen | CYP2D6 | *1, *2, *4, *6, *10, *17, *29, *41, *3, *5, *7, *8, *9, *11, *21, *36, *12, *13, *14, *15, *19, *20, *27, *31, *32, *40, *42, *44, *47, *49, *51, *54, *55, *56 | |
| Cardiovascular disease | Clopidogrel | CYP2C19 | *2, *3, *4, *5, *6, *8, *10, *19, *25, *26, *7, *9, *16, *22, *24, *35, *36, *37 |
| Lovastatin Atorvastatin Simvastatin Pravastatin | SLCO1B1 | *1, *5, *15, *31, *46, *47 | |
| Fluvastatin | SLCO1B1 | *1, *5, *15, *31, *46, *47 | |
| CYP2C9 | *1, *2, *3, *8, *11, *14, *26, *35, *44, *45, *61 | ||
| Warfarin | CYP2C9 | *1, *2, *3 | |
| VKORC1 | rs9923231 | ||
| Hepatitis | Ribavirin | IFNL4 | rs12979860 |
| Psychotropic medications | Amitriptyline | CYP2C19 | *1, *2, *3, *17, *4, *5, *6, *7, *8, *22, *24, *35 |
| CYP2D6 | *1,*2,*4,*9,*10,*17, *161, *156, *144, *143, *132, *129, *124, *119, *114, *109, *101, *100, *99, *91, *81, *69, *62, *60, *59, *56, *55, *54, *52, *51, *50, *49, *47, *45, *44, *42, *41, *40,*36, *35, *31, *29, *21, *19, *18, *15, *14, *13, *12, *11, *8, *7, *6, *5, *3, *27, *32 | ||
| Clomipramine | CYP2C19 | *1, *2, *3, *17, *4, *5, *6, *7, *8, *22, *24, *35 | |
| CYP2D6 | *1,*2,*4,*9,*10,*17, *161, *156, *144, *143, *132, *129, *124, *119, *114, *109, *101, *100, *99, *91, *81, *69, *62, *60, *59, *56, *55, *54, *52, *51, *50, *49, *47, *45, *44, *42, *41, *40,*36, *35, *31, *29, *21, *19, *18, *15, *14, *13, *12, *11, *8, *7, *6, *5, *3, *27, *32 | ||
| Sertraline | CYP2B6 | *6, *9, *7, *8, *12, *13, *18, *19, *20, *43, *42, *41, *40, *39, *38, *37, *36, *34, *28, *26 | |
| CYP2C19 | *2, *3, *35, *26, *25, *24, *22, *19, *16, *10, *9, *8, *7, *6, *5, *4, *17 | ||
| Aripiprazole | CYP2D6 | *4, *6, *114, *42, *40, *38, *36, *31, *21, *20, *19, *18, *13, *12, *11, *8, *7, *5, *3, *15 | |
| Haloperidol | *114, *42, *40, *38, *36, *35, *31, *21, *20, *19, *18, *15, *13, *12, *11, *8, *7, *6, *4, *5, *3, *2, *1 | ||
| Zuclopenthixol decanoate | *114, *42, *41, *40, *38, *36, *35, *31, *29, *21, *20, *19, *18, *17, *15, *14, *13, *12, *11, *10, *9, *8, *7, *6, *5, *4, *3, *2, *1 | ||
| Fluvoxamine | *161, *156, *144, *143, *129, *124, *114, *101, *100, *99, *96, *81, *69, *62, *60, *56, *51, *47, *44, *42, *40, *38, *36, *31, *21, *20, *19, *18, *15, *13, *12, *11, *8, *7, *6, *5, *4, *3 | ||
| Paroxetine | *161, *156, *144, *143, *132, *129, *124, *119, *114, *109, *101, *100, *99, *96, *91, *81, *69, *62, *60, *59, *55, *54, *52, *51, *50, *49, *47, *45, *44, *42, *41, *40, *38, *36, *35, *32, *31, *29, *27, *21, *20, *19, *18, *17, *15, *14, *13, *12, *11, *10, *9, *8, *7, *6, *5, *4, *3, *2, *1 | ||
| Risperidone | *114, *42, *40, *38, *36, *35, *31, *21, *20, *19, *18, *15, *13, *12, *11, *8, *7, *6, *5, *4, *3, *2, *1 | ||
| Bupropion | CYP2D6 | *2, *3, *4, *5, *6, *9, *10, *17, *29, *40, *41 (FDA recommendation for poor metabolizers) | |
| Carbamazepine | HLA-A | *31:01 | |
| HLA-B | *15:02 | ||
| Citalopram, Escitalopram | CYP2C19 | *2, *3, *4, *17, *35, *26, *25, *24, *22, *19, *16, *10, *9, *8, *7, *6, *5, *18 | |
| Phenytoin | CYP2C9 | *2, *3, *8, *11, *14, *26, *35, *44, *45, *61 | |
| HLA-B | *15:02 | ||
| Lamotrigine | HLA-B | *15:02 | |
| Quetiapine | CYP3A4 | *13, *22 | |
| Immunosuppressive medications | Azathioprine | TPMT | *1, *3A, *3B, *3C, *8, *41, |
| NUDT15 | *1, *2, *3, | ||
| Analgesics | Codeine, Tramadol | CYP2D6 | *1,*2, *3,*4,*5,*6,*7,*8,*9,*10,*161, *156, *144, *143, *132, *129, *124, *114, *109, *101, *100, *99, *96, *91, *56, *81, *69, *62, *59, *55, *54, *52, *51, *49, *47, *45,*44, *42, *41, *40, *38, *36, *35, *32, *31, *29, *27, *21, *20, *19, *18, *17, *15, *14, *13, *12, *11 |
| Tuberculosis | Isoniazid | NAT2 | *5, *6, *7, *14, *16 |
| Drug hypersensitivity | Allopurinol | HLA-B | *58:01 |
| ABCG2 | rs2231142 | ||
| Dapsone | G6PD | FDA recommendation | |
| Gastroesophageal reflux disease (GERD) | Omeprazole otc | CYP2C19 | *1, *2, *3, *17, *9, *38, *35, *28, *26, *25, *24, *22, *19, *18, *16, *15, *13, *11, *10, *8, *7, *6, *5, *4 |
| Antiemetics | Tropisetron, Ondansetron | CYP2D6 | *1, *2, *13, *27, *35, *45 |
| Immunodeficiency | Tacrolimus | CYP3A5 | *1, *3 |
| Fungal or yeast infections | Voriconazole | CYP2C19 | *2, *3, *4, *17, *35, *24, *22, *8, *7, *6, *5 |
| PEDIATRIC | |||
| Psychotropic medications | Phenytoin | CYP2C9 | CYP2C9 *1, *2, *3, *8, *9, *11, *14, *26, *35, *44, *45 |
| Immunosuppressive medications | Azathioprine | NUDT15 | *1, *2, *5, *3 |
| TPMT | *1, *3A, *3B, *3C, *22, *34, *41 | ||
| Malignancy | Fluorouracil | DPYD | rs2297595, rs56038477, rs1801158, rs1801160, rs75017182 |
| Mercaptopurine | NUDT15 | *1, *2, *3 *5 | |
| TPMT | *1, *3A, *3B, *3C, *8, *16 | ||
| Methotrexate | MTHFR | rs1801133 | |
| Irinotecan | UGT1A1 | *1, *6, *80+*28, *80+*37 rs4124874 | |
| Inflammatory diseases | Ibuprofen otc | CYP2C9 | *1, *2, *3 *8, *9, *11, *14, *26, *35, *37, *39, *42, *43, *44, *45, *46, *52, *55, *61 |
| Toxic liver disease | Isoniazid | NAT2 | *5, *6, *7, *14, *16 |
| Gastroesophageal reflux disease (GERD) | Omeprazole otc | CYP2C19 | *1, *2, *3, *9, *10, *17, *4, *5, *6, *7, *8, *10, *11, *13, *15, *16, *18, *19, *22, *25, *26, *28, *35, *38 *24 |
| Antiemetics | Ondansetron | CYP2D6 | *1, *2, *4, *10, *35 *14, *17, *27, *29, *33, *34, *39, *45 |
| Hepatitis virus | Ribavirin | IFNL3, IFNL4 | rs12979860, rs8099917 |
| General anaesthetics | Sevoflurane | RYR1 | rs193922809 |
| Halothane, Isoflurane | RYR1 | rs118192161, rs118192162, rs193922772, rs112563513, rs193922816, rs28933397, rs118192122, rs193922747, rs118192176, rs118192177, rs118192175, rs121918592, rs121918593, rs118192172 rs121918594 | |
| CACNA1S | rs1800559, rs772226819 | ||
| Immunosuppressive agents | Tacrolimus | CYP3A4, | *1, *22 |
| CYP3A5 | *1, *3, *6, *7 | ||
| Fungal infections | Voriconazole | CYP2C19 | *1, *2, *3, *17, *4, *5, *6, *7, *8, *9, *10, *16, *17, *19, *22, *24, *25, *26, *35 |
| Cardiovascular medications | Warfarin | CYP2C9 | *1, *2, *3, *8, *11 |
| CYP4F2 | rs2108622 | ||
| VKORC1 | rs9923231, rs7294, rs9934438, rs2359612, rs8050894, rs9923231 | ||
| Drug hypersensitivity | Allopurinol | HLA B | *58:01 |
| DRUG | SPECIALITY PURPOSE | ADVERSE REACTIONS | PHARMGKB GENE | STAR ALLELE | rsID | INDIGEN |
|---|---|---|---|---|---|---|
| Lansoprazole | Gastroesophageal-reflux-disease (GERD) | GI: abdominal pain, cramps, bloating, constipation, diarrhoea, vomiting, mildly elevated AST/ALT CV: hypertension, hypotension, Dermatologic: urticaria, pruritus, Hematologic: thrombocytopenia, leucopenia, leukocytosis, anemia, Endocrine: hyperglycemia, hyperlipidemia | CYP2C19 | *1 | rs3758581 | 0.8889 |
| *2 | rs12769205 | 0.3689 | ||||
| rs4244285 | 0.3678 | |||||
| rs5897349 | 0.0025 | |||||
| *3 | rs4986893 | 0.0064 | ||||
| *8 | rs41291556 | NA | ||||
| rs3758581 | 0.8889 | |||||
| *9 | rs17884712 | NA | ||||
| rs3758581 | 0.8889 | |||||
| *17 | rs12248560 | 0.1436 | ||||
| Pantoprazole | Gastroesophageal-reflux-disease (GERD) | abdominal pain, cramps, bloating, constipation, diarrhea, vomiting, hypertension, hypotension, urticaria, pruritus, thrombocytopenia, leucopenia, leukocytosis, anemia, thrombophlebitis | CYP2C19 | *1 | rs3758581 | 0.8889 |
| *2 | rs12769205 | 0.3689 | ||||
| rs4244285 | 0.3678 | |||||
| rs58973490 | 0.0025 | |||||
| *3 | rs4986893 | 0.0064 | ||||
| *8 | rs41291556 | NA | ||||
| rs3758581 | 0.8889 | |||||
| *9 | rs17884712 | NA | ||||
| rs3758581 | 0.8889 | |||||
| *17 | rs12248560 | 0.1436 | ||||
| phenytoin | Anticonvulsant | Acute, following IV administration: hypotension, bradycardia, ventricular fibrillation, vasodilation; venous irritation; pain, thrombophlebitis, skin rash. Observe IV site carefully. Extravasation may cause tissue inflammation and necrosis. GI side effects: vomiting, constipation. Other: toxic hepatitis, gingival hyperplasia, hyperglycemia and osteoporosis | CYP2C9 | *1 | rs72558189 | 0.001 |
| rs200965026 | 0.0049 | |||||
| rs199523631 | 0.0005 | |||||
| rs1799853 | 0.0186 | |||||
| rs17847037 | 0.0015 | |||||
| rs7900194 | 0.0010 | |||||
| rs2256871 | 0.0103 | |||||
| rs28371685 | 0.0152 | |||||
| rs1057910 | 0.0182 | |||||
| *2 | rs1799853 | 0.0307 | ||||
| *3 | rs1057910 | 0.1093 | ||||
| *5 | rs28371686 | NA | ||||
| *6 | rs9332131 | NA | ||||
| *8 | rs7900194 | 0.0005 | ||||
| *11 | rs28371685 | 0.0029 | ||||
| *13 | rs72558187 | NA | ||||
| *14 | rs72558189 | 0.018 | ||||
| *16 | rs72558192 | NA | ||||
| *29 | rs182132442 | NA | ||||
| *31 | rs57505750 | NA | ||||
| *33 | rs200183364 | NA | ||||
| *37 | rs564813580 | NA | ||||
| *39 | rs762239445 | NA | ||||
| *42 | rs12414460 | NA | ||||
| *43 | rs767576260 | NA | ||||
| *45 | rs199523631 | 0.0015 | ||||
| *50 | NA | NA | ||||
| *52 | rs988617574 | NA | ||||
| *55 | rs1250577724 | NA | ||||
| Gentamicin | Antibiotic | Ototoxicity, Nephrotoxicity | MT-RNR1 | rs267606618 | NA | |
| rs267606619 | NA | |||||
| Tobramycin | Antibiotic | Ototoxicity, Nephrotoxicity | MT-RNR1 | rs267606617 | NA | |
| rs267606619 | NA | |||||
| Amikacin | Antibiotic | Ototoxicity, nephrotoxicity | MT-RNR1 | rs267606617 | NA | |
| Omeprazole | Gastroesophageal-reflux-disease (GERD) | gastrointestinal disturbances such as diarrhoea, abdominal discomfort, and occasionally, an increased risk of infections due to altered gastric pH levels | CYP2C19 | *1 | rs3758581 | 0.8889 |
| *2 | rs12769205 | 0.3689 | ||||
| *2 | rs4244285 | 0.3678 | ||||
| *2 | rs58973490 | 0.0025 | ||||
| *3 | rs4986893 | 0.0064 | ||||
| *9 | rs17884712 | NA | ||||
| *9 | rs3758581 | 0.8889 | ||||
| *10 | rs6413438 | NA | ||||
| *10 | rs3758581 | 0.8889 | ||||
| *17 | rs12248560 | 0.1436 | ||||
| *24 | rs3758581 | 0.8889 | ||||
| *24 | rs118203757 | NA | ||||
| Succinylcholine | Muscle Relaxant | Hyperkalemia, Malignant Hyperthermia, Bradycardia, Increased Intracranial Pressure, Myoglobinuria | RYR1 | rs193922802 | NA | |
| RYR1 | rs193922816 | NA | ||||
| RYR1 | rs112563513 | NA | ||||
| RYR1 | rs121918596 | NA | ||||
| CACNA1S | rs1800559 | NA | ||||
| RYR1 | rs121918592 | NA | ||||
| RYR1 | rs118192122 | NA | ||||
| RYR1 | rs193922772 | NA | ||||
| rs118192163 | NA | |||||
| rs118192177 | NA | |||||
| rs118192176 | NA | |||||
| rs118192124 | NA | |||||
| rs121918595 | NA | |||||
| rs28933397 | NA | |||||
| rs118192178 | NA | |||||
| rs121918593 | NA | |||||
| rs1801086 | NA | |||||
| rs193922807 | NA | |||||
| rs118192168 | NA | |||||
| rs118192172 | NA | |||||
| rs193922876 | NA | |||||
| rs193922764 | NA | |||||
| rs193922818 | NA | |||||
| Epinephrine | Sympathomimetic drug (relaxing muscles)/catecholamines | Breathing problems, irregular heartbeats, pale skin, sweating, nausea, vomiting, dizziness, tremors, headache, restlessness, fear, nervousness, anxiety, excitation | G6PD Deficiency | NA | NA | NA |
| Ibuprofen | Anti inflammatory | Headache, dizziness, drowsiness, fatigue, restless sleep, thirst, sweating, numbness in hands and feet, impaired hearing, blurred vison, eye irritation, fluid retention, ankle swelling, mild allergic reaction, abdominal pain, nausea, vomiting, heat burn, diarrhoea, constipation, frequent urination, bladder irritation, increase risk of heart attack or stroke, bleeding in stomach and bowels, kidney and liver damage, confusion, disorientation, tinnitus, anxiety, paranoia, anaemia, black stools, seizures, coma | CYP2C9 | *1 | rs72558189 | 0.001 |
| rs200965026 | 0.0049 | |||||
| rs199523631 | 0.0005 | |||||
| rs1799853 | 0.0186 | |||||
| rs17847037 | 0.0015 | |||||
| rs7900194 | 0.0010 | |||||
| rs2256871 | 0.0103 | |||||
| rs28371685 | 0.0152 | |||||
| rs1057910 | 0.0182 | |||||
| *2 | rs1799853 | 0.0307 | ||||
| *3 | rs1057910 | 0.1093 | ||||
| *8 | rs7900194 | 0.0005 | ||||
| *9 | rs2256871 | 0.0817 | ||||
| *11 | rs28371685 | 0.0029 | ||||
| *14 | rs72558189 | 0.018 | ||||
| *26 | rs200965026 | 0.0049 | ||||
| *35 | rs72558189 | 0.001 | ||||
| rs1799853 | 0.0186 | |||||
| *37 | rs564813580 | NA | ||||
| *39 | rs762239445 | NA | ||||
| *42 | rs12414460 | NA | ||||
| *43 | rs767576260 | NA | ||||
| *44 | rs200965026 | 0.0049 | ||||
| *45 | rs199523631 | 0.0015 | ||||
| *46 | rs754487195 | NA | ||||
| *52 | rs988617574 | NA | ||||
| *55 | rs1250577724 | NA | ||||
| *61 | rs202201137 | NA | ||||
| rs1799853 | 0.0186 | |||||
| Metoclopramide | Dopamine receptor antagonist | Tardive dyskinesia, diarrhoea, drowsiness, fatigue, muscle pain, restlessness, parkinsonism, somnolence, nausea, vomiting, asthenia, lassitude, depression, hypotension | CYP2D6 | Poor metabolizers (*2, *3, *4, *5, *6, *9, *10, *17, *29, *40, *41) | rs1058164 | 0.568 |
| rs16947 | 0.374 | |||||
| rs1135840 | 0.5646 | |||||
| rs28371725 | 0.1324 | |||||
| rs5030656 | 0.0020 | |||||
| rs5030655 | NA | |||||
| rs28371704 | 0.0884 | |||||
| rs3892097 | 0.1094 | |||||
| rs1058172 | 0.0781 | |||||
| rs1135832 | NA | |||||
| rs1135833 | NA | |||||
| rs35742686 | NA | |||||
| rs61736512 | NA | |||||
| rs59421388 | NA | |||||
| rs1135835 | NA | |||||
| rs1135836 | NA | |||||
| rs74478221 | NA | |||||
| rs766507177 rs1065852 | NA | |||||
| 0.1929 | ||||||
| rs28371703 | 0.0894 | |||||
| rs28371735 | NA | |||||
| rs1135824 | 0.0010 | |||||
| rs72549356 | NA | |||||
| rs28371706 | NA | |||||
| rs28371736 | NA | |||||
| rs747998333 | NA | |||||
| rs75467367 | NA |
| TIER 1* MINIMUM SET | ||||||
|---|---|---|---|---|---|---|
| GENE | ALLELE | rsID | IndiGen | African | Asian (East & South) | Europe |
| CYP2C19 | *2 | rs12769205 | 0.3689 | 0.1967 | 0.3125 & 0.3579 | 0.1451 |
| rs4244285 | 0.3678 | 0.1702 | 0.3125 & 0.3579 | 0.1451 | ||
| rs58973490 | 0.0025 | 0.0008 | 0 & 0 | 0.0040 | ||
| *3 | rs4986893 | 0.0064 | 0.0023 | 0.0556 & 0.0123 | 0.000 | |
| *17 | rs12248560 | 0.1436 | 0.2352 | 0.0149 & 0.136 | 0.2237 | |
| CYP2C9 | *2 | rs1799853 | 0.0307 | 0.0083 | 0.001 & 0.0348 | 0.1243 |
| *3 | rs1057910 | 0.1093 | 0.0023 | 0.0337 & 0.1094 | 0.0726 | |
| *5 | rs28371686 | NA | NA | NA | NA | |
| *6 | rs9332131 | NA | NA | NA | NA | |
| *8 | rs7900194 | 0.0005 | 0.053 | 0.0 & 0.001 | 0.002 | |
| *11 | rs28371685 | 0.0029 | 0.0242 | 0.0 & 0.001 | 0.002 | |
| CYP2D6 | *2 | rs1058164 | 0.568 | NA | NA | NA |
| *4 | rs3892097 | 0.1094 | 0.0605 | 0.002 & 0.1094 | 0.1859 | |
| *5 | NA | NA | NA | NA | NA | |
| *6 | rs5030655 | NA | 0.0008 | 0.0000 | 0.0199 | |
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *9 | rs5030656 | 0.002 | 0.0008 | 0.0000 | 0.0258 | |
| *10 | rs1065852 | 0.1929 | 0.1127 | 0.5714 | 0.2018 | |
| rs1058164 | 0.568 | NA | NA | NA | ||
| *17 | rs1058164 | 0.568 | NA | NA | NA | |
| rs16947 | 0.374 | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *29 | rs61736512 | NA | 0.1097 | 0.0000 | 0.0000 | |
| rs1058164 | 0.568 | NA | NA | NA | ||
| rs16947 | 0.374 | NA | NA | NA | ||
| rs59421388 | NA | 0.1074 | 0.0000 | 0.0000 | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *41 | rs28371725 | 0.1324 | 0.0182 | 0.0377 | 0.9066 | |
| TPMT | *2 | rs1800462 | NA | NA | NA | NA |
| *3A | rs1800460 | 0.0039 | 0.003 | 0.0041 | 0.0278 | |
| rs1142345 | 0.0226 | 0.0666 | 0.0218 & 0.0174 | 0.0288 | ||
| *3B | rs1800460 | 0.0039 | 0.003 | 0.0041 | 0.0278 | |
| *3C | rs1142345 | 0.0226 | 0.0666 | 0.0218 & 0.0174 | 0.0288 | |
| NUDT15 | *3 | rs116855232 | 0.0837 | 0.0008 | 0.0952 & 0.0695 | 0.002 |
| CYP3A4 | *22 | rs35599367 | 0.0083 | 0.0008 | 0.0061 | 0.0497 |
| CYP3A5 | *3 | rs776746 | 0.7059 | NA | NA | NA |
| CYP3A5*6 | rs10264272 | NA | NA | NA | NA | |
| CYP3A5*7 | rs41303343 | NA | NA | NA | NA | |
| TIER 2* OPTIONAL | ||||||
| CYP2C19 | *4 | rs12248560 | 0.1436 | 0.2352 | 0.0149 & 0.136 | 0.2237 |
| *5 | rs3758581 | 0.8889 | NA | NA | NA | |
| *6 | rs72552267 | NA | NA | NA | NA | |
| rs3758581 | 0.8889 | NA | NA | NA | ||
| *7 | rs3758581 | 0.8889 | NA | NA | NA | |
| rs72558186 | NA | NA | NA | NA | ||
| *8 | rs41291556 | NA | NA | NA | NA | |
| rs3758581 | 0.8889 | NA | NA | NA | ||
| *9 | rs17884712 | NA | NA | NA | NA | |
| rs3758581 | 0.8889 | NA | NA | NA | ||
| *10 | rs6413438 | NA | NA | NA | NA | |
| rs3758581 | 0.8889 | NA | NA | NA | ||
| *35 | rs17882687 | NA | NA | NA | NA | |
| rs12769205 | 0.3689 | 0.1967 | 0.3125 & 0.3579 | 0.1451 | ||
| rs3758581 | 0.8889 | NA | NA | NA | ||
| CYP2C9 | *12 | rs9332239 | NA | NA | NA | NA |
| *13 | rs72558187 | NA | NA | NA | NA | |
| *14 | rs72558190 | NA | NA | NA | NA | |
| CYP4F2 | *3 | rs2108622 | 0.4012 | 0.0825 | 0.2143 & 0.4131 | 0.2903 |
| VKORC1 | rs72547529 | NA | NA | NA | NA | |
| rs61742245 | NA | NA | NA | NA | ||
| CYP2C cluster | rs12777823 | 0.3741 | 0.2511 | 0.3145 & 0.362 | 0.1511 | |
| CYP2D6 | *7 | rs5030867 | 0.0073 | NA | 0.0 & 0.0092 | NA |
| *8 | rs1058164 | 0.5680 | NA | NA | NA | |
| rs5030865 | NA | NA | NA | NA | ||
| rs16947 | 0.374 | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *12 | rs5030862 | NA | NA | NA | NA | |
| rs1058164 | 0.5680 | NA | NA | NA | ||
| rs28371710 | NA | NA | NA | NA | ||
| rs16947 | 0.374 | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *14 | rs1058164 | 0.5680 | NA | NA | NA | |
| rs5030865 | NA | NA | NA | NA | ||
| rs16947 | 0.3740 | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *15 | rs28371696 | 0.0074 | 0.0234 | 0.0 & 0.0112 | 0.002 | |
| rs774671100 | NA | NA | NA | NA | ||
| *21 | rs1058164 | 0.568 | NA | NA | NA | |
| rs16947 | 0.374 | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *31 | rs16947 | 0.374 | NA | NA | NA | |
| rs1135840 | 0.5646 | NA | NA | NA | ||
| rs1058164 | 0.5680 | NA | NA | NA | ||
| *40 | rs72549356 | NA | NA | NA | NA | |
| *42 | rs1058164 | 0.5680 | NA | NA | NA | |
| rs16947 | 0.374 | NA | NA | NA | ||
| rs72549346 | NA | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *49 | rs1065852 | 0.1929 | 0.1127 | 0.5714 & 0.1646 | 0.2018 | |
| rs1135822 | NA | NA | NA | NA | ||
| rs1058164 | 0.5680 | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *56 | rs1065852 | 0.1929 | 0.1127 | 0.5714 & 0.1646 | 0.2018 | |
| rs1058164 | 0.568 | NA | NA | NA | ||
| rs16947 | 0.374 | NA | NA | NA | ||
| rs72549347 | NA | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| *59 | rs1058164 | 0.5680 | NA | NA | NA | |
| rs16947 | 0.374 | NA | NA | NA | ||
| rs79292917 | NA | NA | NA | NA | ||
| rs1135840 | 0.5646 | NA | NA | NA | ||
| TPMT | *11 | rs72552738 | NA | NA | NA | NA |
| *29 | rs267607275 | NA | NA | NA | NA | |
| *42 | rs759836180 | NA | NA | NA | NA | |
| NUDT15 | *2 | rs746071566 | NA | NA | NA | NA |
| rs116855232 | 0.0837 | 0.0008 | 0.0952 & 0.0695 | 0.002 | ||
| *4 | rs147390019 | NA | NA | NA | NA | |
| *6 | rs746071566 | NA | NA | NA | NA | |
| *9 | rs746071566 | NA | NA | NA | NA | |
| *14 | rs777311140 | NA | NA | NA | NA | |
| CYP3A4 | *20 | rs67666821 | NA | NA | NA | NA |
| Sl. No | Genes | Drugs | Sl. No | Genes | Drugs |
|---|---|---|---|---|---|
| 1 | CYP2D6 | Metoprolol, Tamoxifen Amitriptyline, Clomipramine Ondansetron, Tropisetron Codeine, Tramadol Bupropion, Aripiprazole Haloperidol, Fluvoxamine Zuclopenthixol decanoate Paroxetine, Risperidone | 13 | NUDT15 | Mercaptopurine Azathioprine |
| 2 | VKORC1 | Warfarin | 14 | DPYD | Fluorouracil Capecitabine |
| 3 | TPMT | Mercaptopurine Azathioprine | 15 | CYP2C9 | Fluvastatin, Warfarin Phenytoin, Ibuprofen |
| 4 | NAT2 | Isoniazid | 16 | CYP2B6 | Sertraline, Efavirenz |
| 5 | HLA-A | Carbamazepine | 17 | CYP2C19 | Clopidogrel, Amitriptyline Clomipramine, Sertraline Citalopram, Escitalopram Omeprazole, Voriconazole |
| 6 | HLA-B | Abacavir, Phenytoin Lamotrigine, Allopurinol | 18 | MTHFR | Methotrexate |
| 7 | G6PD | Dapsone | 19 | IFNL3 | Ribavirin |
| 8 | CYP3A4 | Tacrolimus, Quetiapine | 20 | IFNL4 | Ribavirin |
| 9 | CYP3A5 | Tacrolimus | 21 | CACNA1S | Halothane, Isoflurane |
| 10 | ABCG2 | Allopurinol | 22 | RYR1 | Sevoflurane, Halothane Isoflurane |
| 11 | UGT1A1 | Atazanavir, Irinotecan | 23 | CYP4F2 | Warfarin |
| 12 | SLCO1B1 | Lovastatin, Atorvastatin Simvastatin, Pravastatin Fluvastatin | 24 | MT-RNR1 | Gentamicin, Tobramycin Amikacin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulkarni, S.S.; R, V.; Das, A.; Iyer, G.R. Cataloging Actionable Pharmacogenomic Variants for Indian Clinical Practice: A Scoping Review. J. Xenobiot. 2025, 15, 101. https://doi.org/10.3390/jox15040101
Kulkarni SS, R V, Das A, Iyer GR. Cataloging Actionable Pharmacogenomic Variants for Indian Clinical Practice: A Scoping Review. Journal of Xenobiotics. 2025; 15(4):101. https://doi.org/10.3390/jox15040101
Chicago/Turabian StyleKulkarni, Sacheta Sudhendra, Venkatesh R, Anuradha Das, and Gayatri Rangarajan Iyer. 2025. "Cataloging Actionable Pharmacogenomic Variants for Indian Clinical Practice: A Scoping Review" Journal of Xenobiotics 15, no. 4: 101. https://doi.org/10.3390/jox15040101
APA StyleKulkarni, S. S., R, V., Das, A., & Iyer, G. R. (2025). Cataloging Actionable Pharmacogenomic Variants for Indian Clinical Practice: A Scoping Review. Journal of Xenobiotics, 15(4), 101. https://doi.org/10.3390/jox15040101







