Ecotoxicological Evaluation of Bisphenol A and Alternatives: A Comprehensive In Silico Modelling Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset
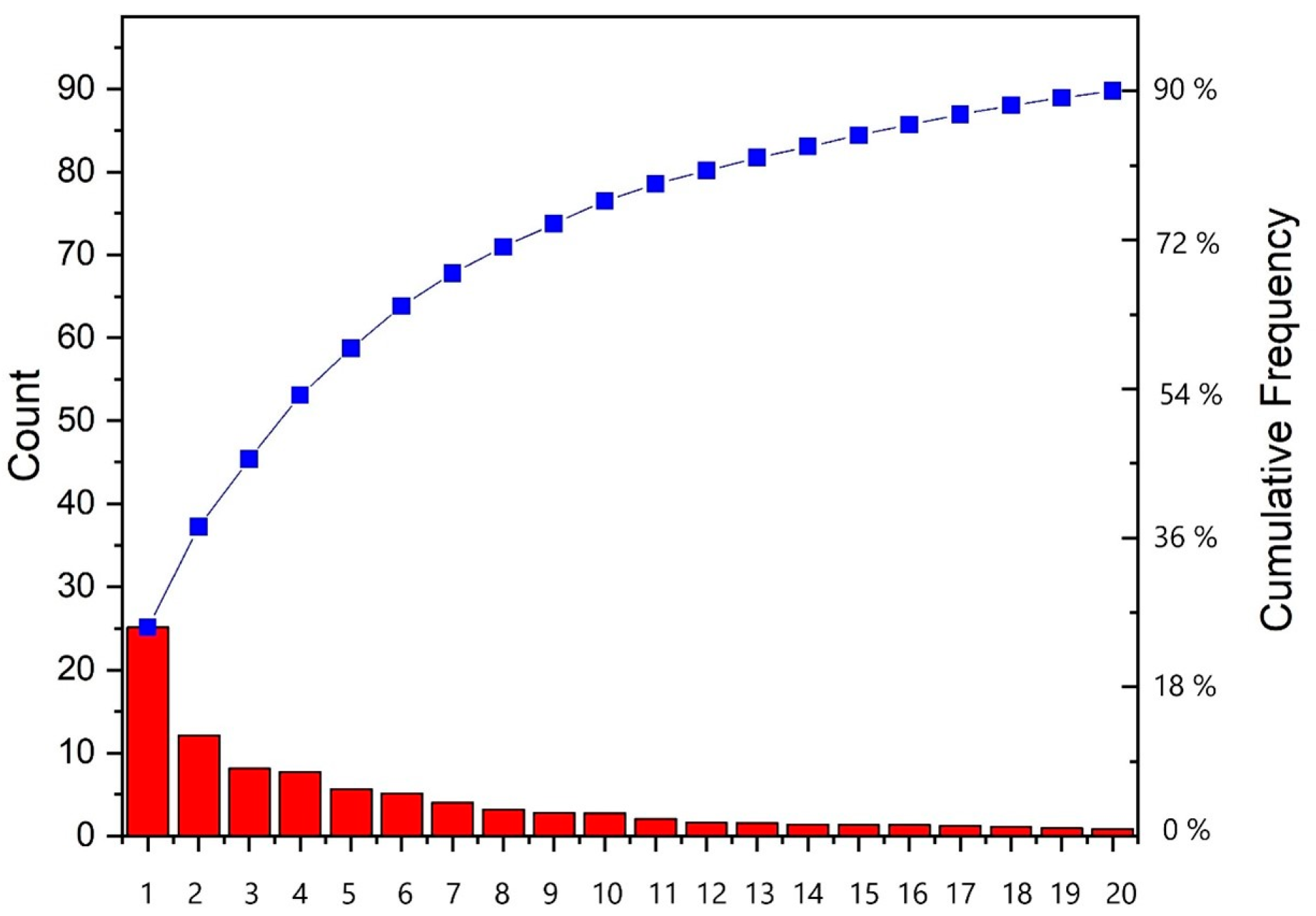
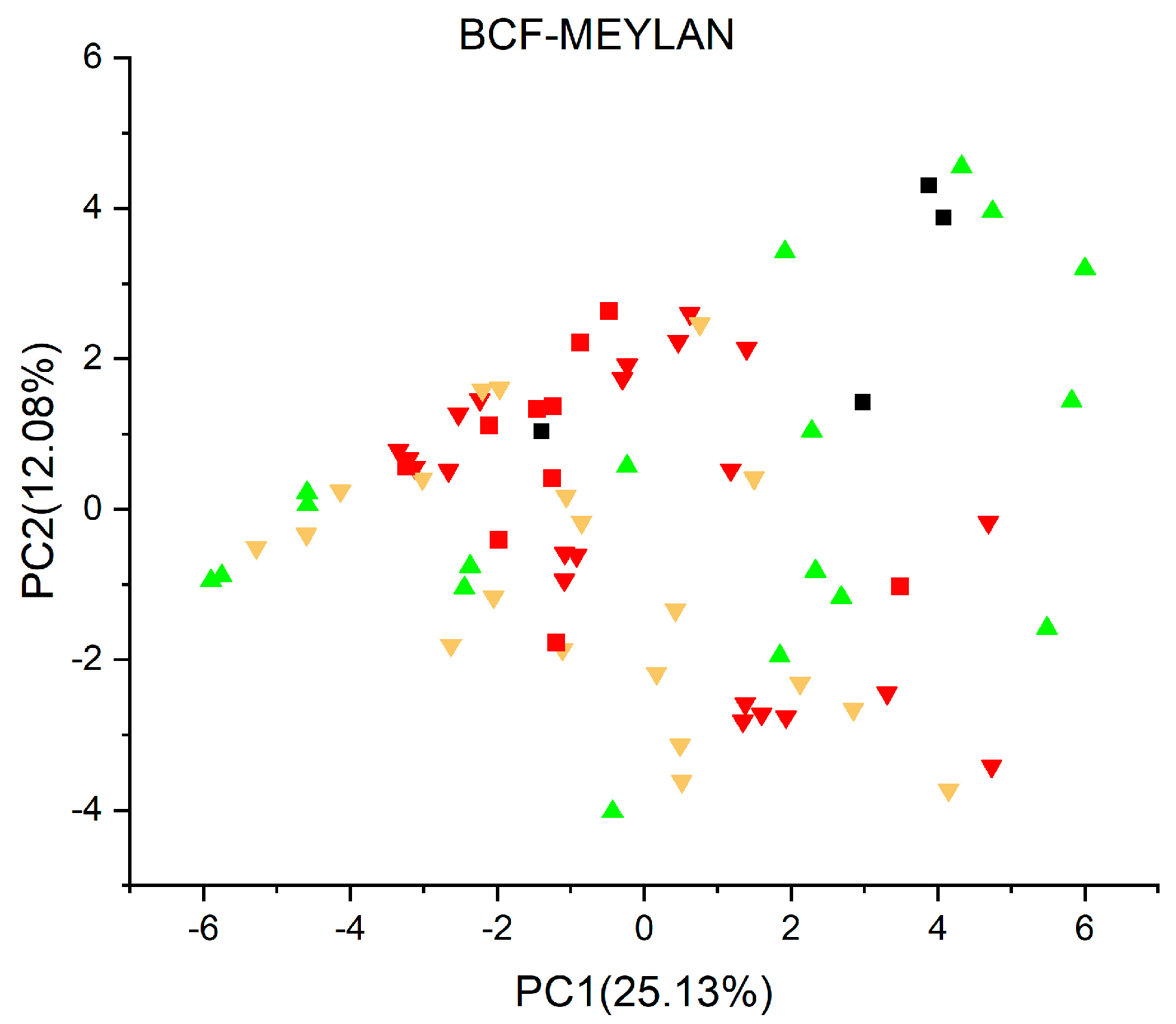
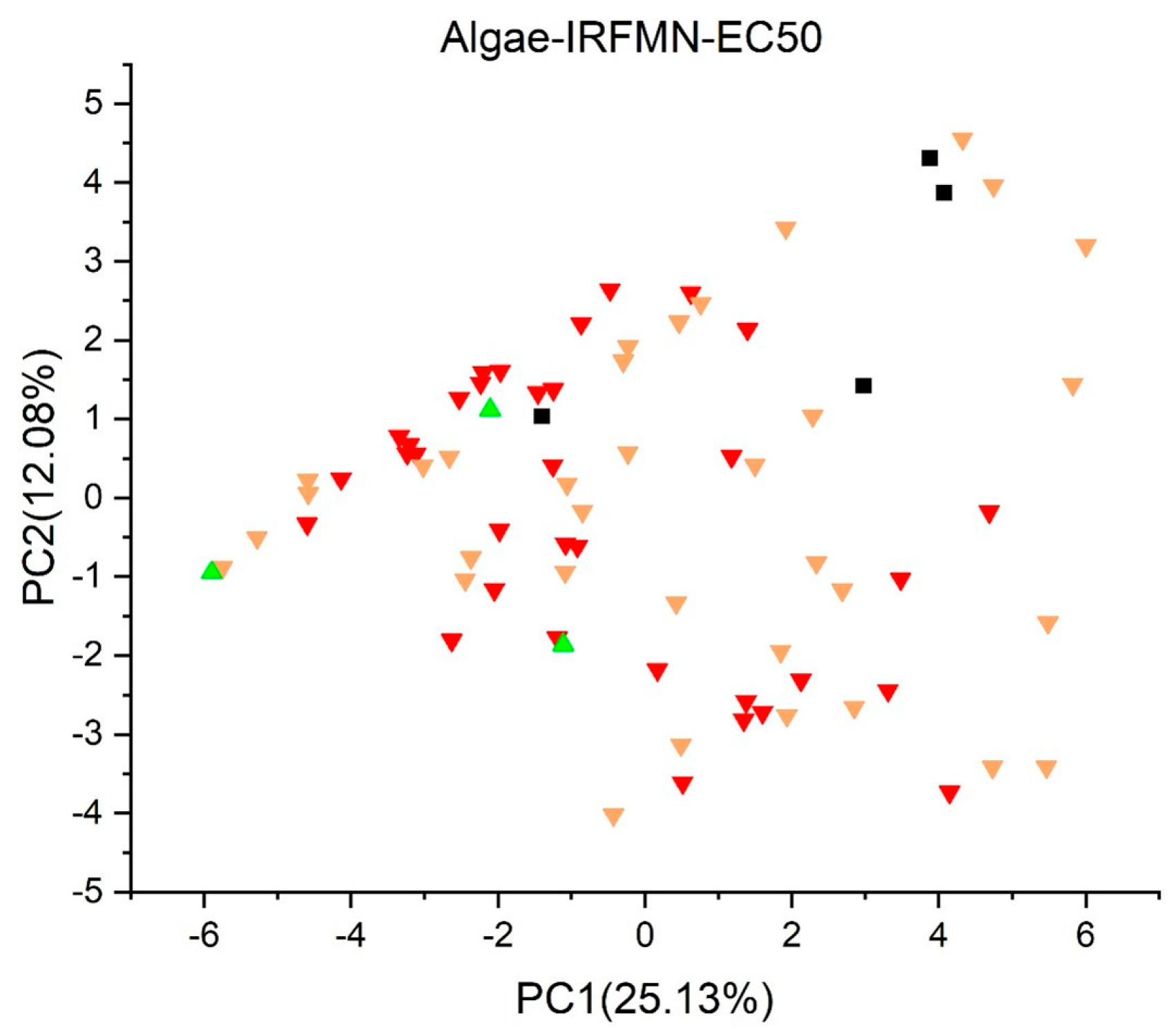
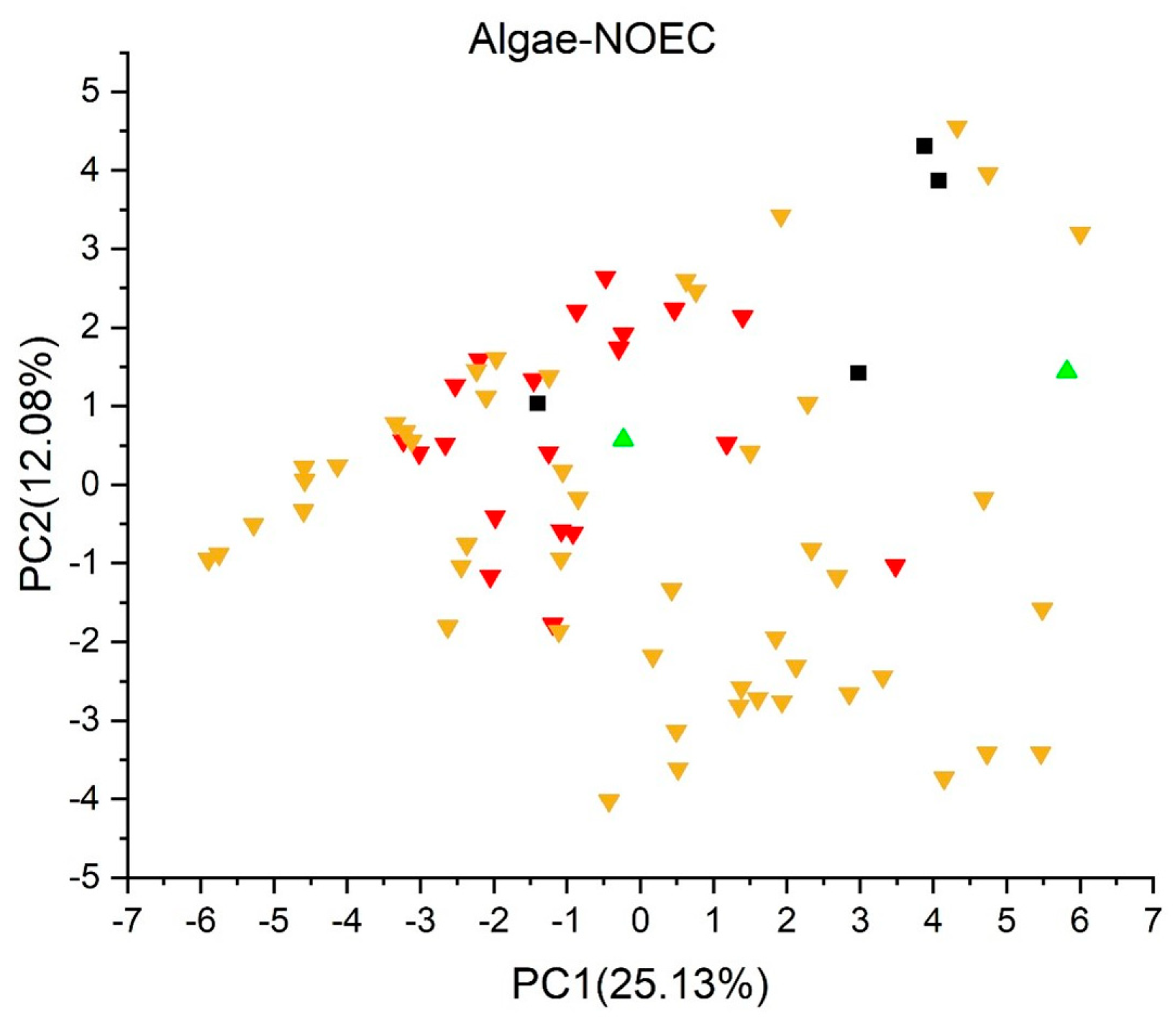
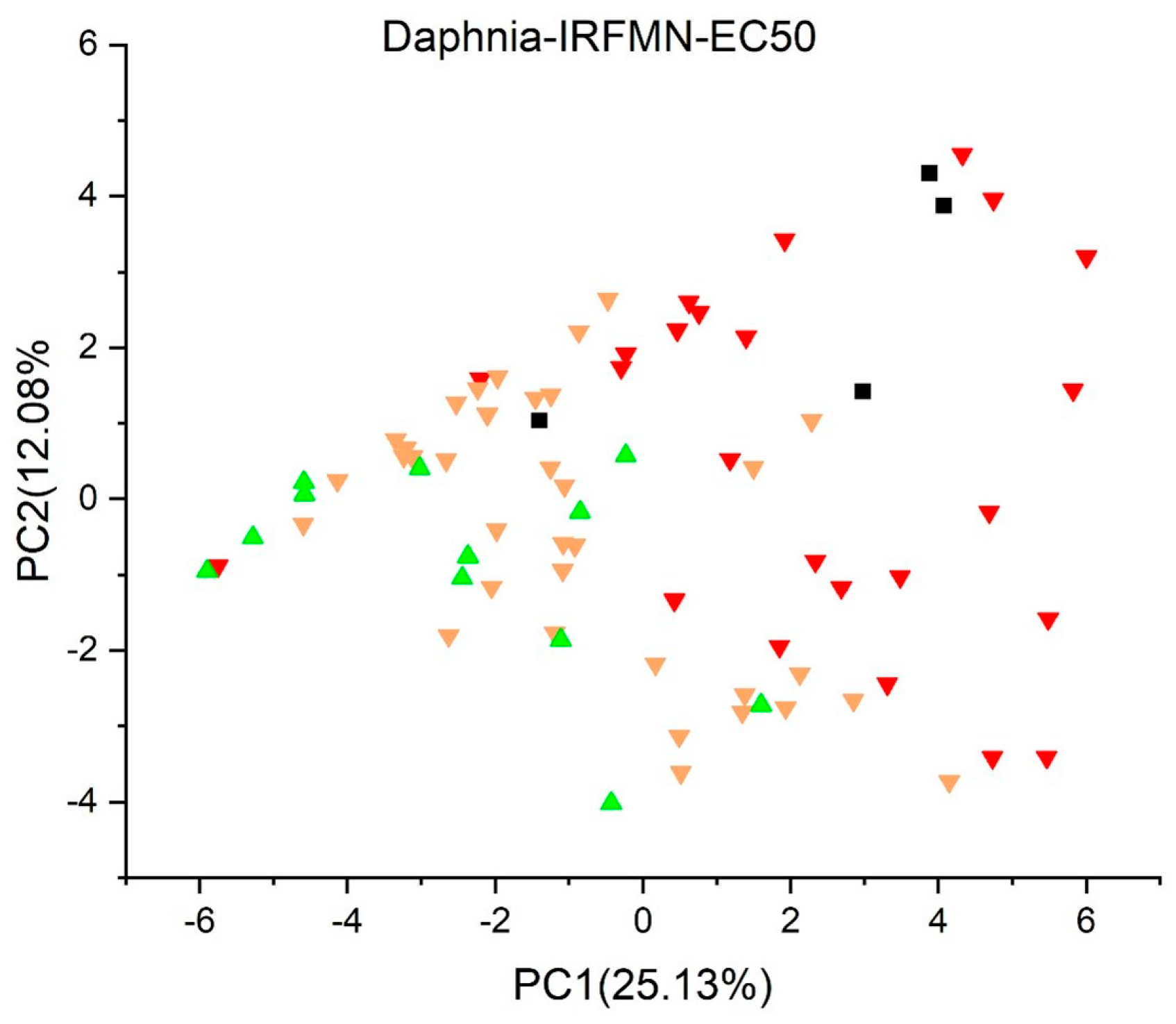
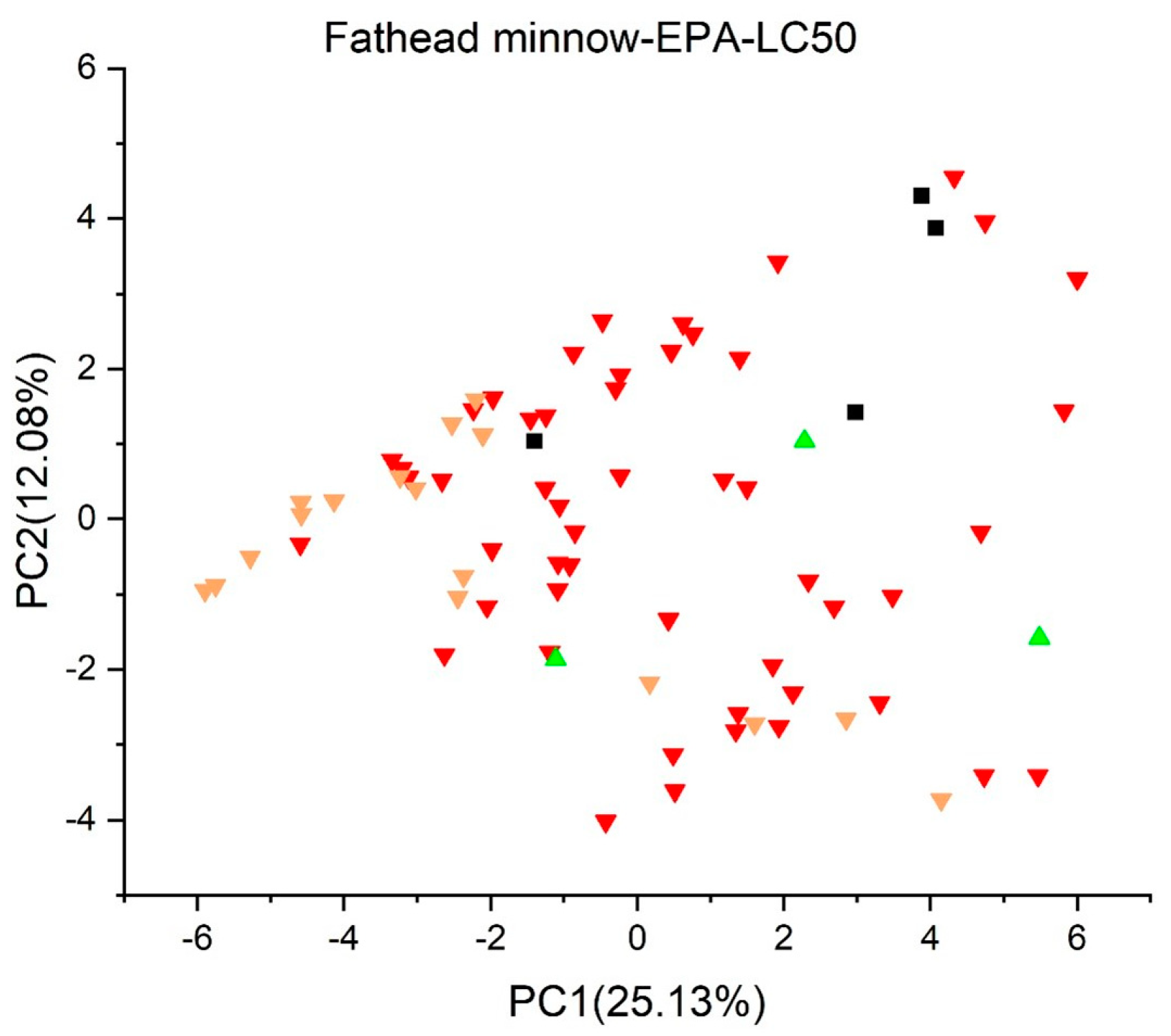
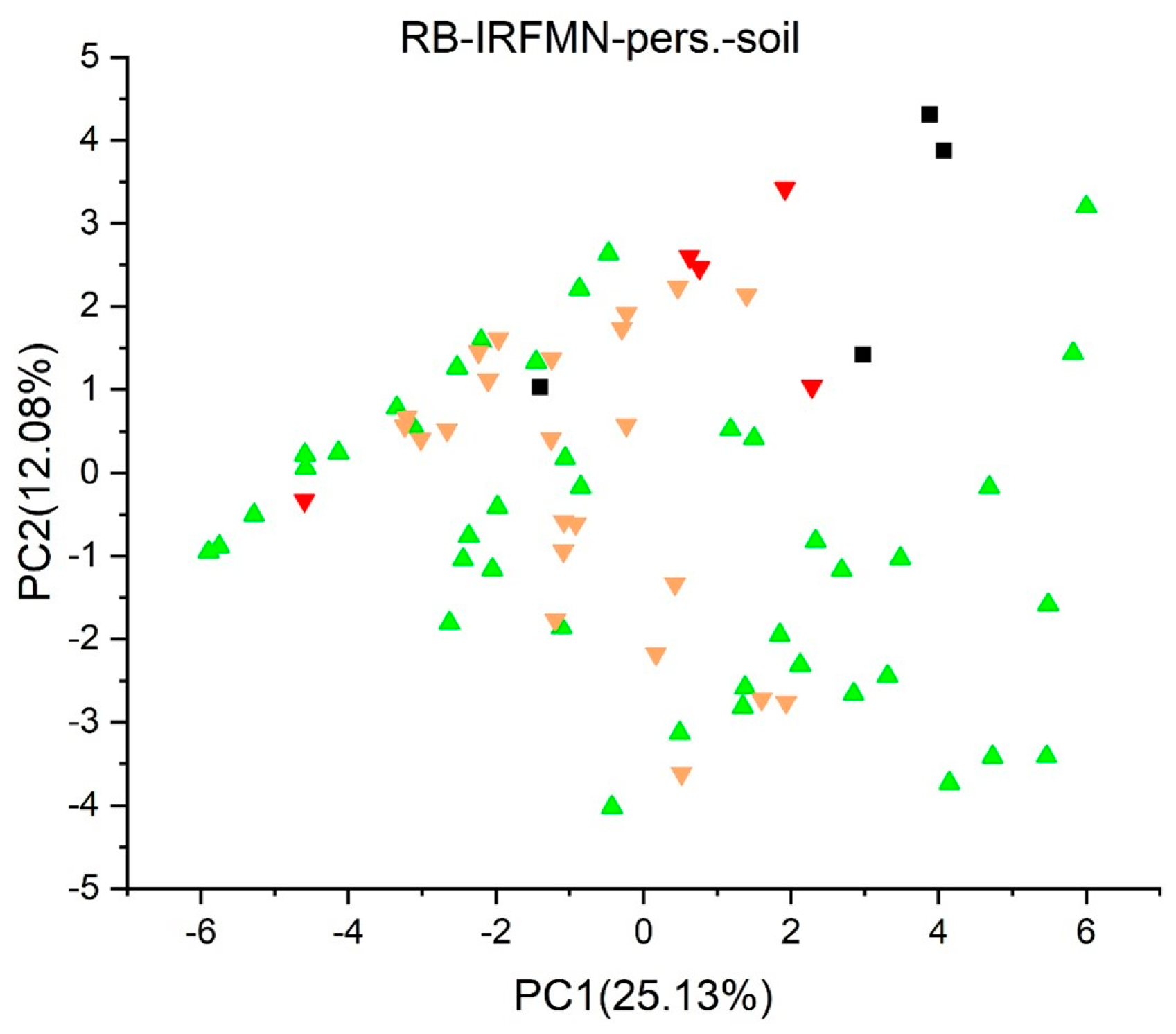
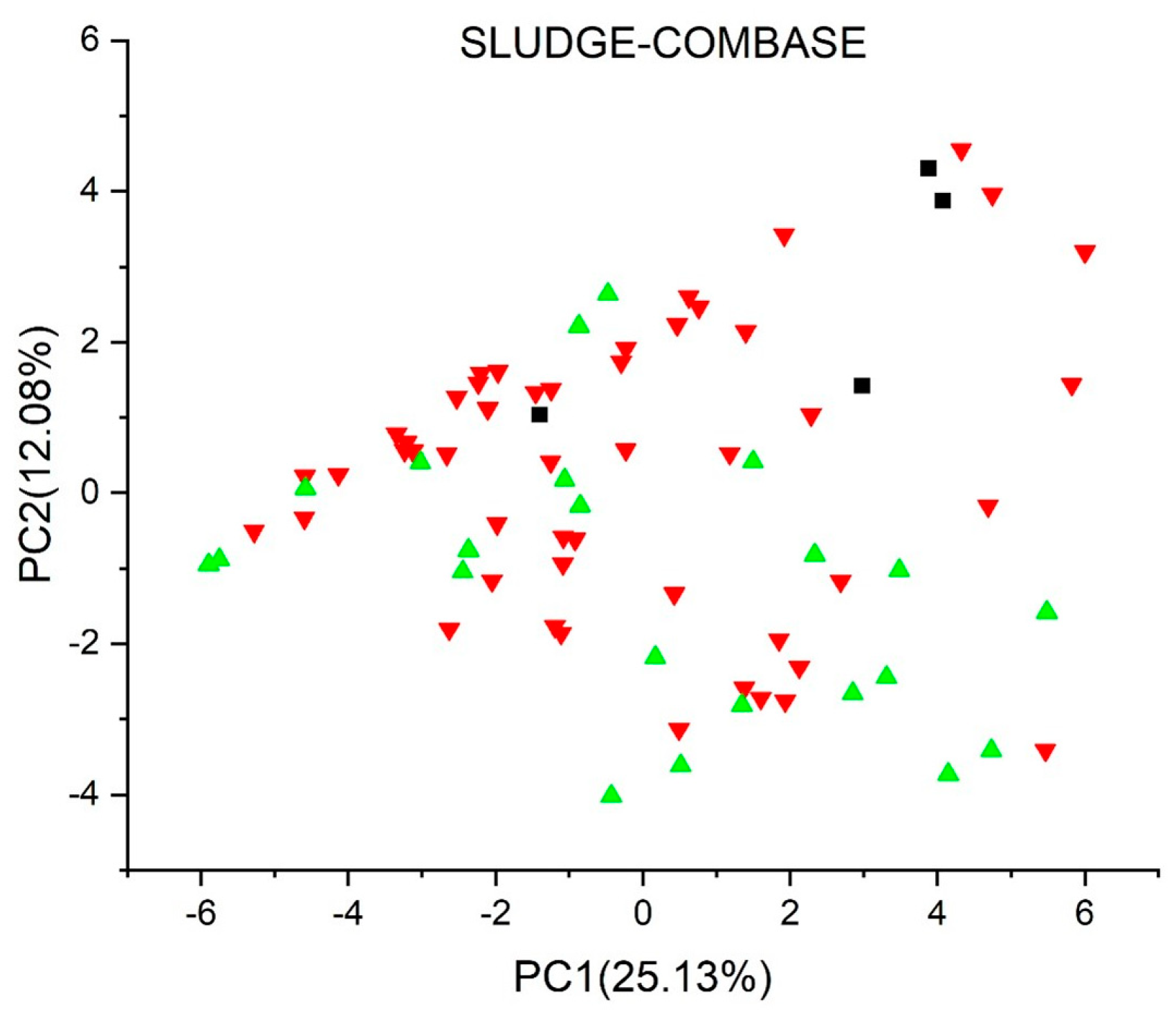
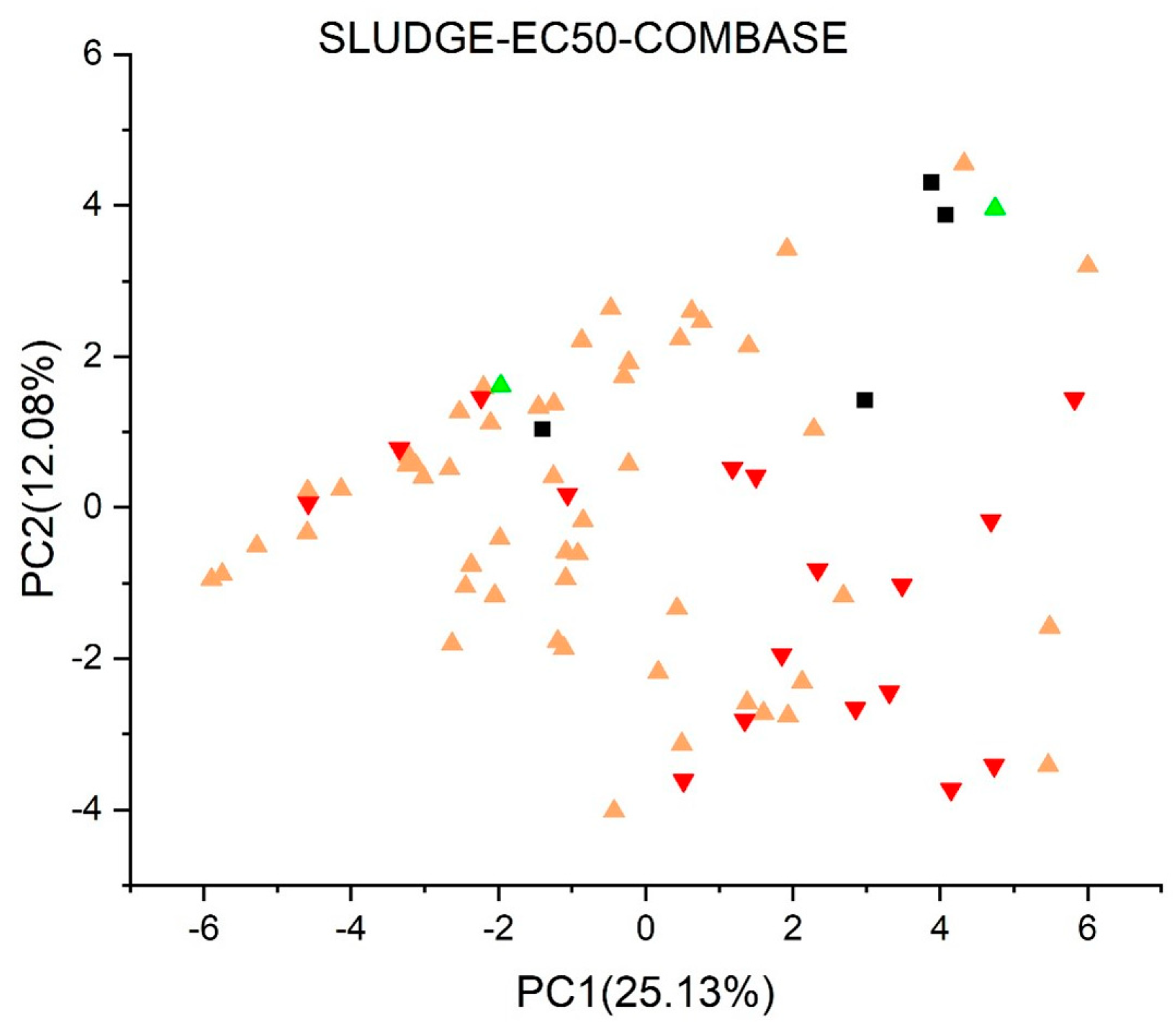
2.2. Principal Component Analysis
2.3. Ecotox Models Implemented in Vega and Their Suitability for Predicting BPA Alternatives
2.3.1. Fish Acute (LC50) Toxicity Model (IRFMN) Version 1.0.1 [34,35]
2.3.2. Fathead Minnow LC50 (96 h) (EPA) Version 1.0.7 [37,38]
2.3.3. Daphnia Acute (EC50) Toxicity Model (IRFMN) Version 1.0.1 [39]
2.3.4. Algae Acute EC50 Toxicity Model (IRFMN) Version 1.0.1 [35,40]
2.3.5. Algae (EC50) Toxicity Model (ProtoQSAR/Combase) Version 1.0.1 [41,42]
2.3.6. Algae Chronic (NOEC) Toxicity Model (IRFMN) Version 1.0.1 [43]
2.3.7. Sludge Classification Toxicity Model for Biocides (ProtoQSAR/COMBASE) Version 1.0.0 [44,45]
2.3.8. Sludge (EC50) Toxicity Version (ProtoQSAR/COMBASE) 1.0.1 [45,46]
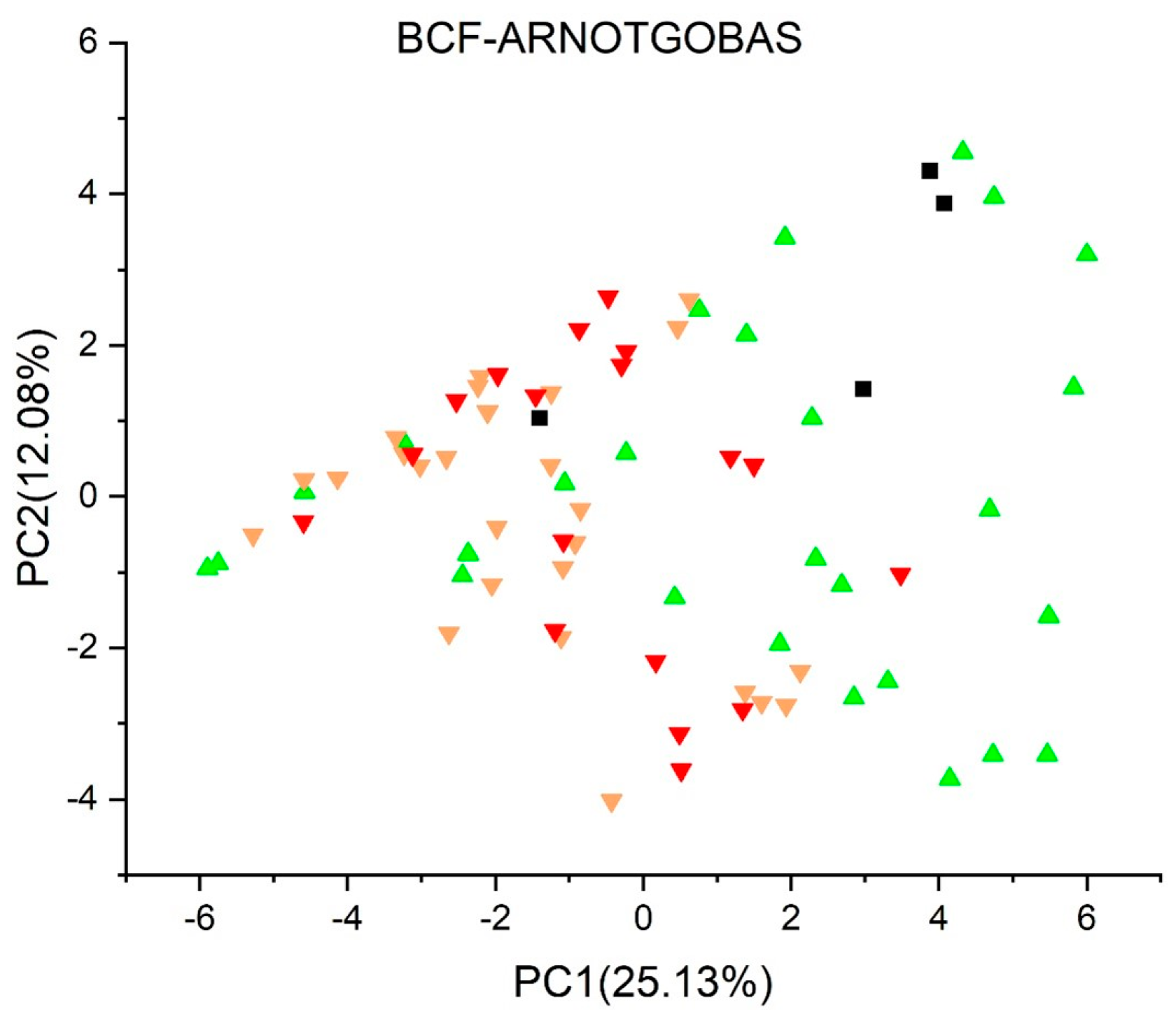
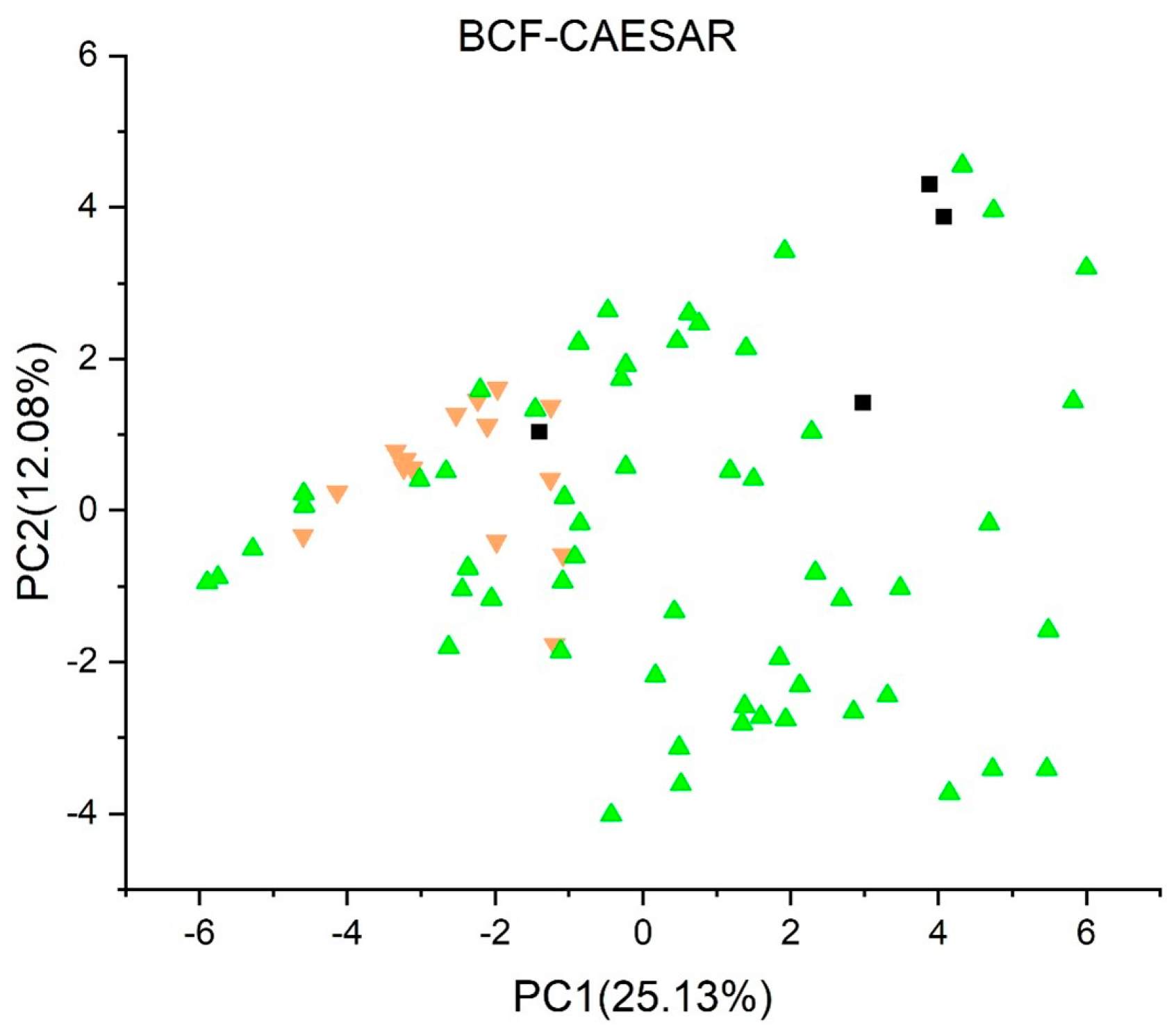
2.3.9. Bioconcentration Factors (BCF) Model (CAESAR) Version 2.1.15 [47,48,49]
2.3.10. Bioconcentration Factors (BCF) Model (Arnot-Gobas) Version 1.0.1 [51,52]
2.3.11. Bioconcentration Factors (BCF) Model (Meylan) Version 1.0.4 [53,54]
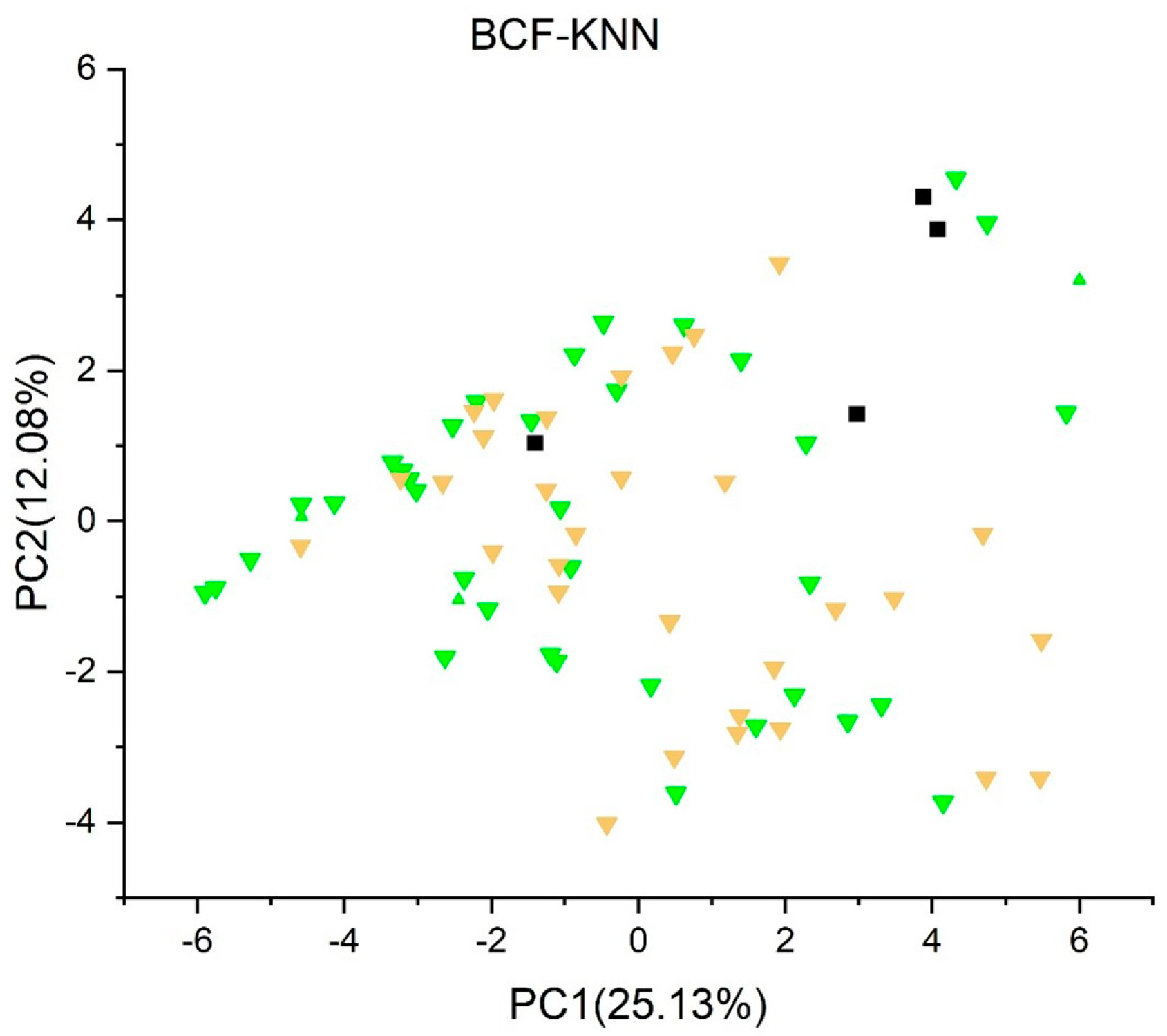
2.3.12. Bioconcentration Factors (BCF) Model (kNN/Read-Across) Version 1.1.1 [55,56]
2.3.13. Persistence (Soil) Quantitative Model (IRFMN) Version 1.0.1 [57]
| Model Name | Biological Model | Endpoint | Bisphenol Derivatives | Reference |
|---|---|---|---|---|
| Fish Acute (LC50) Toxicity model (IRFMN) | Oryzias latipes (Japanese rice fish) | Short-term toxicity to fish. Fish, Acute Toxicity Test | BPA 1, BPF 2 | Toma et al., 2021 [35] https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_FISH_LC50_IRFMN.pdf (accessed on 5 October 2023) |
| Fathead Minnow LC50 96 h (EPA) | Pimephales promelas (Fathead minnow) | Short-term toxicity to fish | BPA, TBBPA 3 | Martin et al., 2001 [38] https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_FATHEAD_LC50_EPA.pdf (accessed on 27 July 2023) |
| Daphnia Acute (EC50) toxicity model (IRFMN) | Daphnia magna | Short-term toxicity to aquatic invertebrates. Acute Immobilization Test | BPA, BPS 4, BPF, BPZ 5, TBBPA | https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_DAPHNIA_EC50_IRFMN.pdf (accessed on 5 October 2023) |
| Algae Acute (EC50) Toxicity model (IRFMN) | Raphidocelis subcapitata (Pseudokirchneriella subcapitata) | Long-term toxicity to aquatic algae and cyanobacteria C.f. OECD TG 201 Freshwater Alga and Cyanobacteria, Growth Inhibition Test | BPA, BPA 2 EO 6, BPF, TBBPA, 4,4′,4″-(ethan-1,1,1-triyl)triphenol | Toma et al., 2021 [35] https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_ALGAE_EC50_IRFMN.pdf (accessed on 27 July 2023) |
| Algae (EC50) Toxicity Model (ProtoQSAR/Combase) | Raphidocelis subcapitata (Pseudokirchneriella subcapitata) | Long-term toxicity to aquatic algae and cyanobacteria C.f. OECD TG 201 Freshwater Alga and Cyanobacteria, Growth Inhibition Test | BPA, BPA 2 EO, BPS, BPF, BPZ, TBBPA, 4,4′,4″-(ethan-1,1,1-triyl)triphenol | Blázquez et al. 2021 [41] https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_ALGAE_EC50_COMBASE.pdf (accessed on 5 October 2023) |
| Algae Chronic (NOEC) Toxicity model (IRFMN) | Raphidocelis subcapitata (Pseudokirchneriella subcapitata) | Long-term toxicity to aquatic algae and cyanobacteria C.f. OECD TG 201 Freshwater Alga and Cyanobacteria, Growth Inhibition Test | BPA, BPA 2EO, BPF, BPZ | https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_ALGAE_NOEC_IRFMN.pdf (accessed on 5 October 2023) |
| Sludge Classification Toxicity model (ProtoQSAR/COMBASE) | Activated sludge | Activated Sludge, Respiration Inhibition Test (OECD 209) | BPS, 4,4′,4″-(ethan-1,1,1-triyl)triphenol | https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_SLUDGE_CLASS_COMBASE.pdf (accessed on 27 July 2023) |
| Sludge (EC50) toxicity (ProtoQSAR/COMBASE) | Activated sludge | Activated Sludge, Respiration Inhibition Test (OECD 209) | 4,4′,4″-(ethan-1,1,1-triyl)triphenol | https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_SLUDGE_EC50_COMBASE.pdf (accessed on 6 October 2023) |
| BCF model (CAESAR) | Cyprinos Carpio and salmonids | BCF fish | BPA, TBBPA | Zhao et al., 2008 [49] https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_BCF_CAESAR.pdf (accessed on 6 October 2023) |
| BCF model (Arnot-Gobas) | Oncorhynchus mykiss (Rainbow trout) | BCF fish | BPA, TBBPA, TBMD 7 | Arnot et al., 2003 [51] https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_BCF_ARTNOTGOBAS.pdf (accessed on 6 October 2023) |
| BCF model (Meylan) | Fish | BCF fish | BPA, TBBPA | Meylan et al., 1999 [54] https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_BCF_MEYLAN.pdf (accessed on 6 Oc-tober 2023) |
| BCF model (kNN/Read-Across) | Fish | BCF fish | BPA, TBBPA | Manganaro et al., 2016 [56] https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_BCF_KNN.pdf (accessed on 6 October 2023) |
| Persistence (soil) quantitative model (IRFMN) | Soil | Biodegradation in soil. Aerobic and Anaerobic Transformation in Soil | TBBPA | https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_PERSISTENCE_SOIL_REG.pdf (accessed on 6 October 2023) |
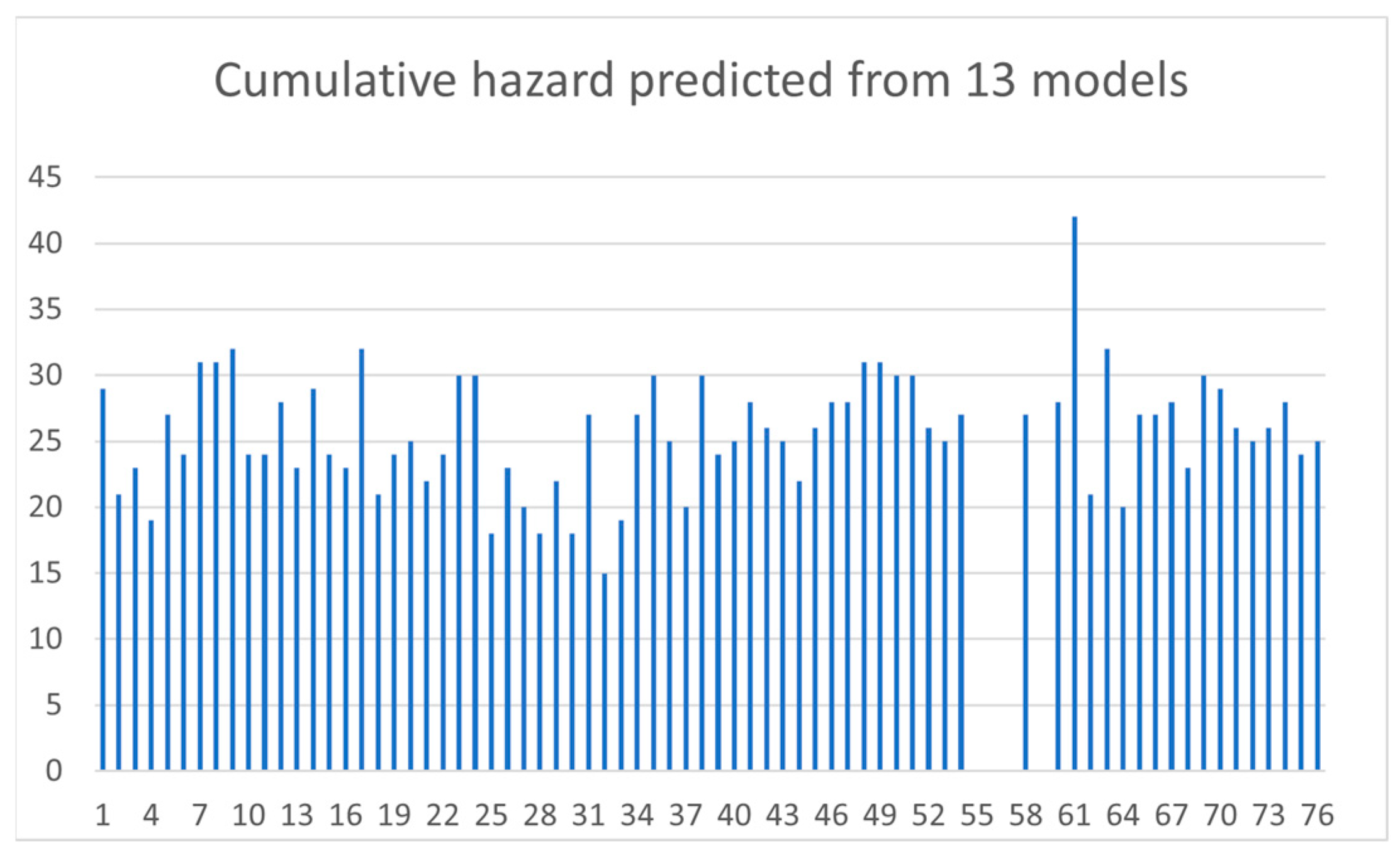
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geens, T.; Aerts, D.; Berthot, C.; Bourguignon, J.-P.; Goeyens, L.; Lecomte, P.; Maghuin-Rogister, G.; Pironnet, A.-M.; Pussemier, L.; Scippo, M.-L.; et al. A review of dietary and non-dietary exposure to bisphenol-A. Food Chem. Toxicol. 2012, 50, 3725–3740. [Google Scholar] [CrossRef] [PubMed]
- Lassen, C.; Brandt, U.K. Migration of Bisphenol A from Cash Register Receipts and Baby Dummies; Environmental Protection Agency: Washington, DC, USA, 2011. [Google Scholar]
- Vandenberg, L.N.; Hauser, R.; Marcus, M.; Olea, N.; Welshons, W.V. Human exposure to bisphenol A (BPA). Reprod. Toxicol. 2007, 24, 139–177. [Google Scholar] [CrossRef]
- Cao, X.-L.; Perez-Locas, C.; Dufresne, G.; Clement, G.; Popovic, S.; Beraldin, F.; Dabeka, R.W.; Feeley, M. Concentrations of bisphenol A in the composite food samples from the 2008 Canadian total diet study in Quebec City and dietary intake estimates. Food Addit. Contam. Part A 2011, 28, 791–798. [Google Scholar] [CrossRef]
- Cunha, S.; Fernandes, J. Assessment of bisphenol A and bisphenol B in canned vegetables and fruits by gas chromatography–mass spectrometry after QuEChERS and dispersive liquid–liquid microextraction. Food Control 2013, 33, 549–555. [Google Scholar] [CrossRef]
- Noonan, G.O.; Ackerman, L.K.; Begley, T.H. Concentration of Bisphenol A in Highly Consumed Canned Foods on the U.S. Market. J. Agric. Food Chem. 2011, 59, 7178–7185. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Xie, H.; Li, X.; Wu, J.; Lin, Y. Bisphenol A induced apoptosis and transcriptome differences of spermatogonial stem cells in vitro. Acta Biochim. Biophys. Sin. 2017, 49, 780–791. [Google Scholar] [CrossRef]
- Harnett, K.G.; Moore, L.G.; Chin, A.; Cohen, I.C.; Lautrup, R.R.; Schuh, S.M. Teratogenicity and toxicity of the new BPA alternative TMBPF, and BPA, BPS, and BPAF in chick embryonic development. Curr. Res. Toxicol. 2021, 2, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Jagne, J.; White, D.; Jefferson, F. Endocrine-Disrupting Chemicals: Adverse Effects of Bisphenol A and Parabens to Women’s Health. Water Air Soil Pollut. 2016, 227, 117385. [Google Scholar] [CrossRef]
- Pouzaud, F.; Thierry-Mieg, M.; Burga, K.; Vérines-Jouin, L.; Fiore, K.; Beausoleil, C.; Michel, C.; Rousselle, C.; Pasquier, E. Concerns related to ED-mediated effects of Bisphenol A and their regulatory consideration. Mol. Cell. Endocrinol. 2018, 475, 92–106. [Google Scholar] [CrossRef]
- Rubin, B.S. Bisphenol A: An endocrine disruptor with widespread exposure and multiple effects. J. Steroid Biochem. Mol. Biol. 2011, 127, 27–34. [Google Scholar] [CrossRef]
- Chen, D.; Kannan, K.; Tan, H.; Zheng, Z.; Feng, Y.-L.; Wu, Y.; Widelka, M. Bisphenol Analogues Other Than BPA: Environmental Occurrence, Human Exposure, and Toxicity—A Review. Environ. Sci. Technol. 2016, 50, 5438–5453. [Google Scholar] [CrossRef] [PubMed]
- Eladak, S.; Grisin, T.; Moison, D.; Guerquin, M.-J.; N’Tumba-Byn, T.; Pozzi-Gaudin, S.; Benachi, A.; Livera, G.; Rouiller-Fabre, V.; Habert, R. A new chapter in the bisphenol A story: Bisphenol S and bisphenol F are not safe alternatives to this compound. Fertil. Steril. 2015, 103, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Rochester, J.R.; Bolden, A.L. Bisphenol S and F: A Systematic Review and Comparison of the Hormonal Activity of Bisphenol A Substitutes. Environ. Health Perspect. 2015, 123, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Takeuchi, S.; Sanoh, S.; Okuda, K.; Kitamura, S.; Uramaru, N.; Sugihara, K.; Yoshinari, K. Profiling of bisphenol A and eight of its analogues on transcriptional activity via human nuclear receptors. Toxicology 2019, 413, 48–55. [Google Scholar] [CrossRef]
- Liu, B.; Lehmler, H.-J.; Sun, Y.; Xu, G.; Sun, Q.; Snetselaar, L.G.; Wallace, R.B.; Bao, W. Association of Bisphenol A and Its Substitutes, Bisphenol F and Bisphenol S, with Obesity in United States Children and Adolescents. Diabetes Metab. J. 2019, 43, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Marx-Stoelting, P.; Rivière, G.; Luijten, M.; Aiello-Holden, K.; Bandow, N.; Baken, K.; Cañas, A.; Castano, A.; Denys, S.; Fillol, C.; et al. A walk in the PARC: Developing and implementing 21st century chemical risk assessment in Europe. Arch. Toxicol. 2023, 97, 893–908. [Google Scholar] [CrossRef]
- Nowak, K.; Jakopin, Ž. In silico profiling of endocrine-disrupting potential of bisphenol analogues and their halogenated transformation products. Food Chem. Toxicol. 2023, 173, 113623. [Google Scholar] [CrossRef]
- EC. Commission Regulation (EU) 2016/2235 of 12 December 2016 amending Annex XVII to Regulation (EC) No 1907/2006 of the European Parliament and of the Council concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH). 2016. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32016R2235&from=EN (accessed on 14 November 2023).
- EFSA. Bisphenol A: EFSA Draft Opinion Proposes Lowering the Tolerable Daily Intake; European Food Safety Authority: Parma, Italy, 2021; Available online: https://www.efsa.europa.eu/en/news/bisphenol-efsa-draft-opinion-proposes-lowering-tolerable-daily-intake (accessed on 14 November 2023).
- Ou-Yang, S.; Lu, J.; Kong, X.; Liang, Z.; Luo, C.; Jiang, H. Computational drug discovery. Acta Pharmacol. Sin. 2012, 33, 1131–1140. [Google Scholar] [CrossRef]
- Xiang, M.; Cao, Y.; Fan, W.; Chen, L.; Mo, Y. Computer-Aided Drug Design: Lead Discovery and Optimization. Comb. Chem. High Throughput Screen. 2012, 15, 328–337. [Google Scholar] [CrossRef]
- EC. Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 Concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH). 2006. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:02006R1907-20161011&from=EN (accessed on 14 November 2023).
- TSCA. Toxic Substances Control Act (TSCA). 2021. Available online: https://www.congress.gov/bill/114th-congress/senate-bill/697/all-info (accessed on 14 November 2023).
- FDA. Use of International Standard ISO 10993-1. “Biological Evaluation of Medical Devices—Part 1: Evaluation and Testing within a Risk Management Process”. Guidance for Industry and Food and Drug Administration Staff. 2016. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/use-international-standard-iso-10993-1-biological-evaluation-medical-devices-part-1-evaluation-and (accessed on 14 November 2023).
- FDA. Premarket Tobacco Product Applications for Electronic Nicotine Delivery Systems. Guidance for Industry. 2016. Available online: https://www.fda.gov/media/97652/download (accessed on 14 November 2023).
- ECHA. Guidance on the Application of the CLP Criteria. Guidance to Regulation (EC) No 1272/2008 on Classification, Labelling and Packaging (CLP) of Substances and Mixtures. 2008. Available online: https://echa.europa.eu/documents/10162/23036412/clp_en.pdf/58b5dc6d-ac2a-4910-9702-e9e1f5051cc5 (accessed on 14 November 2023).
- Freidig, A.; Dekkers, S.; Verwei, M.; Zvinavashe, E.; Bessems, J.; van de Sandt, J. Development of a QSAR for worst case estimates of acute toxicity of chemically reactive compounds. Toxicol. Lett. 2007, 170, 214–222. [Google Scholar] [CrossRef] [PubMed]
- EC. Guidance Document on the Assessment of the Equivalence of Technical Materials of Substances Regulated under Regulation (EC) No 1107/2009 SANCO/10597/2003. 2009. Available online: https://food.ec.europa.eu/system/files/2016-10/pesticides_guidance_equivalence-chem-substances_en.pdf (accessed on 14 November 2023).
- SCCS. Memorandum on the Use of in Silico Methods for Assessment of Chemical Hazard. SCCS/1578/16. In Scientific Committee on Consumer Safety; European Commission: Brussels, Belgium, 2016; Available online: https://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_200.pdf (accessed on 14 November 2023).
- Mauri, A. alvaDesc: A Tool to Calculate and Analyze Molecular Descriptors and Fingerprints. In Ecotoxicological QSARs; Roy, K., Ed.; Springer: New York, NY, USA, 2020; pp. 801–820. [Google Scholar] [CrossRef]
- Mauri, A.; Bertola, M. Alvascience: A New Software Suite for the QSAR Workflow Applied to the Blood–Brain Barrier Permeability. Int. J. Mol. Sci. 2022, 23, 12882. [Google Scholar] [CrossRef] [PubMed]
- Benfenati, E.; Manganaro, A.; Gini, G. VEGA-QSAR: AI inside a platform for predictive toxicology. PAI@ AI* IA 2013, 1107, 21–28. [Google Scholar]
- Benfenati, E.; Colombo, E. Fish Acute (LC50) Toxicity model (IRFMN) (version 1.0.1). In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2022; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_FISH_LC50_IRFMN.pdf (accessed on 5 October 2023).
- Toma, C.; Cappelli, C.I.; Manganaro, A.; Lombardo, A.; Arning, J.; Benfenati, E. New Models to Predict the Acute and Chronic Toxicities of Representative Species of the Main Trophic Levels of Aquatic Environments. Molecules 2021, 26, 6983. [Google Scholar] [CrossRef]
- Results of aquatic toxicity tests of chemicals conducted by Ministry of the Environment in Japan (March 2016). In Series Results of Aquatic Toxicity Tests of Chemicals Conducted by Ministry of the Environment in Japan (March 2016); Ministry of Environment Japan: Tokyo, Japan, 2016; Available online: https://www.env.go.jp/en/chemi/sesaku/aquatic_Mar_2016.pdf (accessed on 2 August 2023).
- Benfenati, E.; Colombo, E. Fathead Minnow LC50 96h (EPA) (version 1.0.8). In QSAR Model Reporting Format (QMRF); Vegahub: 2022. Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_FATHEAD_LC50_EPA.pdf (accessed on 27 July 2023).
- Martin, T.M.; Young, D.M. Prediction of the Acute Toxicity (96-h LC50) of Organic Compounds to the Fathead Minnow (Pimephales promelas) Using a Group Contribution Method. Chem. Res. Toxicol. 2001, 14, 1378–1385. [Google Scholar] [CrossRef]
- Benfenati, E.; Cappelli, C.I.; Toma, C. Daphnia Acute (EC50) toxicity model (IRFMN) v1.0.1. In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2019; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_DAPHNIA_EC50_IRFMN.pdf (accessed on 5 October 2023).
- Benfenati, E.; Gamba, A. Algae Acute (EC50) Toxicity model (IRFMN)—V1.0.1. In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2021; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_ALGAE_EC50_IRFMN.pdf (accessed on 27 July 2023).
- Blázquez, M.; Andreu-Sánchez, O.; Ballesteros, A.; Fernández-Cruz, M.L.; Fito, C.; Gómez-Ganau, S.; Gozalbes, R.; Hernández-Moreno, D.; Julián-Ortiz, D.; Vicente, J.; et al. Computational Tools for the Assessment and Substitution of Biocidal Active Substances of Ecotoxicological Concern. In Chemometrics and Cheminformatics in Aquatic Toxicology; Wiley: New York, NY, USA, 2021; pp. 527–546. [Google Scholar] [CrossRef]
- Gómez-Ganau, S.; Gozalbes, R. Algae (EC50) Toxicity Model (ProtoQSAR/Combase) v.1.0.1. In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2019; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_ALGAE_EC50_COMBASE.pdf (accessed on 5 October 2023).
- Benfenati, E.; Manganaro, A. Algae Chronic (NOEC) Toxicity model (IRFMN)—V.1.0.1. In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2022; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_ALGAE_NOEC_IRFMN.pdf (accessed on 5 October 2023).
- Gómez-Ganau, S. Sludge Classification Toxicity model (ProtoQSAR/COMBASE) (Version 1.0.1). In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2022; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_SLUDGE_CLASS_COMBASE.pdf (accessed on 27 July 2023).
- Gómez-Ganau, S.; Marzo, M.; Gozalbes, R.; Benfenati, E. Computational Approaches to Evaluate Ecotoxicity of Biocides: Cases from the Project COMBASE. In Ecotoxicological QSARs; Roy, K., Ed.; Springer: New York, NY, USA, 2020; pp. 387–404. [Google Scholar] [CrossRef]
- Gómez-Ganau, S. Sludge (EC50) toxicity (ProtoQSAR/COMBASE) (Version 1.0.1). In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2022; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_SLUDGE_EC50_COMBASE.pdf (accessed on 6 October 2023).
- Lombardo, A.; Roncaglioni, A.; Boriani, E.; Milan, C.; Benfenati, E. Assessment and validation of the CAESAR predictive model for bioconcentration factor (BCF) in fish. Chem. Cent. J. 2010, 4, S1. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Boriani, E.; Chana, A.; Roncaglioni, A.; Benfenati, E. Model to predict bioconcentration factors (BCF) v 2.1.15 (CAESAR). In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2022; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_BCF_CAESAR.pdf (accessed on 6 October 2023).
- Zhao, C.; Boriani, E.; Chana, A.; Roncaglioni, A.; Benfenati, E. A new hybrid system of QSAR models for predicting bioconcentration factors (BCF). Chemosphere 2008, 73, 1701–1707. [Google Scholar] [CrossRef]
- Dimitrov, S.; Dimitrova, N.; Parkerton, T.; Comber, M.; Bonnell, M.; Mekenyan, O. Base-line model for identifying the bioaccumulation potential of chemicals. SAR QSAR Environ. Res. 2005, 16, 531–554. [Google Scholar] [CrossRef]
- Arnot, J.A.; Gobas, F.A.P.C. A Generic QSAR for Assessing the Bioaccumulation Potential of Organic Chemicals in Aquatic Food Webs. QSAR Comb. Sci. 2003, 22, 337–345. [Google Scholar] [CrossRef]
- Benfenati, E.; Manganaro, A. BCF model (Arnot-Gobas)—V.1.0.1. In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2020; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_BCF_ARTNOTGOBAS.pdf (accessed on 6 October 2023).
- Meylan, W.M.; Benfenati, E. BCF model (Meylan) v 1.0.4. In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2019; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_BCF_MEYLAN.pdf (accessed on 6 October 2023).
- Meylan, W.M.; Howard, P.H.; Boethling, R.S.; Aronson, D.; Printup, H.; Gouchie, S. Improved method for estimating bioconcentration/bioaccumulation factor from octanol/water partition coefficient. Environ. Toxicol. Chem. 1999, 18, 664–672. [Google Scholar] [CrossRef]
- Manganaro, A.; Benfenati, E. A BCF model (kNN/Read-Across) v 1.1.1. In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2020; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_BCF_KNN.pdf (accessed on 6 October 2023).
- Manganaro, A.; Pizzo, F.; Lombardo, A.; Pogliaghi, A.; Benfenati, E. Predicting persistence in the sediment compartment with a new automatic software based on the k-Nearest Neighbor (k-NN) algorithm. Chemosphere 2016, 144, 1624–1630. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, A.; Benfenati, E.; Manganaro, A. Persistence (soil) quantitative model (IRFMN)—V.1.0.1. In QSAR Model Reporting Format (QMRF); Vegahub: Milan, Italy, 2022; Available online: https://www.vegahub.eu/vegahub-dwn/qmrf/QMRF_PERSISTENCE_SOIL_REG.pdf (accessed on 6 October 2023).
- Gouin, T.; Cousins, I.; Mackay, D. Comparison of two methods for obtaining degradation half-lives. Chemosphere 2004, 56, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, D.; Biganzoli, F.; Todeschini, R.; Consonni, V. Qualitative consensus of QSAR ready biodegradability predictions. Toxicol. Environ. Chem. 2017, 99, 1193–1216. [Google Scholar] [CrossRef]
- ECHA. Bisphenols. In Assessment of Regulatory Needs; ECHA: Helsinki, Finland, 2021; Available online: https://echa.europa.eu/documents/10162/c2a8b29d-0e2d-7df8-dac1-2433e2477b02 (accessed on 14 November 2023).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora Lagares, L.; Vračko, M. Ecotoxicological Evaluation of Bisphenol A and Alternatives: A Comprehensive In Silico Modelling Approach. J. Xenobiot. 2023, 13, 719-739. https://doi.org/10.3390/jox13040046
Mora Lagares L, Vračko M. Ecotoxicological Evaluation of Bisphenol A and Alternatives: A Comprehensive In Silico Modelling Approach. Journal of Xenobiotics. 2023; 13(4):719-739. https://doi.org/10.3390/jox13040046
Chicago/Turabian StyleMora Lagares, Liadys, and Marjan Vračko. 2023. "Ecotoxicological Evaluation of Bisphenol A and Alternatives: A Comprehensive In Silico Modelling Approach" Journal of Xenobiotics 13, no. 4: 719-739. https://doi.org/10.3390/jox13040046
APA StyleMora Lagares, L., & Vračko, M. (2023). Ecotoxicological Evaluation of Bisphenol A and Alternatives: A Comprehensive In Silico Modelling Approach. Journal of Xenobiotics, 13(4), 719-739. https://doi.org/10.3390/jox13040046







