Abstract
Leukemia is a heterogeneous group of hematologic malignancies characterized by distinct genetic and molecular abnormalities. Advancements in genomic technologies have significantly transformed the diagnosis, prognosis, and treatment strategies for leukemia. Among these, next-generation sequencing (NGS) has emerged as a powerful tool, enabling high-resolution genomic profiling that surpasses conventional diagnostic approaches. By providing comprehensive insights into genetic mutations, clonal evolution, and resistance mechanisms, NGS has revolutionized precision medicine in leukemia management. Despite its transformative potential, the clinical integration of NGS presents challenges, including data interpretation complexities, standardization issues, and cost considerations. However, continuous advancements in sequencing platforms and bioinformatics pipelines are enhancing the reliability and accessibility of NGS in routine clinical practice. The expanding role of NGS in leukemia is paving the way for improved risk stratification, targeted therapies, and real-time disease monitoring, ultimately leading to better patient outcomes. This review highlights the impact of NGS on leukemia research and clinical applications, discussing its advantages over traditional diagnostic techniques, key sequencing approaches, and emerging challenges. As precision oncology continues to evolve, NGS is expected to play an increasingly central role in the diagnosis and management of leukemia, driving innovations in personalized medicine and therapeutic interventions.
1. Introduction
Leukemia is a hematologic malignancy characterized by the uncontrolled proliferation of abnormal white blood cells originating from the bone marrow. It is broadly classified into four main types based on the cell lineage affected and the disease progression rate: acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL), and chronic myeloid leukemia (CML) (Table 1) [1]. Acute leukemias (ALL and AML) are characterized by rapid disease progression due to the accumulation of immature, dysfunctional blast cells, whereas chronic leukemias (CLL and CML) progress more slowly, often allowing for longer survival without immediate treatment [2,3,4,5]. ALL primarily affects lymphoid progenitor cells and is the most common leukemia in children, although it can occur in adults. It is further subclassified based on immunophenotyping into B-cell or T-cell ALL [4]. AML, on the other hand, arises from myeloid precursors and is the most prevalent acute leukemia in adults. It is classified into subtypes based on genetic and morphologic features, including recurrent mutations such as FLT3, NPM1, and IDH1/2 [2]. CLL is a slow-growing leukemia affecting mature B lymphocytes and is predominantly found in older adults. It is often asymptomatic in early stages but can progress to a more aggressive form, requiring targeted therapy [5]. CML is defined by the presence of the BCR::ABL1 fusion gene, resulting from the Philadelphia chromosome translocation, which drives the uncontrolled proliferation of myeloid cells [6]. The introduction of tyrosine kinase inhibitors (TKIs) has dramatically improved CML prognosis [7]. The classification of leukemia continues to evolve with advancements in molecular profiling, enabling better risk stratification and personalized treatment approaches [8,9].

Table 1.
Comparison of ALL, AML, CLL, and CML.
Molecular profiling has become an essential tool in the diagnosis and treatment of leukemia, offering detailed insights into the genetic and epigenetic landscape of the disease [10,11]. Unlike conventional diagnostic methods, which primarily rely on cytogenetics, flow cytometry, and histopathological evaluation, molecular profiling enables the identification of specific genetic mutations, chromosomal rearrangements, and epigenetic modifications that define leukemia subtypes [11]. These insights have significantly improved risk stratification, guiding clinicians in selecting appropriate treatment strategies tailored to individual patients. For instance, genomic sequencing has revealed mutations in genes such as FLT3, NPM1, and TP53, which influence prognosis and therapeutic response in AML [12,13]. The integration of molecular profiling in leukemia treatment has also paved the way for targeted therapies, significantly improving patient outcomes [14]. The discovery of the BCR::ABL1 fusion gene in CML, for example, led to the development of TKIs, such as imatinib, which have revolutionized CML management [15]. Similarly, the identification of IDH1/2 and FLT3 mutations in AML has resulted in the development of small-molecule inhibitors targeting these aberrations [16]. These advancements exemplify the shift toward precision medicine, where treatment regimens are customized based on the molecular characteristics of the leukemia subtype rather than a one-size-fits-all approach [17]. Moreover, molecular profiling facilitates real-time disease monitoring and early detection of relapse. Minimal residual disease (MRD) assessment using next-generation sequencing (NGS) and real-time polymerase chain reaction (PCR) techniques allows for the identification of residual leukemic cells that are undetectable by traditional methods [18,19]. This capability is particularly crucial in guiding post-remission therapy decisions and adjusting treatment intensity to prevent relapse [20,21]. The ability to track clonal evolution through serial genomic profiling further enhances personalized treatment approaches, as emerging resistance mutations can be identified and alternative therapies can be considered in a timely manner [22,23]. Despite these significant advancements, challenges remain in the widespread clinical implementation of molecular profiling. The high cost of sequencing technologies, the need for specialized bioinformatics expertise, and the complexity of interpreting vast genomic data pose barriers to routine clinical use. However, continuous advancements in sequencing efficiency, the integration of artificial intelligence in data interpretation, and efforts to standardize molecular testing protocols are expected to improve accessibility and reliability [23,24].
Here, we aim to explore the transformative role of NGS in the diagnosis, classification, and treatment of leukemia, with a particular focus on ALL, AML, CLL, and CML. By comparing NGS with conventional diagnostic methods, this review highlights its advantages in detecting genetic mutations, monitoring minimal residual disease, and guiding targeted therapies. Additionally, it examines the challenges associated with NGS implementation in clinical practice and discusses future directions for integrating this technology into routine leukemia management. Through this comprehensive analysis, the review aims to provide insights into how NGS is shaping the future of precision medicine in leukemia treatment.
2. Conventional Diagnostic Methods in Leukemia
2.1. Bone Marrow Aspiration and Biopsy
Bone marrow aspiration and biopsy are essential diagnostic procedures in the evaluation of leukemia and other hematologic disorders [25]. Bone marrow aspiration involves extracting a liquid sample from the marrow, typically from the iliac crest, to assess cellular morphology, lineage involvement, and blast percentage (Figure 1) [26]. On the other hand, bone marrow biopsy provides a core tissue sample, allowing for the evaluation of marrow architecture, fibrosis, and cellularity. These procedures are frequently performed together to maximize diagnostic accuracy, particularly in cases where aspiration yields insufficient or inconclusive material [26]. Bone marrow aspiration is particularly useful in diagnosing acute leukemias, as it provides clear cytological details of blast morphology, essential for leukemia classification. In cases of chronic leukemias, such as CML or CLL, aspiration allows for the enumeration of abnormal cells and the detection of characteristic cytogenetic abnormalities [27]. However, one major limitation of aspiration is the potential for a “dry tap”, which occurs in conditions with increased marrow fibrosis, such as myelofibrosis or heavily infiltrated leukemic marrow [28,29]. In such cases, bone marrow biopsy becomes invaluable, as it provides a structural overview of the marrow and enables the detection of focal lesions or abnormal infiltration patterns [30]. Despite being the gold standard for leukemia diagnosis, bone marrow aspiration and biopsy are invasive procedures that require skilled personnel to ensure proper sample collection and minimize complications. Discomfort and minor bleeding are common, and in rare cases, infection or hemorrhage may occur [25]. Additionally, sample adequacy can impact diagnostic accuracy, as suboptimal samples may not fully reflect the disease burden. To improve diagnostic efficiency, these procedures are increasingly complemented by molecular and flow cytometric analyses, which provide deeper insights into leukemia pathophysiology and guide targeted treatment strategies [31,32].
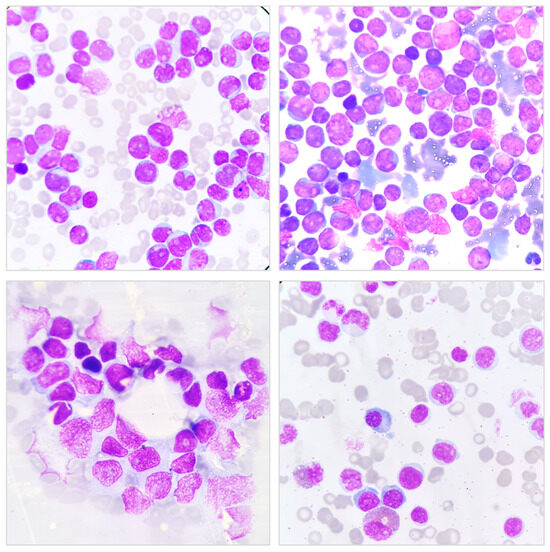
Figure 1.
Bone Marrow Aspiration (BMA) Smear Showing Hematopoietic Cells. Representative microscopic images of a BMA smear stained with Wright–Giemsa stain. The images display various hematopoietic cells, including immature and mature forms, with characteristic nuclear and cytoplasmic features. The purple-stained nuclei contrast against the lighter cytoplasm, aiding in the differentiation of cell lineages. These images are essential for evaluating hematological disorders, including leukemia and other bone marrow pathologies.
2.2. Cytogenetics and Karyotyping
Cytogenetics and karyotyping play a crucial role in the diagnosis and prognostic evaluation of leukemia by identifying chromosomal abnormalities that contribute to disease pathogenesis [33,34]. Karyotyping, a technique that visualizes entire chromosome sets, helps detect numerical and structural chromosomal alterations, including translocations, deletions, duplications, and aneuploidies [35]. These chromosomal aberrations serve as essential biomarkers for risk stratification and treatment planning in various leukemia subtypes. For instance, in CML, the translocation t(9;22)(q34;q11), known as the Philadelphia chromosome, results in the BCR::ABL1 fusion gene, which drives leukemogenesis and is a hallmark of the disease [36]. The identification of this translocation has led to the development of TKIs, such as imatinib, which have transformed CML treatment and significantly improved patient survival. In AML, karyotyping is a key prognostic tool, as specific chromosomal translocations define subgroups with distinct treatment responses and survival outcomes [37]. For example, the t(8;21)(q22;q22) translocation is associated with a favorable prognosis, while complex karyotypes involving three or more chromosomal abnormalities correlate with poor outcomes [38]. Cytogenetic risk stratification is integral to AML treatment decisions, as patients with favorable cytogenetics may benefit from standard chemotherapy, whereas those with high-risk karyotypes may require allogeneic stem cell transplantation [39,40]. Similarly, in ALL, cytogenetic abnormalities such as t(12;21)(p13;q22), which result in the ETV6::RUNX1 fusion, indicate a favorable prognosis, whereas the presence of t(9;22) confers a poor prognosis and necessitates targeted therapy with TKIs [41,42]. Despite its diagnostic and prognostic value, conventional karyotyping has limitations, including the requirement for dividing cells, which may lead to unsuccessful metaphase analysis in cases with low mitotic index. Additionally, karyotyping has limited resolution and may miss submicroscopic genetic alterations [43]. As a result, fluorescence in situ hybridization (FISH) and molecular techniques, such as next-generation sequencing (NGS), are increasingly used alongside cytogenetics to improve diagnostic accuracy. Nevertheless, karyotyping remains a cornerstone in leukemia diagnostics, as it provides a comprehensive overview of chromosomal abnormalities, informs risk stratification, and guides therapeutic decisions [44].
2.3. Fluorescence In Situ Hybridization (FISH)
Fluorescence in situ hybridization (FISH) is a molecular cytogenetic technique widely used in leukemia diagnostics for detecting specific genetic abnormalities, such as gene fusions, deletions, and amplifications [45]. Unlike conventional karyotyping, which requires dividing cells for metaphase analysis, FISH can be performed on interphase nuclei, allowing for the detection of chromosomal abnormalities even in non-proliferating cells. This makes FISH particularly valuable for identifying recurrent cytogenetic alterations associated with different leukemia subtypes [46]. For instance, in CML, FISH is used to detect the BCR::ABL1 fusion gene resulting from the Philadelphia chromosome translocation, a defining characteristic of the disease [47]. Similarly, in AML, FISH plays a crucial role in identifying translocations such as t(8;21), inv(16), and t(15;17), which have significant prognostic and therapeutic implications [2,48]. In CLL, FISH is instrumental in detecting common genetic abnormalities, including deletions at 13q14, 17p13 (TP53), and 11q22 (ATM), as well as trisomy 12. The detection of these abnormalities provides crucial prognostic information, as TP53 and ATM deletions are associated with poor treatment response and aggressive disease progression. Studies have shown that FISH significantly increases the detection rate of cytogenetic abnormalities in CLL, particularly in cases with normal karyotypes, and is essential for refining risk stratification and guiding targeted therapy [49,50,51]. Moreover, FISH has been used to complement standard cytogenetic techniques in cases where conventional karyotyping fails to yield sufficient metaphases, thereby enhancing the overall diagnostic yield [52]. Beyond its diagnostic applications, FISH is also valuable for monitoring MRD and assessing treatment response in leukemia patients. In CML, for example, FISH is often used in combination with quantitative PCR to track the persistence or recurrence of BCR::ABL1 transcripts following TKI therapy. Comparative studies have shown that while PCR is more sensitive for MRD detection, FISH remains a reliable method for assessing disease burden, particularly in resource-limited settings where PCR may not be readily available [53,54].
Despite its advantages, FISH has some limitations, including its reliance on pre-designed probes that target specific genetic abnormalities. This means that FISH cannot comprehensively assess all potential chromosomal aberrations in a single assay, necessitating the use of complementary techniques such as NGS for a more extensive genomic analysis. Furthermore, while FISH provides high sensitivity for known cytogenetic lesions, it does not detect point mutations or small insertions and deletions, which may also have prognostic significance in leukemia. Nonetheless, as an integral part of leukemia diagnostics, FISH continues to provide critical insights into genetic alterations, complementing other cytogenetic and molecular techniques to improve disease classification, prognostication, and treatment selection [55].
2.4. Polymerase Chain Reaction (PCR)
Polymerase chain reaction (PCR) is a powerful molecular diagnostic tool used in leukemia to identify specific genetic mutations associated with disease pathogenesis, prognosis, and treatment response [19,56]. By amplifying targeted DNA or RNA sequences, PCR enables the detection of mutations such as BCR::ABL1 in CML, NPM1 in AML, and IDH1/2 mutations in various leukemia subtypes. These genetic alterations serve as biomarkers for risk stratification and personalized therapy, making PCR an essential component of leukemia diagnostics [57,58]. Several types of PCR techniques are used in leukemia research and clinical practice, each offering unique advantages [59]. Conventional PCR is commonly employed for the qualitative detection of fusion genes, such as PML::RARA in acute promyelocytic leukemia (APL), providing rapid confirmation of molecular abnormalities [60,61]. However, real-time quantitative PCR (RT-qPCR) has become the preferred method for monitoring MRD in leukemia patients. RT-qPCR allows for the quantification of leukemia-associated transcripts, such as BCR::ABL1, with high sensitivity, enabling the detection of disease recurrence at very low levels. This method has been particularly useful in assessing treatment response and guiding therapeutic decisions in CML patients undergoing tyrosine kinase inhibitor therapy [62,63]. Another variant, reverse transcription PCR (RT-PCR), is widely used for detecting gene fusions and aberrant transcript expression in leukemia [63]. RT-PCR is instrumental in diagnosing BCR::ABL1 fusion variants and other translocations in ALL, such as ETV6::RUNX1 and MLL::AF4 [63,64]. Next, nested PCR (NPCR) enhances sensitivity by using two successive rounds of amplification, improving the detection of low-abundance mutations in leukemia patients [65,66]. Interestingly, a study compared the sensitivity of NPCR and RT-qPCR in detecting genetic alterations in acute leukemia patients. It showed that RT-qPCR was more sensitive than NPCR in detecting these alterations. Specifically, RT-qPCR identified 23 cases of BCR::ABL1 and 9 cases of TCF3::PBX1 in ALL patients, whereas detection rates for FLT3-ITD, RUNX1::RUNX1T1, PML::RARA, and CBFB::MYH11 in AML patients were also more efficiently captured by RT-qPCR. The study concluded that RT-qPCR is the superior method for identifying genetic alterations in acute leukemia [67]. Meanwhile, droplet digital PCR (ddPCR) is an emerging technique that offers ultra-sensitive mutation detection by partitioning a sample into thousands of droplets and analyzing them individually. This technology is increasingly used in leukemia research for precise quantification of mutant allele fractions and MRD assessment. Compared to conventional PCR methods, ddPCR provides absolute quantification without the need for standard curves, making it a promising tool for refining leukemia diagnostics and treatment monitoring [68,69]. Despite its advantages, PCR has limitations, including the potential for contamination, false-positive results, and the inability to detect unknown mutations. Additionally, while PCR is highly sensitive for targeted genetic alterations, it does not provide a comprehensive genomic overview, necessitating the integration of NGS for a more complete leukemia mutational profile. Nonetheless, PCR remains a cornerstone of leukemia diagnostics, offering rapid, reliable, and cost-effective detection of clinically relevant mutations that guide treatment decisions [70,71,72].
2.5. Flow Cytometry
Flow cytometry is a widely used immunophenotyping technique that plays a crucial role in leukemia diagnosis, classification, and prognosis. This technology enables the rapid and precise characterization of hematologic malignancies by identifying specific surface and intracellular markers expressed by leukemic cells. By using fluorescently labeled antibodies directed against lineage-specific antigens, flow cytometry allows for the differentiation of leukemia subtypes based on their immunophenotypic profiles. Compared to traditional microscopic examination and cytochemistry, flow cytometry offers higher sensitivity and specificity, making it a cornerstone in modern leukemia diagnostics [73,74]. One of the primary advantages of flow cytometry in leukemia classification is its ability to distinguish between AML and ALL [75]. AML cells typically express markers such as CD13, CD33, and CD117, whereas ALL is characterized by the presence of B-cell markers (CD19, CD22, CD79a) or T-cell markers (CD3, CD7, CD5) [73]. Moreover, flow cytometry can identify mixed phenotype acute leukemia (MPAL), a rare subtype expressing both myeloid and lymphoid markers, which has distinct therapeutic implications [76]. Beyond initial diagnosis, flow cytometry is essential for assessing MRD after chemotherapy. MRD refers to the presence of residual leukemic cells that are undetectable by conventional microscopy but can be quantified using highly sensitive multiparametric flow cytometry. MRD levels have strong prognostic value, guiding treatment decisions and risk stratification. For example, in ALL, patients with undetectable MRD after induction therapy have a significantly lower risk of relapse compared to those with persistent MRD [18,77].
Despite its advantages, flow cytometry has some limitations, including the need for fresh samples and the complexity of data interpretation. Variability in antigen expression due to treatment effects or disease evolution may lead to challenges in MRD detection. Additionally, while flow cytometry provides valuable immunophenotypic data, it does not detect genetic mutations, necessitating complementary molecular techniques such as PCR and NGS for a more comprehensive leukemia assessment [78,79,80]. The comparison of several diagnostic methods in acute and chronic leukemia is shown in Table 2.

Table 2.
Comparison of Diagnostic Methods in Acute and Chronic Leukemia.
2.6. Other Limitations of Conventional Methods
Despite significant advancements in leukemia diagnostics, conventional methods such as karyotyping, FISH, PCR, and flow cytometry have notable limitations that have driven the adoption of NGS. One of the primary shortcomings of traditional cytogenetics and molecular techniques is incomplete genomic coverage. Karyotyping, while useful for detecting large-scale chromosomal abnormalities, fails to identify submicroscopic mutations and cryptic translocations that can significantly impact leukemia prognosis and treatment decisions [81]. Similarly, FISH is limited to targeted analysis of predefined genetic rearrangements, making it ineffective for comprehensive genomic profiling [82]. PCR-based assays are highly sensitive but can only detect known mutations, necessitating multiple assays for comprehensive leukemia classification [83]. Another major limitation of conventional diagnostic approaches is their lower sensitivity compared to targeted NGS, particularly in the detection of MRD and subclonal mutations. Traditional Sanger sequencing, for example, has a detection threshold of around 15–20% variant allele frequency, meaning low-level mutations may go undetected. This limitation is particularly concerning in leukemia cases where early detection of emerging resistant clones is critical for treatment modifications [56]. Studies have shown that NGS-based approaches offer superior sensitivity, enabling the identification of low-abundance mutations that may contribute to disease relapse [84]. Additionally, targeted NGS panels have demonstrated higher concordance rates with traditional cytogenetics while uncovering additional clinically relevant mutations missed by standard methods [85]. Conventional diagnostic methods also lack the ability to provide insights into clonal evolution, a critical factor in leukemia progression and therapeutic resistance. Leukemia is a highly heterogeneous disease that evolves over time, often acquiring new mutations that contribute to treatment resistance and disease relapse [2,86]. Standard techniques such as FISH and PCR offer only a static snapshot of the disease at a given time, failing to capture the dynamic genetic changes that occur throughout treatment. In contrast, NGS enables longitudinal monitoring of clonal evolution by identifying emerging subclones and tracking mutational shifts during therapy. This ability to detect and characterize clonal diversity is essential for adapting treatment strategies and improving patient outcomes [87,88]. The transition from conventional diagnostics to NGS has been driven by these limitations, as NGS provides a more comprehensive, sensitive, and dynamic approach to leukemia characterization [89].
3. Basics, Techniques, and Procedures of NGS
3.1. Basics of NGS
NGS has revolutionized the field of molecular diagnostics, offering high-throughput, comprehensive genomic analysis that surpasses traditional sequencing techniques. Unlike Sanger sequencing, which can only analyze one gene at a time, NGS allows for the simultaneous sequencing of millions of DNA fragments, providing a more efficient and cost-effective method for detecting genetic variations [90]. This advancement has been particularly transformative in leukemia research and clinical management, enabling the identification of novel driver mutations, understanding clonal evolution, and refining risk stratification [87,91,92]. One of the primary advantages of NGS is its ability to detect a wide spectrum of genetic alterations, including single-nucleotide variants (SNVs), insertions and deletions (indels), copy number variations (CNVs), and gene fusions, all within a single assay. This capability is particularly important for leukemia subtypes such as AML and ALL, where genetic heterogeneity influences treatment outcomes [93,94]. By providing detailed molecular profiles, NGS facilitates personalized medicine approaches, allowing clinicians to tailor treatments based on the genetic makeup of an individual’s leukemia [95,96]. Sensitivity is a crucial aspect when comparing NGS to Sanger sequencing. Studies have shown that Sanger sequencing has a variant allele frequency (VAF) detection threshold of approximately 15–20%, meaning it can only reliably detect mutations when they are present in at least 15% of the DNA sample [56]. In contrast, NGS can detect mutations with a VAF as low as 1–5%, significantly enhancing its ability to identify rare mutations and clonal heterogeneity [97]. This is particularly relevant in leukemia, where early detection of emerging resistant clones is critical for adjusting treatment before full-blown relapse occurs. Another advantage of NGS is its ability to provide deeper coverage, meaning it sequences the same genomic region multiple times to improve accuracy [98].
Beyond diagnostics, NGS plays a crucial role in monitoring MRD and tracking clonal evolution over time. Traditional MRD assessment methods, such as flow cytometry and PCR, have limitations in sensitivity and specificity, whereas NGS enables the detection of residual leukemic clones at much lower thresholds. This is particularly relevant in guiding treatment decisions, as early detection of relapse-associated mutations can inform adjustments in therapy, potentially improving patient outcomes [94,99]. Another key advantage of NGS is its ability to uncover previously unrecognized mutations that contribute to treatment resistance. Deep sequencing ensures that even minor subclonal mutations are detected, which is essential for understanding the genetic complexity of leukemia. For example, in CML, NGS can detect compound mutations in the BCR::ABL1 kinase domain that contribute to TKI resistance, whereas Sanger sequencing often fails to distinguish between polyclonal and compound mutations [100,101]. In CLL, NGS has identified mutations in genes such as TP53, NOTCH1, and SF3B1, which are associated with poor prognosis and resistance to conventional chemotherapy. This information allows for more precise risk stratification and the selection of alternative therapeutic strategies, such as targeted inhibitors [87,96].
3.2. Techniques of NGS
NGS encompasses several distinct approaches, each offering unique advantages for leukemia research and clinical diagnostics (Table 3). Whole-genome sequencing (WGS) is the most comprehensive method, as it analyzes the entire genome, including both coding and non-coding regions [102]. This approach is particularly useful for discovering novel mutations, detecting structural variations, and identifying non-coding regulatory elements that may contribute to leukemia pathogenesis [103]. However, the extensive data generated by WGS, along with its high cost and computational demands, have limited its routine clinical use. Despite these challenges, WGS remains a valuable tool for research and precision medicine initiatives, as it provides an unbiased view of the genetic landscape of leukemia [104]. Whole-exome sequencing (WES) offers a more targeted approach by focusing only on the protein-coding regions of the genome, which comprise approximately 1–2% of the entire genome but contain the majority of known disease-causing mutations [102]. WES is a cost-effective alternative to WGS while still capturing clinically relevant mutations in leukemia-related genes [105]. It has been instrumental in identifying key mutations such as NPM1, FLT3, and TP53, which play crucial roles in leukemia prognosis and treatment response. Although WES does not provide insights into non-coding regions or large structural variations, it remains a widely used approach in both research and clinical settings due to its efficiency and high diagnostic yield [106].

Table 3.
Comparison of Different Types of Next-Generation Sequencing (NGS) Approaches.
Targeted gene panels are designed to sequence a predefined set of genes known to be associated with leukemia, providing a cost-effective and clinically applicable solution for mutation detection. These panels offer high sequencing depth, allowing for the identification of low-frequency mutations that may be missed by WES or WGS [102]. Targeted NGS panels are commonly used in clinical diagnostics to detect actionable mutations that can inform treatment decisions, such as IDH1/2 and DNMT3A mutations in AML [107]. Moreover, they reduce data complexity and interpretation challenges compared to broader sequencing approaches, making them highly suitable for routine clinical applications [108]. RNA sequencing (RNA-Seq) is a specialized NGS approach that focuses on the transcriptome, allowing for the detection of fusion genes, alternative splicing events, and gene expression patterns in leukemia [109]. RNA-Seq is particularly useful for identifying oncogenic fusion transcripts, such as BCR::ABL1 in CML or ETV6::RUNX1 in ALL, which may not be detected by DNA-based sequencing methods. Additionally, RNA-Seq provides insights into gene expression profiles that can help classify leukemia subtypes and predict treatment responses. By capturing both known and novel gene fusions, RNA-Seq has become an indispensable tool for leukemia research and diagnostics [110,111,112]. The selection of an NGS approach depends on the specific clinical or research question, balancing cost, sequencing depth, and data complexity. While WGS provides the most comprehensive view, WES and targeted panels offer practical and cost-effective alternatives for clinical applications, and RNA-Seq complements these methods by providing transcriptomic insights [113].
3.3. Step-by-Step Procedures of NGS
The NGS workflow consists of several key steps that ensure accurate and comprehensive genomic analysis in leukemia diagnostics (Figure 2) [114]. The process begins with sample preparation, during which DNA or RNA is extracted from sources such as bone marrow aspirates, peripheral blood, or biopsy samples. The quality and purity of nucleic acids are critical for the success of downstream applications, as degraded or contaminated samples can lead to sequencing artifacts or incomplete coverage [114]. In leukemia, high-quality sample extraction is essential for detecting mutations associated with disease progression and treatment resistance [115]. Following extraction, library preparation involves fragmenting DNA or RNA into small, uniform segments, which are then ligated to sequencing adapters and amplified. This step ensures that sequencing platforms can process the samples efficiently, maximizing coverage and minimizing sequencing bias [114]. Different approaches, such as amplicon-based enrichment or hybrid capture, are used depending on the NGS application. For targeted gene panels in leukemia, amplicon-based enrichment is commonly employed due to its high sensitivity in detecting known mutations with low variant allele frequencies [116].
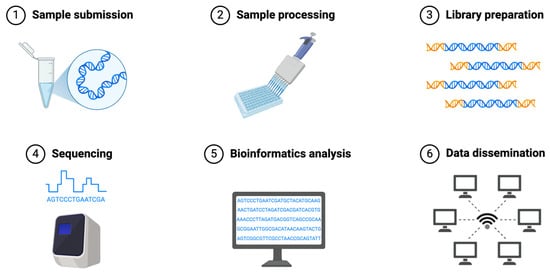
Figure 2.
Common Workflow of Next-Generation Sequencing.
The sequencing step utilizes high-throughput platforms such as Illumina, Oxford Nanopore, and PacBio, each offering distinct advantages (Table 4) [117]. Illumina sequencing, which is widely used in leukemia diagnostics, provides high accuracy and deep coverage, making it ideal for identifying single-nucleotide variants and small insertions or deletions [99,118]. Oxford Nanopore and PacBio long-read sequencing technologies offer additional capabilities, such as resolving complex structural variants and detecting full-length transcript isoforms in RNA-Seq [117]. These advancements enhance leukemia classification and aid in identifying novel fusion genes [118]. Once sequencing is complete, bioinformatics pipelines process the raw data, performing quality control, alignment to a reference genome, variant calling, and annotation. These pipelines use specialized software tools to filter out sequencing errors and distinguish true variants from background noise. Advanced computational approaches are employed to interpret structural variations, gene fusions, and alternative splicing events, which are critical for leukemia diagnosis [114]. For example, WES combined with RNA-Seq enables a comprehensive view of both genomic mutations and transcriptomic changes, improving diagnostic accuracy [114]. The final step in the NGS workflow is clinical interpretation, where identified variants are analyzed for their relevance to leukemia pathogenesis, prognosis, and therapeutic decision-making. This involves cross-referencing detected mutations with established databases such as the Catalogue of Somatic Mutations in Cancer (COSMIC) and ClinVar, which provide insights into the clinical significance of genetic alterations [119,120]. In leukemia, integrating NGS results with clinical and cytogenetic data allows for personalized treatment strategies, including the selection of targeted therapies based on specific driver mutations [95].

Table 4.
Comparison of Illumina, Nanopore, and PacBio NGS Platforms for Leukemia Research.
4. NGS in Acute Lymphoblastic Leukemia (ALL)
NGS plays a crucial role in the diagnosis, risk stratification, and treatment planning of ALL. NGS enables comprehensive genomic profiling, allowing for the identification of novel molecular subtypes, driver mutations, and chromosomal alterations that impact disease progression and therapeutic response [94]. NGS is particularly valuable in identifying cryptic genetic lesions that may not be detected by conventional cytogenetics, such as rare gene fusions or mutations in genes like IKZF1, CRLF2, and JAK2, which influence prognosis and treatment decisions [121]. Additionally, NGS helps in subclassifying ALL into molecularly distinct subtypes, such as Philadelphia chromosome-like (Ph-like) ALL, which requires targeted therapy [122].
Among the most critical genetic abnormalities detected by NGS in ALL are alterations in IKZF1, ETV6::RUNX1, and CRLF2 (Table 5) [123,124]. IKZF1 deletions are a significant genetic aberration in ALL, particularly in B-cell precursor ALL (BCP-ALL), where they are associated with poor prognosis and therapy resistance [124]. These deletions occur in approximately 10–15% of pediatric and adult BCP-ALL cases, with a higher prevalence in BCR::ABL1-positive and Ph-like ALL subtypes [125,126]. Mechanistically, IKZF1 deletions contribute to leukemogenesis by disrupting normal Ikaros function, leading to impaired B-cell differentiation and enhanced leukemic cell proliferation [124]. Studies have shown that patients with IKZF1 deletions exhibit lower event-free survival (EFS) and higher cumulative relapse rates compared to those without the deletion [127,128]. In pediatric BCP-ALL, a higher burden of IKZF1 deletions (>1%) correlates with significantly reduced survival rates [129,130,131]. Furthermore, IKZF1 deletions have been implicated in treatment resistance, particularly to cytarabine, due to reduced drug uptake mechanisms [132]. The ETV6::RUNX1 fusion, resulting from the t(12;21)(p13;q22) translocation, is the most common genetic alteration in childhood BCP-ALL, accounting for approximately 20–25% of cases [133]. This fusion event occurs in utero and is considered an initiating mutation; however, it is insufficient to drive leukemia independently, necessitating additional genetic alterations for full leukemogenesis [134]. Studies in twins and mouse models have demonstrated that ETV6::RUNX1 establishes a preleukemic clone that can persist for years before acquiring secondary mutations that lead to overt leukemia [135,136]. The fusion protein functions as an aberrant transcription factor that disrupts normal hematopoietic differentiation by interfering with RUNX1-mediated gene regulation, leading to transcriptional repression of key developmental genes [137,138]. Despite its role in leukemogenesis, ETV6::RUNX1 is generally associated with a favorable prognosis, with affected patients responding well to standard chemotherapy protocols and exhibiting high survival rates [139]. However, relapse in ETV6::RUNX1-positive ALL remains a concern, often driven by additional genetic events such as deletions in ETV6 or mutations in cell cycle regulators [41]. Meanwhile, CRLF2 rearrangements are a significant genetic alteration in ALL, particularly within the Ph-like B-ALL subtype, a high-risk subgroup of B-ALL that may benefit from TKIs, where they occur in approximately 50% of cases [94,140]. These rearrangements most commonly result from either an IGH-CRLF2 translocation or a P2RY8-CRLF2 fusion, both of which lead to overexpression of the cytokine receptor-like factor 2 (CRLF2) and subsequent activation of the JAK-STAT signaling pathway [141]. Studies have shown that CRLF2 rearrangements frequently co-occur with activating mutations in JAK2, as well as deletions or mutations in IKZF1, which contribute to leukemogenesis and poor treatment response [142]. Patients with CRLF2 rearrangements have been observed to have a significantly increased risk of relapse and inferior survival outcomes compared to those without the alteration, with studies reporting a relapse-free survival rate of only 35.3% at four years [141,142]. Additionally, aberrant activation of downstream pathways such as PI3K/mTOR has been documented in CRLF2-rearranged ALL, highlighting potential therapeutic targets for treatment-resistant cases [143]. Recent studies suggest that patients with CRLF2 rearrangements exhibit resistance to glucocorticoids, a key component of ALL therapy, but this resistance can be overcome with MEK or Akt inhibition, presenting new avenues for targeted treatment [140].

Table 5.
Mutations and Variants Associated with ALL Identified by NGS.
NGS has become an essential tool in risk stratification and treatment decision-making in ALL. The presence of high-risk mutations, such as those in IKZF1, TP53, or the RAS signaling pathway, influences treatment intensity and therapeutic choices [94]. For example, RAS pathway mutations, including NRAS, KRAS, and PTPN11, have been identified as potential drivers of ALL relapse, indicating the need for closer monitoring and possible early intervention with targeted agents [144]. The integration of NGS-based classifiers into clinical risk models has refined traditional prognostic scoring systems by incorporating genetic insights that were previously undetectable by conventional methods. This approach has led to the identification of very high-risk subgroups within T-ALL and B-ALL that require intensified therapy or novel treatment strategies [145]. By enabling early detection of mutations associated with treatment resistance, NGS has facilitated the development of precision medicine approaches in ALL, including the use of targeted inhibitors for kinase-activating lesions in Ph-like ALL [146]. Another impactful application of NGS in ALL is its role in MRD monitoring. MRD, defined as the presence of leukemic cells below the detection threshold of conventional microscopy, is a powerful prognostic indicator used for treatment stratification. NGS-based MRD assays provide greater sensitivity compared to flow cytometry and qPCR, allowing for the detection of one leukemic cell in a million normal cells [147]. This heightened sensitivity has enabled the identification of patients at risk of relapse despite achieving complete remission by conventional methods. Clinical trials have demonstrated that NGS-MRD outperforms multiparametric flow cytometry in detecting residual disease and predicting long-term outcomes, particularly in patients with B-ALL and T-ALL [148]. However, the prognostic value of NGS-based MRD remains an area of active research, with some studies suggesting that MRD positivity detected at ultra-low levels may not always correlate with an increased risk of relapse [149].
5. NGS in Acute Myeloid Leukemia (AML)
NGS has significantly enhanced the understanding of the genomic landscape of AML, revealing key driver mutations such as FLT3, NPM1, and IDH1/2 (Table 6) [150]. The FLT3 mutation, particularly the internal tandem duplication (FLT3-ITD), is one of the most prevalent alterations in AML, occurring in approximately 25–30% of cases [151]. It is associated with aggressive disease progression and poor prognosis [152]. Mutations in NPM1, a nucleophosmin gene, are found in around 30% of adult AML patients and are considered favorable prognostic markers in the absence of FLT3-ITD [153]. Patients with NPM1 mutations but without FLT3-ITD have improved clinical outcomes, likely due to a more favorable response to chemotherapy and higher rates of complete remission [154,155]. In contrast, NPM1 mutations co-occurring with FLT3-ITD neutralize the positive prognostic effect of NPM1, leading to a worse prognosis comparable to FLT3-ITD-only cases [156]. The adverse impact of FLT3-ITD is largely attributed to increased leukemic cell proliferation, resistance to apoptosis, and enhanced aggressiveness of the disease [157]. Studies have shown that in patients with NPM1-mutated AML, those lacking FLT3-ITD exhibit significantly longer disease-free and overall survival compared to those with both mutations [158]. The absence of FLT3-ITD allows NPM1-mutated AML to remain more responsive to standard induction chemotherapy, leading to better remission rates and prolonged survival. Consequently, NPM1-mutated, FLT3-ITD-negative AML is classified as a favorable-risk group in the European LeukemiaNet (ELN) guidelines, whereas FLT3-ITD positivity places patients in an intermediate- or adverse-risk category depending on the allelic ratio and co-occurring mutations [159].

Table 6.
Mutations and Variants Associated with AML Identified by NGS.
Similarly, IDH1 and IDH2 mutations (Table 6) occur in 5–20% of AML cases and play a crucial role in leukemogenesis by altering DNA methylation and cellular metabolism [160]. These mutations affect key metabolic enzymes, leading to the production of the oncometabolite 2-hydroxyglutarate, which disrupts normal DNA methylation and inhibits differentiation of hematopoietic cells, thereby promoting leukemogenesis [161]. IDH1 mutations, primarily affecting codon R132, are more frequently found in cytogenetically normal AML and often co-occur with NPM1 mutations. IDH2 mutations, particularly R140Q and R172K, are also associated with normal karyotype AML but tend to be mutually exclusive with IDH1 mutations [161,162]. The prognostic impact of these mutations varies depending on the specific mutation and co-occurring genetic alterations. IDH1 R132 mutations have been associated with poor prognosis in some studies, particularly when they occur in the absence of FLT3-ITD or NPM1 mutations [163,164]. IDH2 R172 mutations have also been linked to adverse outcomes, while IDH2 R140Q mutations appear to have a more neutral or slightly favorable prognostic effect. Therapeutically, IDH1 and IDH2 inhibitors, such as ivosidenib and enasidenib, respectively, have been developed to target these mutations. These inhibitors work by restoring normal differentiation in leukemic cells and have shown clinical efficacy, particularly in relapsed or refractory AML [165]. Ongoing research is focused on understanding resistance mechanisms to IDH inhibitors, as leukemia stemness and co-occurring mutations, such as RUNX1 and RAS-RTK pathway alterations, can drive resistance to these targeted therapies [166]. Overall, the identification of these mutations through NGS has refined AML classification, prognosis, and personalized treatment approaches. The impact of NGS on personalized therapy in AML is profound, particularly in the selection of targeted inhibitors. FLT3 inhibitors, such as midostaurin and gilteritinib, have shown clinical efficacy in FLT3-mutant AML by inhibiting aberrant tyrosine kinase signaling. Patients with IDH1/2 mutations benefit from specific inhibitors like ivosidenib and enasidenib, which restore normal hematopoietic differentiation [167]. These targeted therapies, guided by NGS-based mutation profiling, have led to improved treatment outcomes and prolonged survival for AML patients. The incorporation of NGS into clinical workflows allows for the rapid detection of actionable mutations, ensuring the timely initiation of personalized treatment regimens [168]. Furthermore, NGS-based stratification has been instrumental in optimizing chemotherapy regimens, as patients harboring concurrent NPM1 and FLT3-ITD mutations may require intensified therapeutic strategies [169].
Beyond diagnosis and treatment selection, NGS plays a critical role in monitoring clonal evolution and treatment resistance in AML. Leukemia is characterized by significant genetic heterogeneity, with subclonal populations evolving in response to therapy. NGS enables the longitudinal tracking of mutational changes, revealing the emergence of resistant clones that drive relapse [86]. For example, patients treated with FLT3 inhibitors may develop secondary mutations in the kinase domain, necessitating alternative therapeutic strategies [170]. Similarly, the persistence or acquisition of IDH1/2 mutations following chemotherapy has been linked to disease relapse, underscoring the importance of dynamic molecular monitoring [171]. NGS-based MRD assessment allows for the detection of residual leukemic clones at ultra-low levels, providing a more sensitive approach to relapse prediction compared to traditional methods [172,173].
6. NGS in Chronic Lymphocytic Leukemia (CLL)
NGS has revolutionized the molecular characterization of CLL by identifying key mutations with significant prognostic and therapeutic implications [87]. Among these, TP53, NOTCH1, and SF3B1 mutations play critical roles in disease progression and treatment resistance (Table 7) [174]. TP53 mutations in CLL are among the most critical genetic alterations, as they are strongly associated with poor prognosis, rapid disease progression, and resistance to standard chemoimmunotherapy. These mutations occur in approximately 5–10% of newly diagnosed CLL cases but are found in over 40% of relapsed or refractory patients, highlighting their role in disease evolution [175]. The majority of TP53 mutations are located within the DNA-binding domain and lead to loss of function, impairing the tumor suppressor’s ability to regulate apoptosis and cell cycle arrest. Notably, TP53 mutations frequently co-occur with 17p deletions, where one allele is lost, and the remaining allele is mutated, further exacerbating disease aggressiveness [176]. However, TP53 mutations can also occur independently of 17p deletion, albeit with similar negative prognostic implications. Studies have demonstrated that CLL patients harboring TP53 mutations exhibit significantly shorter progression-free and overall survival compared to those without these mutations, even when treated with fludarabine-based regimens [177,178]. The presence of TP53 mutations also predicts poor response to standard chemoimmunotherapy, necessitating the use of novel targeted agents such as Bruton’s tyrosine kinase (BTK) inhibitors (ibrutinib, acalabrutinib) and B-cell lymphoma 2 (BCL-2) inhibitors (venetoclax), which have demonstrated superior efficacy in TP53-mutated CLL [179,180]. Due to its strong prognostic and predictive value, TP53 mutation testing is now recommended before initiating treatment in CLL patients, and repeated testing is advised before each line of therapy, as clonal evolution may lead to the emergence of new TP53 mutations over time [181,182,183].

Table 7.
Mutations and Variants Associated with CLL Identified by NGS.
Meanwhile, NOTCH1 mutations (Table 7) occur in approximately 10–15% of newly diagnosed cases and are increasing in prevalence in relapsed or refractory disease [184]. These mutations typically involve frameshift or nonsense alterations in exon 34, leading to the truncation of the PEST domain, which results in prolonged NOTCH1 activation due to impaired degradation of the NOTCH1 intracellular domain (NICD) [185]. The constitutive activation of NOTCH1 signaling promotes leukemic cell survival, proliferation, and resistance to apoptosis, contributing to disease progression. Clinically, NOTCH1 mutations are associated with an aggressive disease course, shorter overall survival, and reduced time to first treatment, making them a strong negative prognostic marker [186]. These mutations frequently co-occur with trisomy 12 and unmutated IGHV, both of which are indicative of a more aggressive CLL phenotype. Importantly, NOTCH1 mutations have been linked to resistance to anti-CD20 monoclonal antibody therapies, such as rituximab and obinutuzumab, potentially explaining inferior responses to chemoimmunotherapy [187]. Moreover, studies suggest a strong association between NOTCH1 mutations and an increased risk of Richter’s transformation, the progression of CLL into aggressive diffuse large B-cell lymphoma (DLBCL), which significantly worsens patient prognosis [188]. Given their clinical relevance, NOTCH1 mutations are now considered an independent prognostic marker in CLL, and testing for these alterations is recommended for risk stratification and treatment decision-making. The development of NOTCH1 inhibitors and γ-secretase inhibitors as potential therapeutic strategies is an active area of research, aiming to counteract the oncogenic effects of NOTCH1 activation in CLL [189]. Next, SF3B1 mutations occur in approximately 10–15% of cases and are increasing in frequency in advanced and treatment-refractory disease [190]. SF3B1 encodes a core component of the spliceosome, which is essential for pre-mRNA splicing, and mutations in this gene lead to widespread alterations in RNA processing. These splicing defects contribute to CLL pathogenesis by disrupting normal gene expression and promoting leukemic cell survival [191,192]. Transcriptomic analyses have shown that SF3B1 mutations affect multiple key pathways, including DNA damage response, telomere maintenance, and NOTCH signaling, further driving disease progression [193]. Clinically, SF3B1 mutations are associated with an aggressive CLL phenotype, characterized by shorter treatment-free survival, poor response to fludarabine-based chemoimmunotherapy, and increased risk of disease progression [194]. Additionally, SF3B1 mutations frequently co-occur with del(11q), which is another high-risk genetic feature in CLL, further reinforcing their association with adverse clinical outcomes [195]. Given their prognostic significance, SF3B1 mutations are now included in molecular risk stratification models for CLL [196]. While standard chemoimmunotherapy is less effective in patients with these mutations, targeted agents such as BTK inhibitors (e.g., ibrutinib) and BCL-2 inhibitors (e.g., venetoclax) have shown better clinical outcomes in SF3B1-mutated CLL [197,198].
Another crucial application of NGS in CLL is the detection of subclonal evolution and treatment resistance [199]. CLL is characterized by significant genetic heterogeneity, with multiple subclonal populations evolving under therapeutic pressure. NGS allows for the tracking of these subclones, revealing dynamic changes in the mutational landscape that contribute to disease relapse [199]. NGS has also significantly influenced therapy selection in CLL, particularly guiding the use of BTK and BCL-2 inhibitors. BTK inhibitors, such as ibrutinib and acalabrutinib, target the B-cell receptor signaling pathway and have shown efficacy in high-risk patients, including those with TP53 aberrations. However, resistance often develops through acquired mutations in BTK (e.g., C481S) and PLCG2, which render the inhibitors less effective [200]. NGS enables early detection of these resistance-associated mutations, allowing for timely treatment adjustments. Similarly, BCL2 inhibitors like venetoclax are highly effective in TP53-mutated CLL, but resistance can emerge due to mutations in BCL2 itself, leading to therapeutic failure [201]. NGS-based monitoring of these mutations has facilitated sequential therapy strategies, optimizing treatment outcomes [202]. Additionally, TP53 and NOTCH1 mutations have been implicated in disease progression despite targeted therapy, highlighting the need for continuous molecular monitoring [203]. The ability of NGS to detect emerging resistant clones before clinical relapse allows for preemptive treatment modifications, such as switching therapy or combining targeted agents to delay disease progression [204].
7. NGS in Chronic Myeloid Leukemia (CML)
NGS has transformed the molecular characterization of CML by improving the detection of BCR::ABL1 fusion variants and resistance mutations [205,206]. The BCR::ABL1 fusion gene, resulting from the t(9;22) chromosomal translocation (Figure 3), is the hallmark of CML and drives disease pathogenesis through constitutive tyrosine kinase activity. While traditional molecular techniques such as qPCR are routinely used for BCR::ABL1 transcript monitoring, they may fail to detect atypical or complex fusion variants [207]. NGS has enabled the identification of previously unrecognized BCR::ABL1 isoforms, facilitating more precise molecular classification and guiding targeted therapy decisions [208]. Atypical BCR::ABL1 mutations in CML represent rare but clinically significant genetic alterations that can impact disease progression and treatment response. While the classic BCR::ABL1 fusion results from the t(9;22) translocation, atypical variants involve uncommon breakpoints, alternative splicing, or additional genetic rearrangements that can lead to unusual fusion transcripts. These atypical mutations often result in differential expression levels or altered tyrosine kinase activity, which may influence sensitivity to TKIs [209]. One example is the e6a2 fusion transcript, which is associated with an aggressive disease course and increased risk of blast crisis, potentially due to the fusion protein’s altered structural and functional properties [210]. Another rare transcript, e18a2, has been reported to cause diagnostic ambiguity due to its atypical splicing pattern, which complicates molecular detection and monitoring [211,212]. Fusion variants arise due to variations in the BCR::ABL1 translocation, resulting in different fusion transcripts that can influence disease progression and response to TKIs. The most common fusion variants include e13a2 (b2a2) and e14a2 (b3a2), both encoding the p210 BCR::ABL1 protein, which is characteristic of classical CML [213]. However, rarer fusion variants such as e13a3 (b2a3) and e14a3 (b3a3) have been identified, which may lead to difficulties in molecular detection using standard PCR assays and require alternative diagnostic techniques like FISH or sequencing [214,215]. Some fusion variants, such as e19a2, are associated with higher platelet counts and unique clinical presentations that differ from typical CML cases [216,217]. Moreover, rare fusions like e6a2 have been linked to an aggressive disease course and poor prognosis, often requiring more intensive treatment strategies [210].
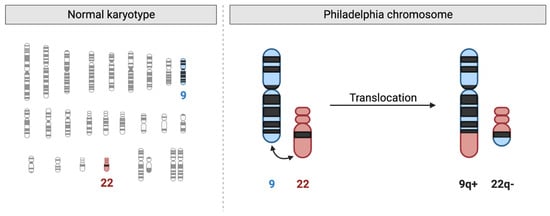
Figure 3.
The Philadelphia Chromosome in Chronic Myeloid Leukemia (CML). On the left, a normal human karyotype is shown, highlighting chromosomes 9 (blue) and 22 (red). On the right, the reciprocal translocation is depicted, where part of chromosome 9q (ABL1 gene) translocates to chromosome 22q, creating the shortened chromosome 22 (22q−, Philadelphia chromosome) and a reciprocal fusion on chromosome 9q (9q+). This translocation results in the BCR::ABL1 fusion gene, which produces an oncogenic tyrosine kinase responsible for the pathogenesis of CML.
Additionally, NGS provides high-resolution detection of BCR::ABL1 kinase domain mutations (Table 8), which are a major cause of resistance to TKIs. While first-line therapy with imatinib is effective in many patients, approximately 30% develop resistance, often due to mutations in the BCR::ABL1 kinase domain [218]. Specific mutations, such as T315I, are known to confer resistance to imatinib and second-generation TKIs like dasatinib and nilotinib. The early detection of such mutations through NGS allows for the strategic selection of alternative inhibitors, such as ponatinib, which retains activity against T315I [219]. Other common mutations include Y253H, E255K/V, and F359V, which are associated with varying degrees of resistance to imatinib, dasatinib, and nilotinib [220]. The localization of these mutations within the kinase domain is critical, as mutations in the P-loop region, such as G250E and E255K, tend to confer high resistance and are associated with worse clinical outcomes [221]. Mutations can emerge under the selective pressure of TKI therapy, leading to clonal evolution and disease progression [222]. Recent advances in targeted therapy, including the development of allosteric inhibitors such as asciminib, offer new therapeutic options for patients with resistant mutations [223]. By identifying low-frequency resistant mutations before clinical relapse, NGS allows for earlier therapeutic intervention and personalized treatment modifications [224].

Table 8.
Mutations and Variants Associated with CML Identified by NGS.
Moreover, NGS has identified compound mutations, where multiple resistance-associated mutations co-exist within the same leukemic clone, leading to complex drug resistance patterns [92]. Studies have shown that compound mutations can emerge through sequential, branching, or parallel routes under selective pressure from TKI therapy, resulting in complex clonal architectures [225]. While third-generation TKIs like ponatinib are effective against most single BCR::ABL1 mutations, certain compound mutations, such as T315I/E255K and T315I/F359V, remain highly resistant even to ponatinib, limiting treatment options [208]. The detection of compound mutations is essential for optimal therapy selection, yet conventional sequencing methods often fail to distinguish between polyclonal mutations (present in separate clones) and true compound mutations (within the same clone) [92]. Advanced techniques such as ddPCR and NGS have improved the ability to accurately identify compound mutations and guide therapeutic decisions [226]. The high frequency of compound mutations in TKI-resistant CML underscores the need for routine molecular monitoring and the development of novel therapeutic approaches, including allosteric inhibitors and combination treatment strategies [227]. Overall, NGS’s ability to sequence entire mutational landscapes has refined therapeutic strategies by guiding treatment escalation, de-escalation, or combination approaches tailored to each patient’s mutational profile [228].
Beyond initial diagnosis and therapy selection, NGS has been instrumental in monitoring disease progression and treatment response in CML. The persistence or re-emergence of BCR::ABL1 transcripts at low levels following treatment is a strong predictor of relapse, and conventional qPCR lacks the sensitivity to detect minor subclones harboring resistance mutations [229]. NGS-based MRD monitoring provides ultra-sensitive detection of leukemic clones, enabling earlier intervention before clinical progression occurs. Studies have shown that NGS can detect emerging TKI-resistant clones months before they become dominant, allowing for proactive treatment adjustments [205]. Another study demonstrated that patients who achieved deep molecular remission (DMR) but still harbored detectable BCR::ABL1 DNA via NGS were more likely to relapse after TKI cessation [230]. It highlights the clinical importance of NGS in MRD monitoring for CML, particularly in patients attempting treatment-free remission (TFR) after TKI therapy. Traditionally, molecular response in CML is assessed using qPCR to measure BCR::ABL1 transcript levels (Table 9). Patients who achieve a DMR, defined as BCR::ABL1 levels below 0.01% (MR4) or undetectable by qPCR (MR4.5 or MR5), are considered potential candidates for TKI discontinuation [231,232]. However, qPCR has a detection limit, and some patients who appear to be in DMR may still harbor leukemic clones at a molecular level that qPCR cannot detect. NGS provides an alternative approach by detecting BCR::ABL1 DNA at very low levels, sometimes beyond the limits of PCR [233,234].

Table 9.
Molecular Goals of CML Treatment Based on qPCR.
Additionally, NGS has been used to uncover secondary genomic alterations beyond BCR::ABL1, such as mutations in SETBP1, TP53, and ASXL1 (Table 8), which contribute to disease acceleration and blast crisis transformation [235]. For instance, SETBP1 mutations in CML, particularly in its atypical form (aCML), have been identified as recurrent genetic alterations associated with poor prognosis and aggressive disease progression. These mutations occur in approximately 24% of aCML cases and are frequently located within codons 858–871, leading to a gain-of-function effect that disrupts ubiquitination, thereby increasing SETBP1 protein levels and promoting leukemic cell proliferation [236,237]. Functionally, SETBP1 mutations result in the inhibition of the tumor suppressor PP2A, leading to enhanced leukemic cell survival and resistance to apoptosis [238]. Clinically, patients with SETBP1 mutations tend to present with higher white blood cell counts and an increased risk of transformation to AML, making this mutation an important prognostic marker [239]. In addition, SETBP1 mutations often co-occur with other high-risk genetic abnormalities, such as -7/del(7q), further contributing to adverse clinical outcomes [240]. Given their significant impact on disease biology, SETBP1 mutations are now considered a key molecular marker in aCML, and their detection may aid in refining risk stratification and guiding more aggressive treatment approaches, including early consideration of allogeneic stem cell transplantation. Meanwhile, TP53 mutations in CML are relatively rare in the chronic phase but become more prevalent in advanced stages, particularly during blast crisis [241]. These mutations are strongly associated with disease progression, therapy resistance, and poor prognosis. Studies have shown that TP53 alterations, including mutations and deletions on chromosome 17p, occur in up to 30% of blast crisis cases, often leading to treatment failure with TKIs [183,242]. Unlike BCR::ABL1 kinase domain mutations, which are the primary mechanism of TKI resistance, TP53 mutations drive genomic instability and promote leukemic transformation. Some cases also exhibit a correlation between TP53 deletions and complex chromosomal rearrangements, further worsening prognosis [243]. Given their role in disease progression, TP53 mutations are considered a marker of clonal evolution and may indicate the need for alternative treatment strategies, such as allogeneic stem cell transplantation or novel targeted therapies. Conversely, TP53 mutations in CLL are more common and occur in approximately 10–20% of cases, with their frequency increasing in relapsed or refractory disease [244]. Unlike in CML, where TP53 mutations are linked to disease progression to blast crisis, in CLL, they are associated with poor response to chemoimmunotherapy, particularly to purine analogs such as fludarabine. While TP53 mutations in both CML and CLL confer poor prognosis, their implications differ; in CML, they mark transformation to a more advanced disease stage, whereas in CLL, they drive resistance to standard therapies and necessitate targeted treatment approaches [245]. Next, AXL mutations in CML are relatively rare, but overexpression of AXL, a receptor tyrosine kinase, has been strongly associated with disease progression and resistance to TKIs [246]. AXL is part of the TAM (Tyro3, AXL, Mer) family of kinases, which plays a role in cell survival, proliferation, and immune evasion. Studies have shown that AXL is upregulated in CML, particularly in cases resistant to imatinib, suggesting a role in acquired resistance mechanisms [246,247]. The overexpression of AXL has been linked to activation of downstream pathways such as PI3K/AKT and STAT5, which contribute to leukemic cell survival and drug resistance. Additionally, AXL appears to regulate the persistence of leukemia stem and progenitor cells, independent of BCR::ABL1 activity, making it a potential therapeutic target even in TKI-resistant cases [248]. Pharmacological inhibition of AXL using BGB324 has demonstrated efficacy in overcoming both TKI-sensitive and TKI-resistant CML cells, including those harboring the T315I mutation, which is resistant to most TKIs except ponatinib [248]. Furthermore, targeting AXL has been shown to decrease leukemia cell proliferation and improve responses to combination therapies, particularly in cases with persistent minimal residual disease [249].
The RAS/MAPK signaling pathway plays a significant role in the pathogenesis of CML, particularly in promoting leukemic cell survival, proliferation, and resistance to apoptosis. This pathway is activated downstream of the BCR::ABL1 fusion protein, which is the hallmark of CML. The BCR::ABL1 oncoprotein constitutively activates multiple signaling pathways, including RAS/MAPK, PI3K/AKT, and JAK/STAT, which collectively drive the uncontrolled growth of leukemic cells [250,251]. The RAS/MAPK cascade is initiated when BCR::ABL1 activates RAS, which subsequently triggers RAF kinases, leading to the phosphorylation of MEK1/2 and ERK1/2. This signaling cascade enhances cell proliferation and inhibits apoptosis, thereby contributing to CML progression. Studies have shown that the activation of this pathway is linked to the resistance of leukemic stem cells to TKIs, such as imatinib, dasatinib, and nilotinib [252]. Additionally, blocking the RAS/MAPK pathway with specific inhibitors such as MEK inhibitors has demonstrated potential in reducing CML cell proliferation and enhancing sensitivity to TKIs [253]. While RAS mutations are rare in chronic-phase CML, they have been reported more frequently in the blast crisis phase, where they contribute to disease progression and drug resistance [254]. The increased activation of the RAS/MAPK pathway in advanced CML suggests that this signaling axis plays a crucial role in leukemic transformation. NRAS and KRAS mutations are relatively rare in the chronic phase but become more prevalent in advanced disease stages, particularly during blast crisis. These mutations play a role in leukemic transformation by driving uncontrolled cell proliferation and survival through constitutive activation of the RAS/MAPK signaling pathway [255,256]. Studies have shown that NRAS mutations are more frequent than KRAS mutations in myeloid malignancies, with HRAS mutations being exceedingly rare. Mutations at codons 12, 13, or 61 of NRAS and KRAS result in a constitutively active GTP-bound state, leading to persistent downstream signaling through RAF-MEK-ERK and PI3K-AKT pathways [257]. This contributes to resistance against apoptosis and enhances leukemic stem cell self-renewal, which may explain why these mutations are more frequently found in patients with blast crisis rather than in the chronic phase. While these mutations are not commonly screened for in early-stage CML, their detection in advanced disease may provide important prognostic insights and influence therapeutic strategies. Overall, these findings support the emerging role of NGS in capturing the full spectrum of genetic evolution in CML, offering a more comprehensive approach to long-term disease management [258].
8. When to Perform NGS in Leukemia Disease Course?
The frequency of NGS testing in leukemia management is still under investigation, with recommendations varying based on disease type, risk stratification, and treatment phase [208]. Current guidelines suggest that NGS should be performed at diagnosis to identify clinically relevant mutations that guide prognosis and treatment decisions, particularly in acute leukemias such as AML and ALL [88]. At diagnosis, targeted NGS panels help classify patients into appropriate risk groups based on mutations in genes such as FLT3, NPM1, TP53, and IDH1/IDH2, enabling personalized therapy selection. Meanwhile, at CML diagnosis, NGS is recommended to detect both BCR::ABL1 fusion transcripts and additional mutations that may impact prognosis and treatment selection. While qPCR is the gold standard for detecting BCR::ABL1, NGS allows for a more comprehensive genetic profile, identifying co-occurring mutations in genes like ASXL1, RUNX1, and TP53, which have been associated with worse outcomes and potential resistance to TKIs [92]. Following diagnosis, the timing of NGS testing varies based on disease course and treatment milestones. For AML, repeat NGS is often recommended at relapse to identify emerging resistance mutations or clonal evolution, which may necessitate changes in therapy, such as switching to targeted inhibitors or considering allogeneic stem cell transplantation [259]. Additionally, in chronic leukemias such as CLL, NGS is useful at treatment initiation and disease progression, particularly for identifying TP53 mutations, which predict poor response to chemoimmunotherapy and favor the use of targeted agents like BTK inhibitors [182,183]. In CML, NGS is also recommended when there is an increase in BCR::ABL1 transcript levels during treatment. Rising transcript levels can indicate emerging resistance, and NGS helps identify resistance mutations within the BCR::ABL1 kinase domain before they become clinically significant [92,208]. Identifying mutations like T315I or compound mutations allows clinicians to switch to appropriate second- or third-generation TKIs before complete treatment failure.
In MRD monitoring, NGS is being explored for ultra-sensitive detection of low-level leukemia cells, particularly post-treatment in AML and ALL. Studies suggest that patients with persistent leukemia-associated mutations detected by NGS, even in the absence of morphological relapse, are at higher risk of recurrence [94]. While qPCR remains the standard for MRD assessment, NGS may be used at key post-treatment time points, such as after induction chemotherapy, before consolidation, and before TFR attempts in CML. Before TKI discontinuation in CML, NGS can play a role in assessing MRD at an ultra-sensitive level. While standard qPCR assesses BCR::ABL1 transcript levels, ultra-deep NGS can detect residual leukemic clones that may predict relapse. Studies have shown that patients who achieve DMR but still harbor detectable BCR::ABL1 DNA by NGS have a higher risk of relapse after stopping TKIs [229]. NGS is also valuable for monitoring patients who have discontinued TKIs due to sustained deep remission. While qPCR is typically used for routine MRD monitoring, periodic NGS testing can help detect very low-level residual clones that could signal impending relapse [259]. Finally, if CML progresses to the accelerated or blast phase, NGS becomes essential for detecting secondary mutations beyond BCR::ABL1. Disease progression is often associated with the accumulation of additional genetic alterations, including mutations in TP53, RUNX1, and SETBP1, which contribute to TKI resistance and poor prognosis [260]. At this stage, NGS guides treatment strategies, including switching TKIs or considering allogeneic stem cell transplantation. Despite these insights, there is no universal agreement on a fixed frequency for NGS testing, as its role depends on leukemia type, disease stage, and available targeted therapies.
9. Challenges and Future Perspectives
9.1. Standardization and Interpretation Challenges
One of the primary challenges in the clinical implementation of NGS in leukemia diagnostics is the lack of standardization in sequencing protocols, data processing, and variant interpretation. Unlike conventional diagnostic methods such as karyotyping and FISH, which have well-established guidelines, NGS technologies vary significantly between laboratories in terms of sequencing platforms, target gene panels, and bioinformatics pipelines. The absence of universal standards complicates the reproducibility of results, making it difficult to compare findings across different institutions [261]. Efforts to establish standardized quality control measures, such as minimum sequencing depth and uniform variant reporting criteria, are crucial for ensuring consistency in NGS-based leukemia diagnostics [91]. The interpretation of NGS data presents another significant hurdle, as the vast number of genetic variants detected often includes mutations of uncertain significance (VUS) [262]. In leukemia, distinguishing between driver mutations, which contribute to disease pathogenesis, and passenger mutations, which have no functional impact, is essential for clinical decision-making. However, many variants identified by NGS have not been well-characterized in existing databases, leading to ambiguity in their clinical relevance. The use of curated genomic databases such as ClinVar and COSMIC, along with functional validation studies, is necessary to refine variant classification [120,263]. Additionally, the integration of artificial intelligence and machine learning models is being explored as a means to automate and enhance variant interpretation, reducing the burden on clinicians and bioinformaticians [264,265].
Regulatory oversight and clinical validation further complicate the widespread adoption of NGS in leukemia diagnostics. The U.S. Food and Drug Administration (FDA) and other regulatory agencies have recognized the need for establishing guidelines to ensure the clinical reliability of NGS-based testing. The FDA’s regulatory approach aims to balance scientific innovation with the need for standardized oversight to protect patient safety. In 2018, the FDA finalized its guidance on NGS-based in vitro diagnostic tests, providing a framework for analytical validation and performance criteria, particularly for identifying genetic variants relevant to leukemia and other diseases [266]. This guidance helps laboratories and test developers establish regulatory-compliant NGS assays for clinical use. Additionally, the FDA has outlined criteria for evaluating the clinical validity of NGS-based tests, incorporating data from recognized genetic variant databases to support the interpretation of genomic findings. One of the key milestones in the FDA’s regulatory efforts was the approval of comprehensive NGS panels for detecting actionable genomic aberrations in cancer, including leukemia. These approvals established a regulatory pathway for NGS-based companion diagnostics, ensuring that these tests meet stringent accuracy and reproducibility standards before being integrated into clinical practice [267]. However, there have been ongoing debates regarding the extent of FDA oversight, particularly concerning laboratory-developed tests (LDTs), which make up a large portion of NGS-based leukemia diagnostics. The FDA’s regulatory approach has been scrutinized for potentially increasing the cost and time required for test development while also ensuring that assays used in clinical decision-making are reliable and reproducible [268]. The FDA has also collaborated with the Centers for Medicare and Medicaid Services (CMS) to align regulatory standards with reimbursement policies, as seen in the decision to grant Medicare coverage for FDA-approved NGS tests used in cancer treatment [269]. This alignment has facilitated broader access to NGS testing for leukemia patients, allowing for more precise molecular profiling and personalized treatment approaches.
The regulation of NGS in leukemia management in Europe is guided by a combination of European Union (EU) directives, national regulations, and recommendations from organizations such as the ELN. The implementation of NGS in clinical practice varies widely across EU countries, with disparities in regulatory policies and reimbursement structures affecting patient access. The European In Vitro Diagnostic Medical Devices Regulation (IVDR), which came into effect in 2022, established stricter requirements for NGS-based diagnostic tests, ensuring they meet high standards of analytical validity and clinical performance. However, despite these regulatory advances, the adoption of NGS in routine leukemia management remains inconsistent due to differences in national guideline implementations and reimbursement policies [270]. The ELN has played a crucial role in integrating NGS into leukemia diagnosis and treatment guidelines. ELN recommendations emphasize the importance of NGS in identifying key mutations for risk stratification and guiding personalized therapies, particularly in AML and CLL [271,272]. However, regulatory frameworks are still evolving, and there is a significant challenge in the widespread implementation of NGS due to the lack of standardized protocols across different countries, leading to variability in how NGS data are interpreted and used in clinical decision-making. Moreover, there is no universally accepted approach for validating NGS assays. Unlike single-gene tests, which undergo rigorous analytical validation, NGS panels cover multiple genes simultaneously, increasing the complexity of quality assurance. Establishing proficiency testing programs and cross-laboratory validation studies will be essential in addressing these concerns and ensuring that NGS results are both accurate and actionable [273]. Future advancements in NGS standardization and interpretation will likely involve greater collaboration between research institutions, regulatory bodies, and clinical laboratories. The development of international guidelines for sequencing quality control, variant classification, and data reporting will facilitate the seamless integration of NGS into leukemia management. Another barrier to NGS adoption in Europe is the uneven reimbursement landscape, which affects patient access to these advanced genomic tests. While some countries, such as Germany and France, have integrated NGS into their national healthcare systems with established reimbursement mechanisms, other EU nations still struggle with financial and logistical barriers that limit NGS availability for leukemia patients [274]. Policymakers and healthcare stakeholders have called for a more harmonized approach to NGS regulation and reimbursement, advocating for multi-stakeholder collaborations to streamline NGS implementation across Europe.
9.2. Other Barriers to Widespread Clinical Application
Beyond regulatory and interpretation challenges, several other barriers hinder the widespread application of NGS in clinical leukemia management. One major limitation is the high cost of NGS testing, which remains a significant financial burden for both healthcare institutions and patients. While the cost of sequencing has decreased over the years, comprehensive leukemia panels, particularly WGS or WES, remain expensive, making them inaccessible in many healthcare settings. A survey of U.S. oncologists and hematologists revealed that limited reimbursement from insurance providers is a primary concern, leading to financial constraints that prevent the routine use of NGS in leukemia diagnostics [275]. Without consistent coverage policies, hospitals and laboratories face difficulties in integrating NGS into standard leukemia care. Another major challenge is the turnaround time for NGS results, which can take days to weeks, depending on the sequencing approach. In acute leukemias, where rapid treatment decisions are crucial, delays in obtaining NGS data may render the information less useful for immediate clinical decision-making [276]. While advances in automation and bioinformatics are improving sequencing efficiency, many institutions still struggle with optimizing workflows to reduce turnaround time [277]. Additionally, the requirement for highly skilled personnel to perform sequencing and analyze complex genomic data remains a barrier, particularly in smaller hospitals and laboratories without dedicated molecular oncology teams. The lack of standardized guidelines for incorporating NGS into leukemia management also contributes to its limited adoption. While organizations like the ELN and the National Comprehensive Cancer Network (NCCN) have provided recommendations on using NGS for risk stratification, there is no universal consensus on when and how often NGS should be performed in leukemia patients [208,271,278]. This uncertainty has led to variations in clinical practice, where some physicians rely on traditional molecular techniques like PCR and FISH rather than adopting NGS-based approaches [89]. Additionally, evidence gaps in clinical utility remain a significant challenge. While NGS has expanded our understanding of leukemia genomics, not all detected mutations have immediate therapeutic implications [279]. Many physicians hesitate to order NGS tests because of concerns about the actionable value of certain mutations, particularly in cases where targeted therapies are not yet available. A survey found that 80.1% of physicians identified the lack of strong evidence linking NGS findings to treatment decisions as a major barrier to routine use [280]. Expanding clinical trials and real-world studies to validate the prognostic and therapeutic significance of emerging leukemia-associated mutations could help bridge this gap.
9.3. Integration of NGS with Other Omics Technologies
The integration of NGS with other omics technologies, such as transcriptomics, proteomics, and metabolomics, is transforming leukemia research by providing a comprehensive understanding of disease biology [102,281]. While NGS primarily focuses on genomic alterations, integrating multiple omics layers allows researchers to analyze gene expression patterns, protein interactions, and metabolic changes that drive leukemia progression. Transcriptomic profiling through RNA-Seq, for instance, helps identify aberrant gene expression signatures and oncogenic fusion transcripts that may not be detected by DNA sequencing alone [282]. This approach has been instrumental in refining leukemia subtypes and uncovering novel therapeutic targets [283]. Additionally, single-cell transcriptomics has provided deeper insights into clonal heterogeneity, enabling the identification of rare leukemia stem cell populations that contribute to relapse and treatment resistance [284]. Proteomics and metabolomics further enhance leukemia characterization by elucidating functional alterations at the protein and metabolic levels. Proteomic studies have identified post-translational modifications that regulate leukemia cell signaling pathways, offering potential drug targets beyond genetic mutations [285]. Mass spectrometry-based proteomics has revealed dysregulated kinase activity in AML, leading to the development of kinase inhibitors tailored to specific protein alterations [286]. Similarly, metabolomics has provided insights into metabolic reprogramming in leukemia, demonstrating that leukemic cells rely on altered energy metabolism for survival and proliferation [287]. The integration of metabolomics with NGS data has been particularly useful in understanding how metabolic vulnerabilities can be exploited for therapeutic intervention, such as targeting amino acid metabolism in leukemia cells [288].
Multi-omics integration also plays a crucial role in overcoming the limitations of NGS in predicting drug responses and treatment resistance. By combining genomics with epigenomics, researchers have identified DNA methylation patterns and histone modifications that influence leukemia progression and therapy resistance [289]. For example, epigenetic alterations in DNMT3A and TET2 have been shown to impact response to hypomethylating agents in AML, highlighting the importance of epigenomic profiling in therapeutic decision-making [290,291]. Additionally, multi-omics approaches have been employed to develop predictive models for leukemia treatment outcomes, incorporating genomic, transcriptomic, and proteomic data to refine risk stratification and personalize therapy [292,293]. Despite its potential, integrating multi-omics data presents significant challenges, including the complexity of data interpretation, the need for standardized analytical frameworks, and the high computational demands of processing large-scale datasets [294]. Advanced bioinformatics tools and machine learning algorithms are being developed to address these challenges, enabling more effective integration of diverse omics datasets [295,296]. The future of leukemia research lies in leveraging multi-omics approaches to achieve a system-level understanding of the disease, paving the way for more precise diagnostics and innovative therapeutic strategies.
9.4. Potential for Real-Time NGS in Clinical Decision-Making
The potential for real-time NGS in clinical decision-making is a rapidly evolving area that could revolutionize leukemia management by providing near-instant genomic insights for treatment selection. Traditional sequencing methods often require days to weeks to process results, delaying critical treatment decisions. However, real-time NGS approaches, particularly with nanopore sequencing technologies, have demonstrated the ability to classify leukemias in minutes, making it feasible to incorporate genomic data into immediate therapeutic strategies [297]. In a recent study, real-time transcriptomic profiling using nanopore sequencing successfully distinguished molecular subtypes of ALL within minutes, highlighting the potential for rapid leukemia diagnosis and classification [24]. One of the most promising applications of real-time NGS is in guiding treatment decisions for patients with relapsed or refractory leukemia. The ability to quickly detect mutations associated with drug resistance, such as FLT3 or TP53 mutations in AML, allows clinicians to adjust therapy before treatment failure occurs. Studies have shown that comprehensive NGS panels can provide actionable genomic data that influence treatment selection, such as identifying patients who may benefit from targeted inhibitors like FLT3 or IDH1/2 inhibitors [298]. Additionally, the integration of real-time NGS into clinical workflows has been proposed as a strategy to refine treatment sequencing patterns in CLL, enabling a more dynamic and personalized approach to therapy selection [299].
Beyond initial diagnosis and therapy selection, real-time NGS is expected to play a key role in monitoring MRD and predicting relapse. Current MRD detection methods, such as flow cytometry and PCR, have limitations in sensitivity and may not capture emerging resistant clones. Real-time NGS offers ultra-sensitive monitoring of leukemia cells, allowing clinicians to detect molecular relapse earlier than conventional methods. A comparative study demonstrated that NGS-based MRD monitoring provided superior sensitivity compared to flow cytometry, underscoring its potential to guide early treatment modifications [89,300]. As sequencing technologies become more efficient and cost-effective, the implementation of real-time NGS for routine leukemia monitoring is expected to enhance patient outcomes by enabling timely therapeutic interventions [102]. Despite its advantages, real-time NGS faces challenges related to data interpretation, cost, and infrastructure requirements. The volume of sequencing data generated in real time necessitates advanced bioinformatics pipelines capable of rapid analysis and clinical annotation [301]. Moreover, integrating real-time sequencing into hospital settings requires significant investment in sequencing platforms and trained personnel. However, ongoing developments in machine learning-driven variant interpretation and cloud-based genomic analysis platforms are expected to mitigate these barriers, making real-time NGS increasingly accessible for clinical use [302]. As these technologies continue to mature, real-time NGS is poised to become a cornerstone of precision medicine, transforming leukemia diagnostics and treatment decision-making.
9.5. Drug Development and Precision Medicine
NGS has become an essential tool in drug development and precision medicine by identifying genetic alterations that can be targeted with novel therapeutics. Traditional drug development relies on broad-spectrum cytotoxic agents, but NGS has shifted this paradigm toward the design of molecularly targeted therapies tailored to specific genetic mutations. In leukemia, this has led to the approval of inhibitors such as FLT3 inhibitors for FLT3-mutant AML and BCL2 inhibitors for CLL. By analyzing the genomic landscape of leukemias, NGS enables the identification of novel drug targets, allowing pharmaceutical companies to develop precision medicines aimed at specific molecular pathways [303]. The application of NGS in genetically stratified clinical trials has revolutionized oncology drug testing, making trials more efficient and outcome-driven. Instead of treating all patients with a one-size-fits-all approach, NGS allows for the enrollment of participants based on their specific mutational profiles, ensuring that only those who are most likely to benefit from a particular therapy receive it. This approach increases the success rates of clinical trials and reduces the exposure of patients to ineffective treatments. For example, the use of NGS-guided patient selection in clinical trials for IDH1 and IDH2 inhibitors in AML has demonstrated improved response rates compared to conventional trial designs [304]. Additionally, real-time genomic monitoring through NGS enables adaptive clinical trial designs, where treatment regimens can be modified in response to emerging resistance mutations [305].
Beyond individual targeted therapies, NGS plays a crucial role in combination drug development, where multiple agents are tested together to overcome resistance mechanisms. Multi-omics data from NGS, transcriptomics, and proteomics have identified synergistic drug combinations that enhance treatment efficacy. For instance, studies integrating NGS and RNA sequencing have revealed that combining BCL2 inhibitors with hypomethylating agents in AML results in greater leukemia cell death than either agent alone. These findings have led to the approval of venetoclax in combination with azacitidine, a treatment now widely used for elderly AML patients who are ineligible for intensive chemotherapy [306]. Despite these advances, significant challenges remain in translating NGS findings into clinically approved therapies. One major barrier is the complexity of interpreting the vast amount of genomic data generated, as not all detected mutations are actionable or have a well-defined therapeutic intervention. Additionally, regulatory agencies such as the U.S. FDA require rigorous validation of NGS-based biomarkers before they can be integrated into drug approval processes. Overcoming these challenges will require further collaboration between researchers, clinicians, and regulatory bodies to standardize genomic data interpretation and integrate NGS findings into routine clinical practice [307]. As precision medicine continues to evolve, the role of NGS in drug discovery and development will expand, leading to more personalized and effective leukemia treatments. The integration of artificial intelligence and machine learning into NGS analysis (Figure 4) is expected to further streamline drug target identification, improving the efficiency of therapeutic development. With ongoing advancements in sequencing technologies, the future of leukemia treatment will increasingly rely on genomics-driven approaches that ensure the right drug is given to the right patient at the right time.
Figure 4.
Potential Application of Artificial Intelligence, Including Machine Learning to Process NGS Data.
10. Conclusions
Next-generation sequencing has revolutionized the diagnosis, prognosis, and treatment of leukemia (Figure 5) by enabling comprehensive genomic profiling with unprecedented accuracy. Compared to conventional diagnostic methods, NGS provides deeper insights into genetic alterations, clonal evolution, and resistance mechanisms, facilitating personalized treatment strategies. While challenges such as data interpretation, standardization, and cost remain, continuous advancements in sequencing technologies and bioinformatics are improving their clinical applicability. As precision medicine evolves, NGS is expected to become an integral tool in leukemia management, driving innovations in targeted therapy and real-time disease monitoring. Future research should focus on optimizing its integration into routine clinical practice, ensuring broader accessibility, and refining its predictive capabilities for improved patient outcomes.
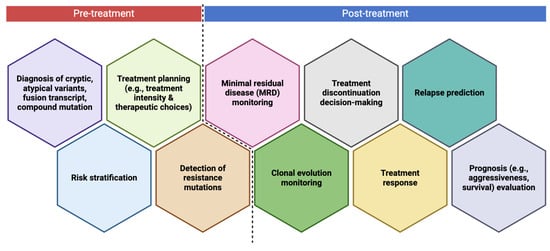
Figure 5.
The Role of Next-Generation Sequencing in Leukemia Management. In the pre-treatment phase, NGS assists in diagnosing cryptic and atypical variants, risk stratification, treatment planning, and detection of resistance mutations. In the post-treatment phase, NGS is utilized for minimal residual disease (MRD) monitoring, clonal evolution monitoring, treatment response assessment, relapse prediction, treatment discontinuation decisions, and prognosis evaluation. These insights contribute to personalized leukemia management and improved patient outcomes.
Author Contributions
Conceptualization, H.S.; methodology, H.S.; validation, H.S.; formal analysis, H.S. and L.P.; investigation, H.S. and L.P.; resources, H.S.; data curation, H.S. and L.P.; writing—original draft preparation, H.S., L.P., F.H.M., M.C.N. and P.Z.R.; writing—review and editing, H.S., L.P., F.H.M., M.C.N. and P.Z.R.; visualization, H.S.; project administration, H.S.; funding acquisition, H.S.; supervision, P.Z.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Chennamadhavuni, A.; Lyengar, V.; Mukkamalla, S.K.R.; Shimanovsky, A. Leukemia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Vakiti, A.; Reynolds, S.B.; Mewawalla, P. Acute Myeloid Leukemia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Debnath, A.; Nath, S. Prognosis and Treatment in Acute Myeloid Leukemia: A Comprehensive Review. Egypt. J. Med. Hum. Genet. 2024, 25, 91. [Google Scholar] [CrossRef]
- Puckett, Y.; Chan, O. Acute Lymphocytic Leukemia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Mukkamalla, S.K.R.; Taneja, A.; Malipeddi, D.; Master, S.R. Chronic Lymphocytic Leukemia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Sampaio, M.M.; Santos, M.L.C.; Marques, H.S.; Gonçalves, V.L.d.S.; Araújo, G.R.L.; Lopes, L.W.; Apolonio, J.S.; Silva, C.S.; Santos, L.K.d.S.; Cuzzuol, B.R.; et al. Chronic Myeloid Leukemia-from the Philadelphia Chromosome to Specific Target Drugs: A Literature Review. World J. Clin. Oncol. 2021, 12, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Guilhot, F.; Hehlmann, R. Long-Term Outcomes of Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia. Blood 2025, 145, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Tazi, Y.; Arango-Ossa, J.E.; Zhou, Y.; Bernard, E.; Thomas, I.; Gilkes, A.; Freeman, S.; Pradat, Y.; Johnson, S.J.; Hills, R.; et al. Unified Classification and Risk-Stratification in Acute Myeloid Leukemia. Nat. Commun. 2022, 13, 4622. [Google Scholar] [CrossRef]
- Wemyss, C.; Jones, E.; Stentz, R.; Carding, S.R. Acute Myeloid Leukaemia and Acute Lymphoblastic Leukaemia Classification and Metabolic Characteristics for Informing and Advancing Treatment. Cancers 2024, 16, 4136. [Google Scholar] [CrossRef]
- Aung, M.M.K.; Mills, M.L.; Bittencourt-Silvestre, J.; Keeshan, K. Insights into the Molecular Profiles of Adult and Paediatric Acute Myeloid Leukaemia. Mol. Oncol. 2021, 15, 2253–2272. [Google Scholar] [CrossRef]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular Profiling for Precision Cancer Therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Kiyoi, H.; Kawashima, N.; Ishikawa, Y. FLT3 Mutations in Acute Myeloid Leukemia: Therapeutic Paradigm beyond Inhibitor Development. Cancer Sci. 2020, 111, 312–322. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Cortes, J.E. Mutations in AML: Prognostic and Therapeutic Implications. Hematol. Am. Soc. Hematol. Educ. Program. 2016, 2016, 348–355. [Google Scholar]
- Downing, J.R. Targeted Therapy in Leukemia. Mod. Pathol. 2008, 21, S2–S7. [Google Scholar] [CrossRef]
- Santos, F.P.S.; Kantarjian, H.; Quintás-Cardama, A.; Cortes, J. Evolution of Therapies for Chronic Myelogenous Leukemia. Cancer J. 2011, 17, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Bewersdorf, J.P.; Shallis, R.M.; Derkach, A.; Goldberg, A.D.; Stein, A.; Stein, E.M.; Marcucci, G.; Zeidan, A.M.; Shimony, S.; DeAngelo, D.J.; et al. Efficacy of FLT3 and IDH1/2 Inhibitors in Patients with Acute Myeloid Leukemia Previously Treated with Venetoclax. Leuk. Res. 2022, 122, 106942. [Google Scholar] [CrossRef] [PubMed]
- Megías-Vericat, J.E.; Martínez-Cuadrón, D.; Solana-Altabella, A.; Montesinos, P. Precision Medicine in Acute Myeloid Leukemia: Where Are We Now and What Does the Future Hold? Expert Rev. Hematol. 2020, 13, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Li, W. Measurable Residual Disease Testing in Acute Leukemia: Technology and Clinical Significance. In Leukemia; Li, W., Ed.; Exon Publications: Brisbane, Australia, 2022; ISBN 978-0-645-33207-0. [Google Scholar]
- Della Starza, I.; De Novi, L.A.; Elia, L.; Bellomarino, V.; Beldinanzi, M.; Soscia, R.; Cardinali, D.; Chiaretti, S.; Guarini, A.; Foà, R. Optimizing Molecular Minimal Residual Disease Analysis in Adult Acute Lymphoblastic Leukemia. Cancers 2023, 15, 374. [Google Scholar] [CrossRef]
- Pulsipher, M.A.; Han, X.; Maude, S.L.; Laetsch, T.W.; Qayed, M.; Rives, S.; Boyer, M.W.; Hiramatsu, H.; Yanik, G.A.; Driscoll, T.; et al. Next-Generation Sequencing of Minimal Residual Disease for Predicting Relapse after Tisagenlecleucel in Children and Young Adults with Acute Lymphoblastic Leukemia. Blood Cancer Discov. 2022, 3, 66–81. [Google Scholar] [CrossRef]
- Medeiros, B.C.; Chan, S.M.; Daver, N.G.; Jonas, B.A.; Pollyea, D.A. Optimizing Survival Outcomes with Post-remission Therapy in Acute Myeloid Leukemia. Am. J. Hematol. 2019, 94, 803–811. [Google Scholar] [CrossRef]
- Duncavage, E.J.; Bagg, A.; Hasserjian, R.P.; DiNardo, C.D.; Godley, L.A.; Iacobucci, I.; Jaiswal, S.; Malcovati, L.; Vannucchi, A.M.; Patel, K.P.; et al. Genomic Profiling for Clinical Decision Making in Myeloid Neoplasms and Acute Leukemia. Blood 2022, 140, 2228–2247. [Google Scholar] [CrossRef]
- Sahoo, O.S.; Aidasani, H.; Nayek, A.; Tripathi, S.; Talukdar, J.; Gul, A.; Kumar, D.; Dhar, R.; Karmakar, S. Role of Next-Generation Sequencing in Revolutionizing Healthcare for Cancer Management. MedComm—Future Med. 2024, 3, e70001. [Google Scholar] [CrossRef]
- Sagniez, M.; Simpson, S.M.; Caron, M.; Rozendaal, M.; Paré, B.; Sontag, T.; Langlois, S.; Rouette, A.; Lavallée, V.-P.; Cellot, S.; et al. Real-Time Molecular Classification of Leukemias. medRxiv 2022. [Google Scholar] [CrossRef]
- Rindy, L.J.; Chambers, A.R. Bone Marrow Aspiration and Biopsy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Jain, S.; Sharma, R. Laboratory Evaluation of Bone Marrow. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Percival, M.-E.; Lai, C.; Estey, E.; Hourigan, C.S. Bone Marrow Evaluation for Diagnosis and Monitoring of Acute Myeloid Leukemia. Blood Rev. 2017, 31, 185–192. [Google Scholar] [CrossRef]
- Humphries, J.E. Dry Tap Bone Marrow Aspiration: Clinical Significance. Am. J. Hematol. 1990, 35, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Astle, J.M.; Xu, M.L.; Friedman, T.; Brown, E. Limitations of Poor Bone Marrow Aspirations (for an Accurate Diagnosis) despite the Multimodal Analytical Era: A Longitudinal Retrospective Study. Am. J. Hematol. 2017, 92, E600–E602. [Google Scholar] [CrossRef] [PubMed]
- Donald, S.; Kakkar, N. Dry Tap on Bone Marrow Aspiration: A Red Flag. J. Hematop. 2021, 14, 125–130. [Google Scholar] [CrossRef]
- Azharuddin, M.; Alvi, U.; Rao, S.; Fatima, A.; Uplaonkar, S. Comparative Evaluation of Simultaneous Bone Marrow Aspiration and Bone Marrow Biopsy in Diagnosis of Leukemic Patients—An Institutional Study. Glob. J. Res. Anal. 2023, 12, 54–56. [Google Scholar] [CrossRef]
- Helbig, D.; Quesada, A.E.; Xiao, W.; Roshal, M.; Tallman, M.S.; Knorr, D.A. Spontaneous Remission in a Patient With Acute Myeloid Leukemia Leading to Undetectable Minimal Residual Disease. J. Hematol. 2020, 9, 18–22. [Google Scholar]
- Balciuniene, J.; Ning, Y.; Lazarus, H.M.; Aikawa, V.; Sherpa, S.; Zhang, Y.; Morrissette, J.J.D. Cancer Cytogenetics in a Genomics World: Wedding the Old with the New. Blood Rev. 2024, 66, 101209. [Google Scholar] [CrossRef]
- Jain, H.; Shetty, D. Role of Cytogenetics and Fluorescence In Situ Hybridization in the Laboratory Workup of Acute Myeloid Leukemias. Indian J. Med. Paediatr. Oncol. 2023, 44, 543–553. [Google Scholar] [CrossRef]
- Ozkan, E.; Lacerda, M.P. Genetics, Cytogenetic Testing And Conventional Karyotype. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Kang, Z.-J.; Liu, Y.-F.; Xu, L.-Z.; Long, Z.-J.; Huang, D.; Yang, Y.; Liu, B.; Feng, J.-X.; Pan, Y.-J.; Yan, J.-S.; et al. The Philadelphia Chromosome in Leukemogenesis. Chin. J. Cancer 2016, 35, 48. [Google Scholar] [CrossRef]
- Luquet, I.; Bidet, A.; Cuccuini, W.; Lafage-Pochitaloff, M.; Mozziconacci, M.-J.; Terré, C. Cytogenetics in the Management of Acute Myeloid Leukemia: An Update by the Groupe Francophone de Cytogénétique Hématologique (GFCH). Ann. Biol. Clin. 2016, 74, 535–546. [Google Scholar] [CrossRef]
- Mrózek, K. Acute Myeloid Leukemia with a Complex Karyotype. Semin. Oncol. 2008, 35, 365–377. [Google Scholar] [CrossRef]
- Hou, H.A.; Lin, C.C.; Chou, W.C.; Liu, C.Y.; Chen, C.Y.; Tang, J.L.; Lai, Y.J.; Tseng, M.H.; Huang, C.F.; Chiang, Y.C.; et al. Integration of Cytogenetic and Molecular Alterations in Risk Stratification of 318 Patients with de Novo Non-M3 Acute Myeloid Leukemia. Leukemia 2014, 28, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Boscaro, E.; Urbino, I.; Catania, F.M.; Arrigo, G.; Secreto, C.; Olivi, M.; D’Ardia, S.; Frairia, C.; Giai, V.; Freilone, R.; et al. Modern Risk Stratification of Acute Myeloid Leukemia in 2023: Integrating Established and Emerging Prognostic Factors. Cancers 2023, 15, 3512. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Chang, L.; Zhu, X. Pathogenesis of ETV6/RUNX1-Positive Childhood Acute Lymphoblastic Leukemia and Mechanisms Underlying Its Relapse. Oncotarget 2017, 8, 35445–35459. [Google Scholar] [CrossRef] [PubMed]
- Baranger, L.; Cuccuini, W.; Lefebvre, C.; Luquet, I.; Perot, C.; Radford, I.; Lafage-Pochitaloff, M. Cytogenetics in the Management of Children and Adult Acute Lymphoblastic Leukemia (ALL): An Update by the Groupe Francophone de Cytogénétique Hématologique (GFCH). Ann. Biol. Clin. 2016, 74, 547–560. [Google Scholar] [CrossRef]
- Bridge, J.A. Advantages and Limitations of Cytogenetic, Molecular Cytogenetic, and Molecular Diagnostic Testing in Mesenchymal Neoplasms. J. Orthop. Sci. 2008, 13, 273–282. [Google Scholar] [CrossRef]
- He, R.; Wiktor, A.E.; Hanson, C.A.; Ketterling, R.P.; Kurtin, P.J.; Van Dyke, D.L.; Litzow, M.R.; Howard, M.H.; Reichard, K.K. Conventional Karyotyping and Fluorescence In Situ Hybridization. Am. J. Clin. Pathol. 2015, 143, 873–878. [Google Scholar] [CrossRef]
- Ratan, Z.A.; Zaman, S.B.; Mehta, V.; Haidere, M.F.; Runa, N.J.; Akter, N. Application of Fluorescence In Situ Hybridization (FISH) Technique for the Detection of Genetic Aberration in Medical Science. Cureus 2017, 9, e1325. [Google Scholar] [CrossRef]
- Wolff, D.J.; Bagg, A.; Cooley, L.D.; Dewald, G.W.; Hirsch, B.A.; Jacky, P.B.; Rao, K.W.; Rao, P.N. Guidance for Fluorescence in Situ Hybridization Testing in Hematologic Disorders. J. Mol. Diagn. 2007, 9, 134–143. [Google Scholar] [CrossRef]
- Mark, H.F.L.; Sokolic, R.A.; Mark, Y. Conventional Cytogenetics and FISH in the Detection of BCR/ABL Fusion in Chronic Myeloid Leukemia (CML). Exp. Mol. Pathol. 2006, 81, 1–7. [Google Scholar] [CrossRef]
- UZ, B.; Eliaçık, E.; Işık, A.; Aksu, S.; Büyükaşık, Y.; Haznedaroğlu, İ.C.; Göker, H.; Sayınalp, N.; Özcebe, O.İ. Co-Expression of t(15;17) and t(8;21) in a Case of Acute Promyelocytic Leukemia: Review of the Literature. Turk. J. Haematol. 2013, 30, 400–404. [Google Scholar] [CrossRef]
- Abdool, A.; Donahue, A.C.; Wohlgemuth, J.G.; Yeh, C.-H. Detection, Analysis and Clinical Validation of Chromosomal Aberrations by Multiplex Ligation-Dependent Probe Amplification in Chronic Leukemia. PLoS ONE 2010, 5, e15407. [Google Scholar] [CrossRef] [PubMed]
- Quijano, S.; López, A.; Rasillo, A.; Sayagués, J.M.; Barrena, S.; Sánchez, M.L.; Teodosio, C.; Giraldo, P.; Giralt, M.; Pérez, M.C.; et al. Impact of Trisomy 12, Del(13q), Del(17p), and Del(11q) on the Immunophenotype, DNA Ploidy Status, and Proliferative Rate of Leukemic B-Cells in Chronic Lymphocytic Leukemia. Cytom. Part B Clin. Cytom. 2008, 74B, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.G.; Van Dyke, D.L. Analysis of Common Abnormalities Seen in Chronic Lymphocytic Leukemia Using Fluorescence In Situ Hybridization. In Chronic Lymphocytic Leukemia; Malek, S.N., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 1881, pp. 35–49. ISBN 978-1-4939-8875-4. [Google Scholar]
- Jing, Y.; Lin, S.; Wang, F.-T.; Yu, J.-W.; Jiang, F. Detection of Gene Abnormalities in 43 Cases of Chronic Lymphocytic Leukemia by Fluorescence in Situ Hybridization. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2018, 26, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.C.S.; Egan, D.; Radich, J.P. Molecular Monitoring of Chronic Myeloid Leukemia: Present and Future. Expert Rev. Mol. Diagn. 2016, 16, 1083–1091. [Google Scholar] [CrossRef]
- Ali, J.; Khan, S.A.; Rauf, S.-E.-; Ayyub, M.; Ali, N.; Afridi, N.K. Comparative Analysis of Fluorescence In Situ Hybridization and Real Time Polymerase Chain Reaction in Diagnosis of Chronic Myeloid Leukemia. J. Coll. Physicians Surg. Pak. 2017, 27, 26–29. [Google Scholar]
- Shakoori, A.R. Fluorescence In Situ Hybridization (FISH) and Its Applications. In Chromosome Structure and Aberrations; Bhat, T.A., Wani, A.A., Eds.; Springer: New Delhi, India, 2017; pp. 343–367. ISBN 978-81-322-3671-9. [Google Scholar]
- Qin, D. Molecular Testing for Acute Myeloid Leukemia. Cancer Biol. Med. 2022, 19, 4–13. [Google Scholar] [CrossRef]
- Taylor, J.; Xiao, W.; Abdel-Wahab, O. Diagnosis and Classification of Hematologic Malignancies on the Basis of Genetics. Blood 2017, 130, 410–423. [Google Scholar] [CrossRef]
- Quattrocchi, A.; Cappelli, L.V.; De Simone, G.; De Marinis, E.; Gentile, M.; Gasperi, T.; Pulsoni, A.; Ascenzi, P.; Nervi, C. Biomarkers in Acute Myeloid Leukemia: From State of the Art in Risk Classification to Future Challenges of RNA Editing as Disease Predictor and Therapy Target. Asp. Mol. Med. 2023, 2, 100023. [Google Scholar] [CrossRef]
- Khehra, N.; Padda, I.S.; Swift, C.J. Polymerase Chain Reaction (PCR). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Testa, U.; Pelosi, E. Function of PML-RARA in Acute Promyelocytic Leukemia. Adv. Exp. Med. Biol. 2024, 1459, 321–339. [Google Scholar] [CrossRef]
- Shigeto, S.; Matsuda, K.; Yamaguchi, A.; Sueki, A.; Uehara, M.; Sugano, M.; Uehara, T.; Honda, T. Rapid Diagnosis of Acute Promyelocytic Leukemia with the PML-RARA Fusion Gene Using a Combination of Droplet-Reverse Transcription-Polymerase Chain Reaction and Instant-Quality Fluorescence in Situ Hybridization. Clin. Chim. Acta 2016, 453, 38–41. [Google Scholar] [CrossRef][Green Version]
- Press, R.D.; Kamel-Reid, S.; Ang, D. BCR-ABL1 RT-qPCR for Monitoring the Molecular Response to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia. J. Mol. Diagn. 2013, 15, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Salmon, M.; White, H.E.; Zizkova, H.; Gottschalk, A.; Motlova, E.; Cerveira, N.; Colomer, D.; Coriu, D.; Franke, G.N.; Gottardi, E.; et al. Impact of BCR::ABL1 Transcript Type on RT-qPCR Amplification Performance and Molecular Response to Therapy. Leukemia 2022, 36, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, T.; Gökbuget, N.; Schwartz, S.; Fischer, L.; Hubert, D.; Sindram, A.; Hoelzer, D.; Thiel, E. Clinical Features and Prognostic Implications of TCF3-PBX1 and ETV6-RUNX1 in Adult Acute Lymphoblastic Leukemia. Haematologica 2010, 95, 241–246. [Google Scholar] [CrossRef]
- Guo, J.Q.; Lin, H.; Kantarjian, H.; Talpaz, M.; Champlin, R.; Andreeff, M.; Glassman, A.; Arlinghaus, R.B. Comparison of Competitive-Nested PCR and Real-Time PCR in Detecting BCR-ABL Fusion Transcripts in Chronic Myeloid Leukemia Patients. Leukemia 2002, 16, 2447–2453. [Google Scholar] [CrossRef]
- Frenoy, N.; Chabli, A.; Sol, O.; Goldschmit, E.; Lemonnier, M.P.; Misset, J.L.; Debuire, B. Application of a New Protocol for Nested PCR to the Detection of Minimal Residual Bcr/Abl Transcripts. Leukemia 1994, 8, 1411–1414. [Google Scholar]
- Pessoa, F.M.C.d.P.; de Oliveira, M.B.; Barreto, I.V.; Machado, A.K.d.C.; de Oliveira, D.S.; Ribeiro, R.M.; Medeiros, J.C.; Maciel, A.d.R.; Silva, F.A.C.; Gurgel, L.A.; et al. Nested-PCR vs. RT-qPCR: A Sensitivity Comparison in the Detection of Genetic Alterations in Patients with Acute Leukemias. DNA 2024, 4, 285–299. [Google Scholar] [CrossRef]
- Ip, B.B.K.; Wong, A.T.C.; Law, J.H.Y.; Au, C.H.; Ma, S.Y.; Chim, J.C.S.; Liang, R.H.S.; Leung, A.Y.H.; Wan, T.S.K.; Ma, E.S.K. Application of Droplet Digital PCR in Minimal Residual Disease Monitoring of Rare Fusion Transcripts and Mutations in Haematological Malignancies. Sci. Rep. 2024, 14, 6400. [Google Scholar] [CrossRef]
- Chen, X.; Liu, L.; Zhang, A.; Yi, M.; Lan, Y.; Zheng, Z.; Zhang, L.; Liu, X.; Chang, L.; Zou, Y.; et al. Droplet Digital PCR for Genetic Mutations Monitoring Predicts Relapse Risk in Pediatric Acute Myeloid Leukemia. Int. J. Hematol. 2022, 116, 669–677. [Google Scholar] [CrossRef]
- Bilous, N.I.; Abramenko, I.V.; Chumak, A.A.; Dyagil, I.S.; Martina, Z.V. Detection of notch1 C.7544_7545delct mutation in chronic lymphocytic leukemia using conventional and real-time polymerase chain reaction. Exp. Oncol. 2016, 38, 112–116. Available online: http://dspace.nbuv.gov.ua/handle/123456789/138001 (accessed on 15 March 2025).
- Garibyan, L.; Avashia, N. Research Techniques Made Simple: Polymerase Chain Reaction (PCR). J. Investig. Dermatol. 2013, 133, e6. [Google Scholar] [CrossRef]
- Ghannam, M.G.; Varacallo, M.A. Biochemistry, Polymerase Chain Reaction. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Li, W. Flow Cytometry in the Diagnosis of Leukemias. In Leukemia; Li, W., Ed.; Exon Publications: Brisbane, Australia, 2022; ISBN 978-0-645-33207-0. [Google Scholar]
- Cai, Q.; Lan, H.; Yi, D.; Xian, B.; Zidan, L.; Li, J.; Liao, Z. Flow Cytometry in Acute Myeloid Leukemia and Detection of Minimal Residual Disease. Clin. Chim. Acta 2025, 564, 119945. [Google Scholar] [CrossRef]
- Ally, F.; Chen, X. Acute Myeloid Leukemia: Diagnosis and Evaluation by Flow Cytometry. Cancers 2024, 16, 3855. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, R.; Nair, R.A.; Jacob, P.M.; Nair Anila, K.A.R.; Prem, S.; Binitha, R.; Kusumakumary, P. Flow Cytometric Analysis of Mixed Phenotype Acute Leukemia: Experience from a Tertiary Oncology Center. Indian J. Pathol. Microbiol. 2015, 58, 181. [Google Scholar] [CrossRef]
- Kruse, A.; Abdel-Azim, N.; Kim, H.N.; Ruan, Y.; Phan, V.; Ogana, H.; Wang, W.; Lee, R.; Gang, E.J.; Khazal, S.; et al. Minimal Residual Disease Detection in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2020, 21, 1054. [Google Scholar] [CrossRef]
- Chen, X.; Cherian, S. Acute Myeloid Leukemia Immunophenotyping by Flow Cytometric Analysis. Clin. Lab. Med. 2017, 37, 753–769. [Google Scholar] [CrossRef]
- Wang, X.M. Advances and Issues in Flow Cytometric Detection of Immunophenotypic Changes and Genomic Rearrangements in Acute Pediatric Leukemia. Transl. Pediatr. 2014, 3, 149–155. [Google Scholar] [CrossRef]
- Kala, P.S.; Zubair, M. Flow Cytometry Blood Cell Identification. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Smith, A.C.; Hoischen, A.; Raca, G. Cytogenetics Is a Science, Not a Technique! Why Optical Genome Mapping Is So Important to Clinical Genetic Laboratories. Cancers 2023, 15, 5470. [Google Scholar] [CrossRef]
- Wiedmeier-Nutor, J.E.; McCabe, C.E.; O’Brien, D.R.; Jessen, E.; Bonolo de Campos, C.; Boddicker, N.J.; Griffin, R.; Allmer, C.; Rabe, K.G.; Cerhan, J.R.; et al. Utility of Targeted Sequencing Compared to FISH for Detection of Chronic Lymphocytic Leukemia Copy Number Alterations. Cancers 2024, 16, 2450. [Google Scholar] [CrossRef]
- Koutsi, A.; Vervesou, E.-C. Diagnostic Molecular Techniques in Haematology: Recent Advances. Ann. Transl. Med. 2018, 6, 242. [Google Scholar] [CrossRef]
- Oduro, K.A.; Spivey, T.; Moore, E.M.; Meyerson, H.; Yoest, J.; Tomlinson, B.; Beck, R.; Alouani, D.; Sadri, N. Clonal Dynamics and Relapse Risk Revealed by High-Sensitivity FLT3-Internal Tandem Duplication Detection in Acute Myeloid Leukemia. Mod. Pathol. 2024, 37, 100534. [Google Scholar] [CrossRef]
- Mareschal, S.; Palau, A.; Lindberg, J.; Ruminy, P.; Nilsson, C.; Bengtzén, S.; Engvall, M.; Eriksson, A.; Neddermeyer, A.; Marchand, V.; et al. Challenging Conventional Karyotyping by Next-Generation Karyotyping in 281 Intensively Treated Patients with AML. Blood Adv. 2021, 5, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Vosberg, S.; Greif, P.A. Clonal Evolution of Acute Myeloid Leukemia from Diagnosis to Relapse. Genes Chromosomes Cancer 2019, 58, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Vicente, A.E.; Bikos, V.; Hernández-Sánchez, M.; Malcikova, J.; Hernández-Rivas, J.-M.; Pospisilova, S. Next-Generation Sequencing in Chronic Lymphocytic Leukemia: Recent Findings and New Horizons. Oncotarget 2017, 8, 71234–71248. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.M.; Llop, M.; Sargas, C.; Pedrola, L.; Panadero, J.; Hervás, D.; Cervera, J.; Such, E.; Ibáñez, M.; Ayala, R.; et al. Clinical Utility of a Next-Generation Sequencing Panel for Acute Myeloid Leukemia Diagnostics. J. Mol. Diagn. 2019, 21, 228–240. [Google Scholar] [CrossRef]
- Madaci, L.; Farnault, L.; Abbou, N.; Gabert, J.; Venton, G.; Costello, R. Impact of Next-Generation Sequencing in Diagnosis, Prognosis and Therapeutic Management of Acute Myeloid Leukemia/Myelodysplastic Neoplasms. Cancers 2023, 15, 3280. [Google Scholar] [CrossRef]
- Prieto-Conde, M.I.; Corchete, L.A.; García-Álvarez, M.; Jiménez, C.; Medina, A.; Balanzategui, A.; Hernández-Ruano, M.; Maldonado, R.; Sarasquete, M.E.; Alcoceba, M.; et al. A New Next-Generation Sequencing Strategy for the Simultaneous Analysis of Mutations and Chromosomal Rearrangements at DNA Level in Acute Myeloid Leukemia Patients. J. Mol. Diagn. 2020, 22, 60–71. [Google Scholar] [CrossRef]
- Bacher, U.; Shumilov, E.; Flach, J.; Porret, N.; Joncourt, R.; Wiedemann, G.; Fiedler, M.; Novak, U.; Amstutz, U.; Pabst, T. Challenges in the Introduction of Next-Generation Sequencing (NGS) for Diagnostics of Myeloid Malignancies into Clinical Routine Use. Blood Cancer J. 2018, 8, 113. [Google Scholar] [CrossRef]
- Soverini, S.; Abruzzese, E.; Bocchia, M.; Bonifacio, M.; Galimberti, S.; Gozzini, A.; Iurlo, A.; Luciano, L.; Pregno, P.; Rosti, G.; et al. Next-Generation Sequencing for BCR-ABL1 Kinase Domain Mutation Testing in Patients with Chronic Myeloid Leukemia: A Position Paper. J. Hematol. Oncol. 2019, 12, 131. [Google Scholar] [CrossRef]
- Haferlach, T. Advancing Leukemia Diagnostics: Role of Next Generation Sequencing (NGS) in Acute Myeloid Leukemia. Hematol. Rep. 2020, 12, 8957. [Google Scholar] [CrossRef]
- Coccaro, N.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. Next-Generation Sequencing in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2019, 20, 2929. [Google Scholar] [CrossRef]
- Chen, W.-L.; Guo, Z.; Hou, Z.-Z.; Chen, Y.-J. Research Progress of Next Generation Sequencing in Acute Leukemia—Review. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2023, 31, 612–615. Available online: https://pubmed.ncbi.nlm.nih.gov/37096544/#:~:text=NGS%20reveals%20the%20genetic%20characteristics,predict%20disease%20recurrence%20by%20detecting (accessed on 23 March 2025). [PubMed]
- Bourbon, E.; Chabane, K.; Mosnier, I.; Bouvard, A.; Thonier, F.; Ferrant, E.; Michallet, A.-S.; Poulain, S.; Hayette, S.; Sujobert, P.; et al. Next-CLL, a New Next-Generation Sequencing–Based Method for Assessment of IGHV Gene Mutational Status in Chronic Lymphoid Leukemia. J. Mol. Diagn. 2023, 25, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Menon, V.; Brash, D.E. Next-Generation Sequencing Methodologies To Detect Low-Frequency Mutations: Catch Me If You Can. Mutat. Res. Rev. Mutat. Res. 2023, 792, 108471. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, H.; Zhang, V.W.; Tian, X.; Feng, Y.; Wang, G.; Gorman, E.; Wang, H.; Lutz, R.E.; Schmitt, E.S.; et al. Capture-Based High-Coverage NGS: A Powerful Tool to Uncover a Wide Spectrum of Mutation Types. Genet. Med. 2016, 18, 513–521. [Google Scholar] [CrossRef]
- Levine, R.L.; Valk, P.J.M. Next-Generation Sequencing in the Diagnosis and Minimal Residual Disease Assessment of Acute Myeloid Leukemia. Haematologica 2019, 104, 868–871. [Google Scholar] [CrossRef]
- Chaudhary, P.; Chaudhary, S.; Patel, F.; Patel, S.; Vaishnani, T.; Trivedi, N.; Patel, D.; Sonagara, T.; Hirapara, A.; Vyas, K.; et al. Validation of a Novel NGS Based BCR::ABL1 Kinase Domain Mutation Detection Assay in Indian Cohort. Sci. Rep. 2024, 14, 15745. [Google Scholar] [CrossRef]
- Bhatwadekar, S.S.; Shah, P. Mutational Phasing: Clinical Relevance in Tyrosine Kinase Domain Mutations Using Next Generation Sequencing in Chronic Myeloid Leukemia. Blood 2018, 132, 4269. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Ryan, S.L.; Peden, J.F.; Kingsbury, Z.; Schwab, C.J.; James, T.; Polonen, P.; Mijuskovic, M.; Becq, J.; Yim, R.; Cranston, R.E.; et al. Whole Genome Sequencing Provides Comprehensive Genetic Testing in Childhood B-Cell Acute Lymphoblastic Leukaemia. Leukemia 2023, 37, 518–528. [Google Scholar] [CrossRef]
- Nannini, M.; Pantaleo, M.A. Next Generation Sequencing (NGS) in Oncology: Lights and Shadows. CBN 2016, 4, 17–19. [Google Scholar] [CrossRef]
- Ljungström, V.; Cortese, D.; Young, E.; Pandzic, T.; Mansouri, L.; Plevova, K.; Ntoufa, S.; Baliakas, P.; Clifford, R.; Sutton, L.-A.; et al. Whole-Exome Sequencing in Relapsing Chronic Lymphocytic Leukemia: Clinical Impact of Recurrent RPS15 Mutations. Blood 2016, 127, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, B.; Tekin, M.; Mahdieh, N. The Promise of Whole-Exome Sequencing in Medical Genetics. J. Hum. Genet. 2014, 59, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Debarri, H.; Lebon, D.; Roumier, C.; Cheok, M.; Marceau-Renaut, A.; Nibourel, O.; Geffroy, S.; Helevaut, N.; Rousselot, P.; Gruson, B.; et al. IDH1/2 but Not DNMT3A Mutations Are Suitable Targets for Minimal Residual Disease Monitoring in Acute Myeloid Leukemia Patients: A Study by the Acute Leukemia French Association. Oncotarget 2015, 6, 42345–42353. [Google Scholar] [CrossRef]
- Miller, E.M.; Patterson, N.E.; Zechmeister, J.M.; Bejerano-Sagie, M.; Delio, M.; Patel, K.; Ravi, N.; Quispe-Tintaya, W.; Maslov, A.; Simmons, N.; et al. Development and Validation of a Targeted next Generation DNA Sequencing Panel Outperforming Whole Exome Sequencing for the Identification of Clinically Relevant Genetic Variants. Oncotarget 2017, 8, 102033–102045. [Google Scholar] [CrossRef]
- Ahmed, F.; Zhong, J. Advances in DNA/RNA Sequencing and Their Applications in Acute Myeloid Leukemia (AML). Int. J. Mol. Sci. 2024, 26, 71. [Google Scholar] [CrossRef]
- Petiti, J.; Pignochino, Y.; Schiavon, A.; Giugliano, E.; Berrino, E.; Giordano, G.; Itri, F.; Dragani, M.; Cilloni, D.; Lo Iacono, M. Comprehensive Molecular Profiling of NPM1-Mutated Acute Myeloid Leukemia Using RNAseq Approach. Int. J. Mol. Sci. 2024, 25, 3631. [Google Scholar] [CrossRef]
- Kim, J.C.; Chan-Seng-Yue, M.; Ge, S.; Zeng, A.G.X.; Ng, K.; Gan, O.I.; Garcia-Prat, L.; Flores-Figueroa, E.; Woo, T.; Zhang, A.X.W.; et al. Transcriptomic Classes of BCR-ABL1 Lymphoblastic Leukemia. Nat. Genet. 2023, 55, 1186–1197. [Google Scholar] [CrossRef]
- Borad, M.J.; Egan, J.; Champion, M.; Hunt, K.; McWilliams, R.; McCullough, A.; Aldrich, J.; Nasser, S.; Liang, W.; Barrett, M.; et al. Abstract CT112: Implementation of CLIA Enabled Integrated Whole Genome (WGS)/Exome (WES)/Transcriptome (RNAseq) next-Gen Sequencing to Identify Therapeutically Relevant Targets in Advanced Cancer Patients. Cancer Res. 2015, 75, CT112. [Google Scholar] [CrossRef]
- Ramarao-Milne, P.; Kondrashova, O.; Patch, A.-M.; Nones, K.; Koufariotis, L.T.; Newell, F.; Addala, V.; Lakis, V.; Holmes, O.; Leonard, C.; et al. Comparison of Actionable Events Detected in Cancer Genomes by Whole-Genome Sequencing, in Silico Whole-Exome and Mutation Panels. ESMO Open 2022, 7, 100540. [Google Scholar] [CrossRef]
- Li, J.; Batcha, A.M.N.; Grüning, B.; Mansmann, U.R. An NGS Workflow Blueprint for DNA Sequencing Data and Its Application in Individualized Molecular Oncology. Cancer Inform. 2016, 14, 87–107. [Google Scholar] [CrossRef]
- Vicente-Garcés, C.; Esperanza-Cebollada, E.; Montesdeoca, S.; Torrebadell, M.; Rives, S.; Dapena, J.L.; Català, A.; Conde, N.; Camós, M.; Vega-García, N. Technical Validation and Clinical Utility of an NGS Targeted Panel to Improve Molecular Characterization of Pediatric Acute Leukemia. Front. Mol. Biosci. 2022, 9, 854098. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.R. Target Enrichment Approaches for Next-Generation Sequencing Applications in Oncology. Diagnosis 2022, 12, 1539. [Google Scholar] [CrossRef]
- Espinosa, E.; Bautista, R.; Larrosa, R.; Plata, O. Advancements in Long-Read Genome Sequencing Technologies and Algorithms. Genomics 2024, 116, 110842. [Google Scholar] [CrossRef]
- Ramesh, A.; Koo, S.; Kang, S.J.; Ghosal, A.; Alarcon, F.; Gyuris, T.; Jung, S.C.; Magnan, C.; Nam, H.; Thomas, B.B.; et al. A Novel NGS-Based Simultaneous Detection of DNA and RNA Biomarkers Using Total Nucleic Acid (TNA) for Acute Lymphocytic Leukemia (ALL). Blood 2021, 138, 1875. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Sondka, Z.; Dhir, N.B.; Carvalho-Silva, D.; Jupe, S.; Madhumita, N.; McLaren, K.; Starkey, M.; Ward, S.; Wilding, J.; Ahmed, M.; et al. COSMIC: A Curated Database of Somatic Variants and Clinical Data for Cancer. Nucleic Acids Res. 2024, 52, D1210–D1217. [Google Scholar] [CrossRef]
- Gil, J.V.; Such, E.; Sargas, C.; Simarro, J.; Miralles, A.; Pérez, G.; De Juan, I.; Palanca, S.; Avetisyan, G.; Santiago, M.; et al. Design and Validation of a Custom Next-Generation Sequencing Panel in Pediatric Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2023, 24, 4440. [Google Scholar] [CrossRef]
- Inaba, H.; Azzato, E.M.; Mullighan, C.G. Integration of Next-Generation Sequencing to Treat Acute Lymphoblastic Leukemia with Targetable Lesions: The St. Jude Children’s Research Hospital Approach. Front. Pediatr. 2017, 5, 258. [Google Scholar] [CrossRef]
- Zaliova, M.; Kotrova, M.; Bresolin, S.; Stuchly, J.; Stary, J.; Hrusak, O.; Te Kronnie, G.; Trka, J.; Zuna, J.; Vaskova, M. ETV6/RUNX1-like Acute Lymphoblastic Leukemia: A Novel B-Cell Precursor Leukemia Subtype Associated with the CD27/CD44 Immunophenotype. Genes Chromosomes Cancer 2017, 56, 608–616. [Google Scholar] [CrossRef]
- Paolino, J.; Tsai, H.K.; Harris, M.H.; Pikman, Y. IKZF1 Alterations and Therapeutic Targeting in B-Cell Acute Lymphoblastic Leukemia. Biomedicines 2024, 12, 89. [Google Scholar] [CrossRef]
- Gupta, D.G.; Varma, N.; Sreedharanunni, S.; Abdulkadir, S.A.; Naseem, S.; Sachdeva, M.U.S.; Binota, J.; Bose, P.; Malhotra, P.; Khadwal, A.; et al. ‘Evaluation of Adverse Prognostic Gene Alterations & MRD Positivity in BCR::ABL1-like B-Lineage Acute Lymphoblastic Leukaemia Patients, in a Resource-Constrained Setting. Br. J. Cancer 2023, 129, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Alghandour, R.; Sakr, D.H.; Shaaban, Y. Philadelphia-like Acute Lymphoblastic Leukemia: The Journey from Molecular Background to the Role of Bone Marrow Transplant—Review Article. Ann. Hematol. 2023, 102, 1287–1300. [Google Scholar] [CrossRef] [PubMed]
- Dörge, P.; Meissner, B.; Zimmermann, M.; Möricke, A.; Schrauder, A.; Bouquin, J.-P.; Schewe, D.; Harbott, J.; Teigler-Schlegel, A.; Ratei, R.; et al. IKZF1 Deletion Is an Independent Predictor of Outcome in Pediatric Acute Lymphoblastic Leukemia Treated According to the ALL-BFM 2000 Protocol. Haematologica 2013, 98, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, R.P.; Waanders, E.; van der Velden, V.H.J.; van Reijmersdal, S.V.; Venkatachalam, R.; Scheijen, B.; Sonneveld, E.; van Dongen, J.J.M.; Veerman, A.J.P.; van Leeuwen, F.N.; et al. IKZF1 Deletions Predict Relapse in Uniformly Treated Pediatric Precursor B-ALL. Leukemia 2010, 24, 1258–1264. [Google Scholar] [CrossRef]
- Stanulla, M.; Cavé, H.; Moorman, A.V. IKZF1 Deletions in Pediatric Acute Lymphoblastic Leukemia: Still a Poor Prognostic Marker? Blood 2020, 135, 252–260. [Google Scholar] [CrossRef]
- Huang, Z.; Jia, Y.; Ruan, G.; Zuo, Y.; Wu, J.; Lu, A.; Xue, Y.; Cheng, Y.; Zhang, L. Quantitative Analysis of IKZF1 Gene Deletions in Pediatric B-Cell Precursor Acute Lymphoblastic Leukemia: Higher Levels Are Associated with a Poorer Prognosis. Pediatr. Hematol. Oncol. 2022, 39, 243–253. [Google Scholar] [CrossRef]
- Clappier, E.; Grardel, N.; Bakkus, M.; Rapion, J.; Moerloose, B.; Kastner, P.; Caye, A.; Vivent, J.; Costa, V.; Ferster, A.; et al. IKZF1 Deletion Is an Independent Prognostic Marker in Childhood B-Cell Precursor Acute Lymphoblastic Leukemia, and Distinguishes Patients Benefiting from Pulses during Maintenance Therapy: Results of the EORTC Children’s Leukemia Group Study 58951. Leukemia 2015, 29, 2154–2161. [Google Scholar] [CrossRef]
- Vervoort, B.M.T.; Butler, M.; Grünewald, K.J.T.; van Ingen Schenau, D.S.; Tee, T.M.; Lucas, L.; Huitema, A.D.R.; Boer, J.M.; Bornhauser, B.C.; Bourquin, J.; et al. IKZF1 Gene Deletions Drive Resistance to Cytarabine in B-Cell Precursor Acute Lymphoblastic Leukemia. Haematologica 2024, 109, 3904–3917. [Google Scholar] [CrossRef]
- von Goessel, H.; Jacobs, U.; Semper, S.; Krumbholz, M.; Langer, T.; Keller, T.; Schrauder, A.; van der Velden, V.H.J.; van Dongen, J.J.M.; Harbott, J.; et al. Cluster Analysis of Genomic ETV6–RUNX1 (TEL–AML1) Fusion Sites in Childhood Acute Lymphoblastic Leukemia. Leuk. Res. 2009, 33, 1082–1088. [Google Scholar] [CrossRef]
- Schäfer, D.; Olsen, M.; Lähnemann, D.; Stanulla, M.; Slany, R.; Schmiegelow, K.; Borkhardt, A.; Fischer, U. Five Percent of Healthy Newborns Have an ETV6-RUNX1 Fusion as Revealed by DNA-Based GIPFEL Screening. Blood 2018, 131, 821–826. [Google Scholar] [CrossRef]
- van der Weyden, L.; Giotopoulos, G.; Rust, A.G.; Matheson, L.S.; van Delft, F.W.; Kong, J.; Corcoran, A.E.; Greaves, M.F.; Mullighan, C.G.; Huntly, B.J.; et al. Modeling the Evolution of ETV6-RUNX1-Induced B-Cell Precursor Acute Lymphoblastic Leukemia in Mice. Blood 2011, 118, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Hernández, G.; Hauer, J.; Martín-Lorenzo, A.; Schäfer, D.; Bartenhagen, C.; García-Ramírez, I.; Auer, F.; González-Herrero, I.; Ruiz-Roca, L.; Gombert, M.; et al. Infection Exposure Promotes ETV6-RUNX1 Precursor B-Cell Leukemia via Impaired H3K4 Demethylases. Cancer Res. 2017, 77, 4365–4377. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.S.; Francis, A.; Turkistany, S.; Shukla, D.; Wong, A.; Batista, C.R.; DeKoter, R.P. ETV6-RUNX1 Interacts with a Region in SPIB Intron 1 to Regulate Gene Expression in Pre-B-Cell Acute Lymphoblastic Leukemia. Exp. Hematol. 2019, 73, 50–63.e2. [Google Scholar] [CrossRef]
- Zerella, J.R.; Homan, C.C.; Arts, P.; Brown, A.L.; Scott, H.S.; Hahn, C.N. Transcription Factor Genetics and Biology in Predisposition to Bone Marrow Failure and Hematological Malignancy. Front. Oncol. 2023, 13, 1183318. [Google Scholar] [CrossRef]
- Lee, J.W.; Kim, S.; Jang, P.-S.; Chung, N.-G.; Jeong, D.-C.; Kim, M.; Cho, B.; Kim, H.-K. Outcome and Prognostic Factors for ETV6/RUNX1 Positive Pediatric Acute Lymphoblastic Leukemia Treated at a Single Institution in Korea. Cancer Res. Treat. 2017, 49, 446–453. [Google Scholar] [CrossRef][Green Version]
- Meyer, L.K.; Delgado-Martin, C.; Maude, S.L.; Shannon, K.M.; Teachey, D.T.; Hermiston, M.L. CRLF2 Rearrangement in Ph-like Acute Lymphoblastic Leukemia Predicts Relative Glucocorticoid Resistance That Is Overcome with MEK or Akt Inhibition. PLoS ONE 2019, 14, e0220026. [Google Scholar] [CrossRef]
- Tasian, S.K.; Loh, M.L. Understanding the Biology of CRLF2-Overexpressing Acute Lymphoblastic Leukemia. Crit. Rev. Oncog. 2011, 16, 13–24. [Google Scholar]
- Harvey, R.C.; Mullighan, C.G.; Chen, I.-M.; Wharton, W.; Mikhail, F.M.; Carroll, A.J.; Kang, H.; Liu, W.; Dobbin, K.K.; Smith, M.A.; et al. Rearrangement of CRLF2 Is Associated with Mutation of JAK Kinases, Alteration of IKZF1, Hispanic/Latino Ethnicity, and a Poor Outcome in Pediatric B-Progenitor Acute Lymphoblastic Leukemia. Blood 2010, 115, 5312–5321. [Google Scholar] [CrossRef]
- Tasian, S.K.; Doral, M.Y.; Borowitz, M.J.; Wood, B.L.; Chen, I.-M.; Harvey, R.C.; Gastier-Foster, J.M.; Willman, C.L.; Hunger, S.P.; Mullighan, C.G.; et al. Aberrant STAT5 and PI3K/mTOR Pathway Signaling Occurs in Human CRLF2-Rearranged B-Precursor Acute Lymphoblastic Leukemia. Blood 2012, 120, 833–842. [Google Scholar] [CrossRef]
- Péterffy, B.; Krizsán, S.; Egyed, B.; Bedics, G.; Benard-Slagter, A.; Palit, S.; Erdélyi, D.J.; Müller, J.; Nagy, T.; Hegyi, L.L.; et al. Molecular Profiling Reveals Novel Gene Fusions and Genetic Markers for Refined Patient Stratification in Pediatric Acute Lymphoblastic Leukemia. Mod. Pathol. 2025, 38, 100741. [Google Scholar] [CrossRef]
- Simonin, M.; Vasseur, L.; Lengliné, E.; Lhermitte, L.; Cabannes-Hamy, A.; Balsat, M.; Schmidt, A.; Dourthe, M.-E.; Touzart, A.; Graux, C.; et al. NGS-Based Stratification Refines the Risk Stratification in T-ALL and Identifies a Very-High-Risk Subgroup of Patients. Blood 2024, 144, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Sherali, N.; Hamadneh, T.; Aftab, S.; Alfonso, M.S.; Tsouklidis, N. Integration of Next-Generation Sequencing in Diagnosing and Minimal Residual Disease Detection in Patients With Philadelphia Chromosome-Like Acute Lymphoblastic Leukemia. Cureus 2020, 12, e10696. [Google Scholar] [CrossRef] [PubMed]
- Ashouri, K.; Nittur, V.; Ginosyan, A.A.; Hwang, J.; Adnani, B.; Chen, D.; Savitala-Damerla, L.; Schiff, K.; Chaudhary, P.; Kovach, A.E.; et al. Concordance of Next-Generation Sequencing and Multiparametric Flow Cytometry Methods for Detecting Measurable Residual Disease in Adult Acute Lymphoblastic Leukemia: Optimizing Prediction of Clinical Outcomes From a Single-Center Study. Clin. Lymphoma Myeloma Leuk. 2024, 24, e59–e66.e2. [Google Scholar] [CrossRef] [PubMed]
- Paolino, J.D.; Harris, M.H.; Stevenson, K.E.; Koch, V.; Cole, P.D.; Gennarini, L.M.; Kahn, J.M.; Kelly, K.M.; Kirsch, I.; Michon, B.; et al. Performance of Next Generation Sequencing for Minimal Residual Disease Detection for Pediatric Patients with Acute Lymphoblastic Leukemia: Results from the Prospective Clinical Trial DFCI 16-001. Blood 2021, 138, 3485. [Google Scholar] [CrossRef]
- Krstevska Bozhinovikj, E.; Matevska-Geshkovska, N.; Staninova Stojovska, M.; Gjorgievska, E.; Jovanovska, A.; Ridova, N.; Panovska Stavridis, I.; Kocheva, S.; Dimovski, A. Presence of Minimal Residual Disease Determined by Next-Generation Sequencing Is Not a Reliable Prognostic Biomarker in Children with Acute Lymphoblastic Leukemia. Leuk. Lymphoma 2025, 1–8. [Google Scholar] [CrossRef]
- Woolbright, W.C.; Wong, V.; Severson, E.; Kuo, D.J. The Application of Next-Generation Sequencing Tumor Molecular Profiling in the Diagnosis and Management of a Case of Acute Myelogenous Leukemia With MLL-PTD in a Pediatric Heart Transplant Recipient. J. Pediatr. Hematol./Oncol. 2021, 43, e246–e249. [Google Scholar] [CrossRef]
- Mosquera Orgueira, A.; Peleteiro Raíndo, A.; Cid López, M.; Antelo Rodríguez, B.; Díaz Arias, J.Á.; Ferreiro Ferro, R.; Alonso Vence, N.; Bendaña López, Á.; Abuín Blanco, A.; Bao Pérez, L.; et al. Gene Expression Profiling Identifies FLT3 Mutation-like Cases in Wild-Type FLT3 Acute Myeloid Leukemia. PLoS ONE 2021, 16, e0247093. [Google Scholar] [CrossRef]
- Tao, S.; Wang, C.; Chen, Y.; Deng, Y.; Song, L.; Shi, Y.; Ling, L.; Ding, B.; He, Z.; Yu, L. Prognosis and Outcome of Patients with Acute Myeloid Leukemia Based on FLT3-ITD Mutation with or without Additional Abnormal Cytogenetics. Oncol. Lett. 2019, 18, 6766–6774. [Google Scholar] [CrossRef]
- Jain, P.; Kantarjian, H.; Patel, K.; Faderl, S.; Garcia-Manero, G.; Benjamini, O.; Borthakur, G.; Pemmaraju, N.; Kadia, T.; Daver, N.; et al. Mutated Nucleophosmin-1 (NPM1) in Patients with Acute Myeloid Leukemia (AML) in Remission and Relapse. Leuk. Lymphoma 2014, 55, 1337–1344. [Google Scholar] [CrossRef]
- Becker, H.; Marcucci, G.; Maharry, K.; Radmacher, M.D.; Mrózek, K.; Margeson, D.; Whitman, S.P.; Wu, Y.-Z.; Schwind, S.; Paschka, P.; et al. Favorable Prognostic Impact of NPM1 Mutations in Older Patients With Cytogenetically Normal De Novo Acute Myeloid Leukemia and Associated Gene- and MicroRNA-Expression Signatures: A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2010, 28, 596–604. [Google Scholar] [CrossRef]
- Baek, D.W.; Lee, J.M.; Kim, J.-H.; Cho, H.J.; Ham, J.-Y.; Suh, J.-S.; Sohn, S.-K.; Moon, J.H. Favorable Long-Term Survival Using Consolidation Chemotherapy without Allogeneic Hematopoietic Cell Transplantation for Acute Myeloid Leukemia with Wild-Type NPM1 without FLT3-ITD. Blood Res. 2019, 54, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Issa, G.C.; Bidikian, A.; Venugopal, S.; Konopleva, M.; DiNardo, C.D.; Kadia, T.M.; Borthakur, G.; Jabbour, E.; Pemmaraju, N.; Yilmaz, M.; et al. Clinical Outcomes Associated with NPM1 Mutations in Patients with Relapsed or Refractory AML. Blood Adv. 2022, 7, 933–942. [Google Scholar] [CrossRef]
- Sallmyr, A.; Fan, J.; Datta, K.; Kim, K.-T.; Grosu, D.; Shapiro, P.; Small, D.; Rassool, F. Internal Tandem Duplication of FLT3 (FLT3/ITD) Induces Increased ROS Production, DNA Damage, and Misrepair: Implications for Poor Prognosis in AML. Blood 2008, 111, 3173–3182. [Google Scholar] [CrossRef] [PubMed]
- Dunna, N.R.; Rajappa, S.; Digumarti, R.; Vure, S.; Kagita, S.; Damineni, S.; Rao, V.R.; Yadav, S.K.; Ravuri, R.R.; Satti, V. Fms like Tyrosine Kinase (FLT3) and Nucleophosmin 1 (NPM1) Mutations in de Novo Normal Karyotype Acute Myeloid Leukemia (AML). Asian Pac. J. Cancer Prev. 2010, 11, 1811–1816. [Google Scholar]
- Lazana, I.; Tsirigotis, P.; Konstantellos, I.; Chondropoulos, S.; Stamouli, M.; Gkirkas, K.; Baltadakis, I.; Theodorou, E.; Karaolidou, F.; Tzenou, T.; et al. The Prognostic Impact of Other Somatic Mutations and Cytogenetic Abnormalities in Patients with NPM1 and/or FLT3-ITD Mutated AML in the Era of FLT3-Inhibitors: Real-Life Data from the Greek AML Registry. Blood 2024, 144, 5956. [Google Scholar] [CrossRef]
- Yang, H.; Ye, D.; Guan, K.-L.; Xiong, Y. IDH1 and IDH2 Mutations in Tumorigenesis: Mechanistic Insights and Clinical Perspectives. Clin. Cancer Res. 2012, 18, 5562–5571. [Google Scholar] [CrossRef]
- Rakheja, D.; Konoplev, S.; Medeiros, L.J.; Chen, W. IDH Mutations in Acute Myeloid Leukemia. Human Pathol. 2012, 43, 1541–1551. [Google Scholar] [CrossRef]
- Lachowiez, C.A.; Reville, P.K.; Kantarjian, H.; Jabbour, E.; Borthakur, G.; Daver, N.; Issa, G.; Furudate, K.; Tanaka, T.; Pierce, S.; et al. Contemporary Outcomes in -Mutated Acute Myeloid Leukemia: The Impact of Co-Occurring Mutations and Venetoclax-Based Treatment. Am. J. Hematol. 2022, 97, 1443–1452. [Google Scholar] [CrossRef]
- Salem, D.; El-Aziz, S.A.; El-Menshawy, N.; Abouzeid, T.; Ebrahim, M. Prevalence and Prognostic Value of IDH1 R132 Mutation in Newly Diagnosed AML Egyptian Patients with Normal Karyotype. Indian J. Hematol. Blood Transfus. 2017, 33, 49–55. [Google Scholar] [CrossRef]
- Abbas, S.; Lugthart, S.; Kavelaars, F.G.; Schelen, A.; Koenders, J.E.; Zeilemaker, A.; van Putten, W.J.L.; Rijneveld, A.W.; Löwenberg, B.; Valk, P.J.M. Acquired Mutations in the Genes Encoding IDH1 and IDH2 Both Are Recurrent Aberrations in Acute Myeloid Leukemia: Prevalence and Prognostic Value. Blood 2010, 116, 2122–2126. [Google Scholar] [CrossRef]
- Issa, G.C.; DiNardo, C.D. Acute Myeloid Leukemia with IDH1 and IDH2 Mutations: 2021 Treatment Algorithm. Blood Cancer J. 2021, 11, 107. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Morita, K.; DiNardo, C.D.; Furudate, K.; Tanaka, T.; Yan, Y.; Patel, K.P.; MacBeth, K.J.; Wu, B.; Liu, G.; et al. Leukemia Stemness and Co-Occurring Mutations Drive Resistance to IDH Inhibitors in Acute Myeloid Leukemia. Nat. Commun. 2021, 12, 2607. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, A.; Zarychta, J.; Lejman, M.; Latoch, E.; Zawitkowska, J. Clinical Implications of Isocitrate Dehydrogenase Mutations and Targeted Treatment of Acute Myeloid Leukemia with Mutant Isocitrate Dehydrogenase Inhibitors—Recent Advances, Challenges and Future Prospects. Int. J. Mol. Sci. 2024, 25, 7916. [Google Scholar] [CrossRef] [PubMed]
- Castelli, G.; Pelosi, E.; Testa, U. Targeted Therapies in the Treatment of Adult Acute Myeloid Leukemias: Current Status and Future Perspectives. Int. J. Hematol. Oncol. 2016, 5, 143–164. [Google Scholar] [CrossRef]
- Yang, F.; Anekpuritanang, T.; Press, R.D. Clinical Utility of Next-Generation Sequencing in Acute Myeloid Leukemia. Mol. Diagn. Ther. 2020, 24, 1–13. [Google Scholar] [CrossRef]
- Fedorov, K.; Maiti, A.; Konopleva, M. Targeting FLT3 Mutation in Acute Myeloid Leukemia: Current Strategies and Future Directions. Cancers 2023, 15, 2312. [Google Scholar] [CrossRef]
- Ok, C.Y.; Loghavi, S.; Sui, D.; Wei, P.; Kanagal-Shamanna, R.; Yin, C.C.; Zuo, Z.; Routbort, M.J.; Tang, G.; Tang, Z.; et al. Persistent IDH1/2 Mutations in Remission Can Predict Relapse in Patients with Acute Myeloid Leukemia. Haematologica 2019, 104, 305–311. [Google Scholar] [CrossRef]
- Jain, G.; Thakral, D.; Sahoo, R.K.; Kumar, I.; Vashishtha, S.; Verma, P.; Gupta, R. Next Generation Sequencing Guided Treatment Modulation and Prognosis in Acute Myeloid Leukemia: Case Vignettes. Am. J. Blood Res. 2020, 10, 134–144. [Google Scholar]
- Kim, J.J.; Jang, J.E.; Lee, H.A.; Park, M.R.; Kook, H.W.; Lee, S.-T.; Choi, J.R.; Min, Y.H.; Shin, S.; Cheong, J.-W. Development of a Next-Generation Sequencing-Based Gene Panel Test to Detect Measurable Residual Disease in Acute Myeloid Leukemia. Ann. Lab. Med. 2023, 43, 328–336. [Google Scholar] [CrossRef]
- Chiaretti, S.; Marinelli, M.; Del Giudice, I.; Bonina, S.; Piciocchi, A.; Messina, M.; Vignetti, M.; Rossi, D.; Di Maio, V.; Mauro, F.R.; et al. NOTCH1, SF3B1, BIRC3 and TP53 Mutations in Patients with Chronic Lymphocytic Leukemia Undergoing First-Line Treatment: Correlation with Biological Parameters and Response to Treatment. Leuk. Lymphoma 2014, 55, 2785–2792. [Google Scholar] [CrossRef]
- Lazarian, G.; Tausch, E.; Eclache, V.; Sebaa, A.; Bianchi, V.; Letestu, R.; Collon, J.-F.; Lefebvre, V.; Gardano, L.; Varin-Blank, N.; et al. TP53 Mutations Are Early Events in Chronic Lymphocytic Leukemia Disease Progression and Precede Evolution to Complex Karyotypes. Int. J. Cancer 2016, 139, 1759–1763. [Google Scholar] [CrossRef] [PubMed]
- Zenz, T.; Vollmer, D.; Trbusek, M.; Smardova, J.; Benner, A.; Soussi, T.; Helfrich, H.; Heuberger, M.; Hoth, P.; Fuge, M.; et al. TP53 Mutation Profile in Chronic Lymphocytic Leukemia: Evidence for a Disease Specific Profile from a Comprehensive Analysis of 268 Mutations. Leukemia 2010, 24, 2072–2079. [Google Scholar] [CrossRef] [PubMed]
- Brieghel, C.; Aarup, K.; Torp, M.H.; Andersen, M.A.; Yde, C.W.; Tian, X.; Wiestner, A.; Ahn, I.E.; Niemann, C.U. Clinical Outcomes in Patients with Multi-Hit TP53 Chronic Lymphocytic Leukemia Treated with Ibrutinib. Clin. Cancer Res. 2021, 27, 4531–4538. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.; Martinez, P.; Wade, R.; Hockley, S.; Oscier, D.; Matutes, E.; Dearden, C.E.; Richards, S.M.; Catovsky, D.; Morgan, G.J. Mutational Status of the TP53 Gene as a Predictor of Response and Survival in Patients with Chronic Lymphocytic Leukemia: Results from the LRF CLL4 Trial. J. Clin. Oncol. 2011, 29, 2223–2229. [Google Scholar] [CrossRef]
- Robak, T. TP53 Mutations in Chronic Lymphocytic Leukemia. Acta Haematol. Pol. 2021, 52, 75–76. [Google Scholar] [CrossRef]
- Fürstenau, M.; Eichhorst, B. Novel Agents in Chronic Lymphocytic Leukemia: New Combination Therapies and Strategies to Overcome Resistance. Cancers 2021, 13, 1336. [Google Scholar] [CrossRef]
- De Luca, G.; Cerruti, G.; Lastraioli, S.; Conte, R.; Ibatici, A.; Di Felice, N.; Morabito, F.; Monti, P.; Fronza, G.; Matis, S.; et al. The Spectrum of Subclonal TP53 Mutations in Chronic Lymphocytic Leukemia: A next Generation Sequencing Retrospective Study. Hematol. Oncol. 2022, 40, 962–975. [Google Scholar] [CrossRef]
- Malcikova, J.; Tausch, E.; Rossi, D.; Sutton, L.A.; Soussi, T.; Zenz, T.; Kater, A.P.; Niemann, C.U.; Gonzalez, D.; Davi, F.; et al. ERIC Recommendations for TP53 Mutation Analysis in Chronic Lymphocytic Leukemia—Update on Methodological Approaches and Results Interpretation. Leukemia 2018, 32, 1070–1080. [Google Scholar] [CrossRef]
- Campo, E.; Cymbalista, F.; Ghia, P.; Jäger, U.; Pospisilova, S.; Rosenquist, R.; Schuh, A.; Stilgenbauer, S. TP53 Aberrations in Chronic Lymphocytic Leukemia: An Overview of the Clinical Implications of Improved Diagnostics. Haematologica 2018, 103, 1956–1968. [Google Scholar] [CrossRef]
- Hoofd, C.; Huang, S.J.; Gusscott, S.; Lam, S.; Wong, R.; Johnston, A.; Ben-Neriah, S.; Steidl, C.; Scott, D.W.; Bruyere, H.; et al. Ultrasensitive Detection of NOTCH1 c.7544_7545delCT Mutations in Chronic Lymphocytic Leukemia by Droplet Digital PCR Reveals High Frequency of Subclonal Mutations and Predicts Clinical Outcome in Cases with Trisomy 12. J. Mol. Diagn. 2020, 22, 571–578. [Google Scholar] [CrossRef]
- Willander, K.; Dutta, R.K.; Ungerbäck, J.; Gunnarsson, R.; Juliusson, G.; Fredrikson, M.; Linderholm, M.; Söderkvist, P. NOTCH1 Mutations Influence Survival in Chronic Lymphocytic Leukemia Patients. BMC Cancer 2013, 13, 274. [Google Scholar] [CrossRef]
- Jelloul, F.Z.; Yang, R.; Garces, S.; Kanagal-Shamanna, R.; Ok, C.Y.; Loghavi, S.; Routbort, M.J.; Zuo, Z.; Yin, C.C.; Floyd, K.; et al. Landscape of NOTCH1 Mutations and Co-Occurring Biomarker Alterations in Chronic Lymphocytic Leukemia. Leuk. Res. 2022, 116, 106827. [Google Scholar] [CrossRef] [PubMed]
- Villamor, N.; Conde, L.; Martínez-Trillos, A.; Cazorla, M.; Navarro, A.; Beà, S.; López, C.; Colomer, D.; Pinyol, M.; Aymerich, M.; et al. NOTCH1 Mutations Identify a Genetic Subgroup of Chronic Lymphocytic Leukemia Patients with High Risk of Transformation and Poor Outcome. Leukemia 2013, 27, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Rasi, S.; Fabbri, G.; Spina, V.; Fangazio, M.; Forconi, F.; Marasca, R.; Laurenti, L.; Bruscaggin, A.; Cerri, M.; et al. Mutations of NOTCH1 Are an Independent Predictor of Survival in Chronic Lymphocytic Leukemia. Blood 2012, 119, 521–529. [Google Scholar] [CrossRef]
- Pozzo, F.; Bittolo, T.; Tissino, E.; Zucchetto, A.; Bomben, R.; Polcik, L.; Dannewitz Prosseda, S.; Hartmann, T.N.; Gattei, V. Multiple Mechanisms of NOTCH1 Activation in Chronic Lymphocytic Leukemia: NOTCH1 Mutations and Beyond. Cancers 2022, 14, 2997. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, S.; Chen, S.; Chen, G.; Zhang, R.; Li, J.; Qu, J. SF3B1 Mutation Is a Prognostic Factor in Chronic Lymphocytic Leukemia: A Meta-Analysis. Oncotarget 2017, 8, 69916–69923. [Google Scholar] [CrossRef]
- Cazzola, M.; Rossi, M.; Malcovati, L. Biologic and Clinical Significance of Somatic Mutations of SF3B1 in Myeloid and Lymphoid Neoplasms. Blood 2013, 121, 260–269. [Google Scholar] [CrossRef]
- Wan, Y.; Wu, C.J. SF3B1 Mutations in Chronic Lymphocytic Leukemia. Blood 2013, 121, 4627–4634. [Google Scholar] [CrossRef]
- Wang, L.; Brooks, A.N.; Fan, J.; Wan, Y.; Gambe, R.; Li, S.; Hergert, S.; Yin, S.; Freeman, S.S.; Levin, J.Z.; et al. Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell 2016, 30, 750–763. [Google Scholar] [CrossRef]
- Rossi, D.; Bruscaggin, A.; Spina, V.; Rasi, S.; Khiabanian, H.; Messina, M.; Fangazio, M.; Vaisitti, T.; Monti, S.; Chiaretti, S.; et al. Mutations of the SF3B1 Splicing Factor in Chronic Lymphocytic Leukemia: Association with Progression and Fludarabine-Refractoriness. Blood 2011, 118, 6904–6908. [Google Scholar] [CrossRef]
- Wang, L.; Lawrence, M.S.; Wan, Y.; Stojanov, P.; Sougnez, C.; Stevenson, K.; Werner, L.; Sivachenko, A.; DeLuca, D.S.; Zhang, L.; et al. SF3B1 and Other Novel Cancer Genes in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2011, 365, 2497–2506. [Google Scholar] [CrossRef] [PubMed]
- Sud, A.; Parry, E.M.; Wu, C.J. The Molecular Map of CLL and Richter’s Syndrome. Semin. Hematol. 2024, 61, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Zou, Y.-X.; Gu, D.-L.; Zhu, H.-C.; Zhu, H.-Y.; Wang, L.; Liang, J.-H.; Xia, Y.; Wu, J.-Z.; Shao, C.-L.; et al. SF3B1 Mutation Predicts Unfavorable Treatment-Free Survival in Chinese Chronic Lymphocytic Leukemia Patients. Ann. Transl. Med. 2019, 7, 176. [Google Scholar] [CrossRef]
- Alanazi, M.A.; Kwa, F.A.A.; Omar, M.M.A.; Antonipillai, J.; Jackson, D.E. Efficacy and Challenges Involving Combination Therapies in CLL. Drug Discov. Today 2024, 29, 104243. [Google Scholar] [CrossRef]
- Nadeu, F.; Delgado, J.; Royo, C.; Baumann, T.; Stankovic, T.; Pinyol, M.; Jares, P.; Navarro, A.; Martín-García, D.; Beà, S.; et al. Clinical Impact of Clonal and Subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM Mutations in Chronic Lymphocytic Leukemia. Blood 2016, 127, 2122–2130. [Google Scholar] [CrossRef]
- Tam, C.S.; Balendran, S.; Blombery, P. Novel Mechanisms of Resistance in CLL: Variant BTK Mutations in Second-Generation and Noncovalent BTK Inhibitors. Blood 2025, 145, 1005–1009. [Google Scholar] [CrossRef]
- Lovell, A.R.; Sawyers, J.; Bose, P. An Update on the Efficacy of Venetoclax for Chronic Lymphocytic Leukemia. Expert Opin. Pharmacother. 2023, 24, 1307–1316. [Google Scholar] [CrossRef]
- Machnicki, M.M.; Górniak, P.; Pepek, M.; Szymczyk, A.; Iskierka-Jażdżewska, E.; Steckiewicz, P.; Bluszcz, A.; Rydzanicz, M.; Hus, M.; Ploski, R.; et al. Predictive Significance of Selected Gene Mutations Identified Using Next Generation Sequencing in Relapsed and Refractory Chronic Lymphocytic Leukemia Patients Treated with Ibrutinib. Blood 2019, 134, 5456. [Google Scholar] [CrossRef]
- Brown, J.; Mashima, K.; Fernandes, S.; Naeem, A.; Shupe, S.; Fardoun, R.; Davids, M. Mutations Detected in Real World Clinical Sequencing during BTK Inhibitor Treatment in CLL. Res. Sq. 2024, rs.3.rs-3837426. [Google Scholar] [CrossRef]
- Sun, C.; Mali, R.; Kositsky, R.; Tian, X.; Tomczak, H.; Nuttall, B.; White, R.; Rule, S.; Munugalavadla, V.; Wiestner, A. Extended Follow-up and Resistance Mutations in CLL Patients Treated with Acalabrutinib. Blood 2023, 142, 1891. [Google Scholar] [CrossRef]
- Soverini, S.; Bavaro, L.; De Benedittis, C.; Martelli, M.; Iurlo, A.; Orofino, N.; Sica, S.; Sorà, F.; Lunghi, F.; Ciceri, F.; et al. Prospective Assessment of NGS-Detectable Mutations in CML Patients with Nonoptimal Response: The NEXT-in-CML Study. Blood 2020, 135, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Branford, S.; Shanmuganathan, N. NGS in CML—New Standard Diagnostic Procedure? Hemasphere 2019, 3, 48–50. [Google Scholar] [CrossRef] [PubMed]
- Jean, J.; Sukhanova, M.; Dittmann, D.; Gao, J.; Jennings, L.J. A Novel BCR::ABL1 Variant Detected with Multiple Testing Modalities. Case Rep. Hematol. 2024, 2024, 8486267. [Google Scholar] [CrossRef]
- Cross, N.C.P.; Ernst, T.; Branford, S.; Cayuela, J.-M.; Deininger, M.; Fabarius, A.; Kim, D.D.H.; Machova Polakova, K.; Radich, J.P.; Hehlmann, R.; et al. European LeukemiaNet Laboratory Recommendations for the Diagnosis and Management of Chronic Myeloid Leukemia. Leukemia 2023, 37, 2150–2167. [Google Scholar] [CrossRef]
- Sahoo, T.; Hastie, A.; Ortega, A.; Mylavarapu, A.; Matthews, B.; Hauenstein, J.; Chaubey, A. 91. Atypical BCR::ABL1 Rearrangements Identified by Optical Genome Mapping in Patients with Chronic Myeloid Leukemia. Cancer Genet. 2023, 278–279, 28. [Google Scholar] [CrossRef]
- Crampe, M.; Haslam, K.; Groarke, E.; Kelleher, E.; O’Shea, D.; Conneally, E.; Langabeer, S.E. Chronic Myeloid Leukemia with an E6a2 BCR-ABL1 Fusion Transcript: Cooperating Mutations at Blast Crisis and Molecular Monitoring. Case Rep. Hematol. 2017, 2017, 9071702. [Google Scholar] [CrossRef]
- Molica, M.; Abruzzese, E.; Breccia, M. Prognostic Significance of Transcript-Type BCR—ABL1 in Chronic Myeloid Leukemia. Mediterr. J. Hematol. Infect. Dis. 2020, 12, e2020062. [Google Scholar] [CrossRef]
- Pretzsch, T.; Progscha, S.; Burmeister, T. Diagnostic Ambiguity Caused by an Atypical E18a2 BCR::ABL1 Transcript in a Chronic Myeloid Leukemia Patient. Case Rep. Hematol. 2024, 2024, 9439134. [Google Scholar] [CrossRef]
- Hanfstein, B.; Lauseker, M.; Hehlmann, R.; Saussele, S.; Erben, P.; Dietz, C.; Fabarius, A.; Proetel, U.; Schnittger, S.; Haferlach, C.; et al. Distinct Characteristics of E13a2 versus E14a2 BCR-ABL1 Driven Chronic Myeloid Leukemia under First-Line Therapy with Imatinib. Haematologica 2014, 99, 1441–1447. [Google Scholar] [CrossRef]
- Jinawath, N.; Norris-Kirby, A.; Smith, B.D.; Gocke, C.D.; Batista, D.A.; Griffin, C.A.; Murphy, K.M. A Rare E14a3 (B3a3) BCR-ABL Fusion Transcript in Chronic Myeloid Leukemia: Diagnostic Challenges in Clinical Laboratory Practice. J. Mol. Diagn. 2009, 11, 359–363. [Google Scholar] [CrossRef]
- Leske, I.B.; Hantschel, O. The E13a3 (B2a3) and E14a3 (B3a3) BCR::ABL1 Isoforms Are Resistant to Asciminib. Leukemia 2024, 38, 2041–2045. [Google Scholar] [CrossRef] [PubMed]
- Langabeer, S.E.; McCarron, S.L.; Kelly, J.; Krawczyk, J.; McPherson, S.; Perera, K.; Murphy, P.T. Chronic Myeloid Leukemia with E19a2 BCR-ABL1 Transcripts and Marked Thrombocytosis: The Role of Molecular Monitoring. Case Rep. Hematol. 2012, 2012, 458716. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tutulan-Cunita, A.C.; Chirieac, S.M.; Mocanu, G.; Luca, C.; Costache, M.; Lungeanu, A.; Arghir, A. Variant E19a2 BCR-ABL1 Fusion Transcript in Typical Chronic Myeloid Leukemia. Clin. Lab. 2011, 57, 785–788. [Google Scholar] [PubMed]
- Limsuwanachot, N.; Rerkamnuaychoke, B.; Niparuck, P.; Singdong, R.; Kongruang, A.; Hirunpatrawong, P.; Siriyakorn, T.; Yenchitsomanus, P.; Siriboonpiputtana, T. A Customized Mass Array Panel for BCR::ABL1 Tyrosine Kinase Domain Mutation Screening in Chronic Myeloid Leukemia. J. Mass Spectrom. Adv. Clin. Lab 2023, 28, 122–132. [Google Scholar] [CrossRef]
- Tanneeru, K.; Guruprasad, L. Ponatinib Is a Pan-BCR-ABL Kinase Inhibitor: MD Simulations and SIE Study. PLoS ONE 2013, 8, e78556. [Google Scholar] [CrossRef]
- Dmytrenko, I.; Minchenko, J.; Dyagil, I. Characteristics of BCR/ABL1 Kinase Domain Mutations in the Patients with Chronic Myeloid Leukemia Who Are Primary Resistant to the Imatinib Therapy. Biosystems 2019, 11, 27–33. [Google Scholar] [CrossRef]
- Jabbour, E.; Parikh, S.A.; Kantarjian, H.; Cortes, J. Chronic Myeloid Leukemia—Mechanisms of Resistance and Treatment. Hematol. Oncol. Clin. N. Am. 2011, 25, 981–995. [Google Scholar] [CrossRef]
- Alves, R.; Gonçalves, A.C.; Rutella, S.; Almeida, A.M.; De Las Rivas, J.; Trougakos, I.P.; Sarmento Ribeiro, A.B. Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia—From Molecular Mechanisms to Clinical Relevance. Cancers 2021, 13, 4820. [Google Scholar] [CrossRef]
- Manley, P.W.; Barys, L.; Cowan-Jacob, S.W. The Specificity of Asciminib, a Potential Treatment for Chronic Myeloid Leukemia, as a Myristate-Pocket Binding ABL Inhibitor and Analysis of Its Interactions with Mutant Forms of BCR-ABL1 Kinase. Leuk. Res. 2020, 98, 106458. [Google Scholar] [CrossRef]
- Yan, Z.; Shi, L.; Li, W.; Liu, W.; Galderisi, C.; Spittle, C.; Li, J. A Novel Next-Generation Sequencing Assay for the Identification of BCR::ABL1 Transcript Type and Accurate and Sensitive Detection of TKI-Resistant Mutations. J. Appl. Lab. Med. 2024, 9, 886–900. [Google Scholar] [CrossRef]
- Khorashad, J.S.; Kelley, T.W.; Szankasi, P.; Mason, C.C.; Soverini, S.; Adrian, L.T.; Eide, C.A.; Zabriskie, M.S.; Lange, T.; Estrada, J.C.; et al. BCR-ABL1 Compound Mutations in Tyrosine Kinase Inhibitor–Resistant CML: Frequency and Clonal Relationships. Blood 2013, 121, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Vannuffel, P.; Bavaro, L.; Nollet, F.; Aynaci, A.; Martelli, M.; Devos, H.; De Rop, C.; Soverini, S. Droplet Digital PCR Phasing (DROP-PHASE): A Novel Method for Straightforward Detection of BCR-ABL1 Compound Mutations in Tyrosine Kinase Inhibitors Resistant Chronic Myeloid Leukemia (CML) and Acute Lymphoblastic Leukemia (ALL). Blood 2019, 134, 4660. [Google Scholar] [CrossRef]
- Iqbal, Z.; Akram, A.M.; Akhtar, T.; Khalid, M.; Aziz, Z.; Aleem, A.; Gill, A.T.; Khalid, A.M.; Alanazi, A.; Shah, I.H.; et al. High Frequencies of Compound BCR-ABL Mutations and Their Association with Imatinib Resistant, Disease Progression and Late Chronic Phase Disease in Pakistani Chronic Myeloid Leukemia Patients Necessitate the Inclusion of Molecular Testing in Routine Clinical Settings. Blood 2015, 126, 5167. [Google Scholar] [CrossRef]
- Machova Polakova, K.; Kulvait, V.; Benesova, A.; Linhartova, J.; Klamova, H.; Jaruskova, M.; De Benedittis, C.; Haferlach, T.; Baccarani, M.; Martinelli, G.; et al. Next-Generation Deep Sequencing Improves Detection of BCR-ABL1 Kinase Domain Mutations Emerging under Tyrosine Kinase Inhibitor Treatment of Chronic Myeloid Leukemia Patients in Chronic Phase. J. Cancer Res. Clin. Oncol. 2015, 141, 887–899. [Google Scholar] [CrossRef]
- Branford, S.; Apperley, J.F. Measurable Residual Disease in Chronic Myeloid Leukemia. Haematologica 2022, 107, 2794–2809. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, X.; Zhou, X.; Cheng, Z.; Hu, Y. Exploration of Treatment-free Remission in CML, Based on Molecular Monitoring. Cancer Med. 2023, 13, e6849. [Google Scholar] [CrossRef]
- Bernardi, S.; Cavalleri, A.; Mutti, S.; Garuffo, L.; Farina, M.; Leoni, A.; Iurlo, A.; Bucelli, C.; Toffoletti, E.; Di Giusto, S.; et al. Digital PCR (dPCR) Is Able to Anticipate the Achievement of Stable Deep Molecular Response in Adult Chronic Myeloid Leukemia Patients: Results of the DEMONSTRATE Study. Ann. Hematol. 2025, 104, 207–217. [Google Scholar] [CrossRef]
- Abruzzese, E.; Bocchia, M.; Trawinska, M.M.; Raspadori, D.; Bondanini, F.; Sicuranza, A.; Pacelli, P.; Re, F.; Cavalleri, A.; Farina, M.; et al. Minimal Residual Disease Detection at RNA and Leukemic Stem Cell (LSC) Levels: Comparison of RT-qPCR, d-PCR and CD26+ Stem Cell Measurements in Chronic Myeloid Leukemia (CML) Patients in Deep Molecular Response (DMR). Cancers 2023, 15, 4112. [Google Scholar] [CrossRef]
- Park, H.; Kim, I.; Kim, H.-J.; Shin, D.-Y.; Lee, S.-Y.; Kwon, O.-H.; Kim, D.-Y.; Lee, K.-H.; Ahn, J.-S.; Park, J.; et al. Ultra-Deep Sequencing Mutation Analysis of the BCR/ABL1 Kinase Domain in Newly Diagnosed Chronic Myeloid Leukemia Patients. Leuk. Res. 2021, 111, 106728. [Google Scholar] [CrossRef]
- Grossmann, V.; Wild, M.; Schnittger, S.; Kern, W.; Haferlach, T.; Haferlach, C.; Kohlmann, A. Ultra-Deep Next-Generation Sequencing (NGS) Detects BCR-ABL1 Kinase Domain Mutations with High Sensitivity and Allows to Monitor the Composition of Distinct Subclones During Tyrosine Kinase Inhibitor Treatment. Blood 2010, 116, 2268. [Google Scholar] [CrossRef]
- Talati, C.; Isenalumhe, L.; Kuykendall, A.; Shah, B.D.; Chavez, J.C.; Hussaini, M.O.; Pinilla-Ibarz, J.; Sweet, K.L. Role of Somatic Mutations in Clonal Evolution of Chronic Myeloid Leukemia from Chronic Phase to Blast Phase. Blood 2017, 130, 1587. Available online: https://ashpublications.org/blood/article/130/Supplement%201/1587/79741/Role-of-Somatic-Mutations-in-Clonal-Evolution-of (accessed on 23 March 2025).
- Gotlib, J. How I Treat Atypical Chronic Myeloid Leukemia. Blood 2017, 129, 838–845. [Google Scholar] [CrossRef] [PubMed]
- Piazza, R.; Valletta, S.; Winkelmann, N.; Redaelli, S.; Spinelli, R.; Pirola, A.; Antolini, L.; Mologni, L.; Donadoni, C.; Papaemmanuil, E.; et al. Recurrent SETBP1 Mutations in Atypical Chronic Myeloid Leukemia. Nat. Genet. 2013, 45, 18–24. [Google Scholar] [CrossRef]
- Makishima, H.; Yoshida, K.; Nguyen, N.; Przychodzen, B.; Sanada, M.; Okuno, Y.; Ng, K.P.; Gudmundsson, K.O.; Vishwakarma, B.A.; Jerez, A.; et al. Somatic SETBP1 Mutations in Myeloid Malignancies. Nat. Genet. 2013, 45, 942–946. [Google Scholar] [CrossRef]
- Wang, L.; Du, F.; Zhang, H.-M.; Wang, H.-X. Evaluation of a Father and Son with Atypical Chronic Myeloid Leukemia with SETBP1 Mutations and a Review of the Literature. Braz. J. Med. Biol. Res. 2015, 48, 583–587. [Google Scholar] [CrossRef]
- Makishima, H. Somatic SETBP1 Mutations in Myeloid Neoplasms. Int. J. Hematol. 2017, 105, 732–742. [Google Scholar] [CrossRef]
- Kojima, K.; Ishizawa, J.; Andreeff, M. Pharmacological Activation of Wild-Type P53 in the Therapy of Leukemia. Exp. Hematol. 2016, 44, 791–798. [Google Scholar] [CrossRef]
- Neubauer, A.; He, M.; Schmidt, C.A.; Huhn, D.; Liu, E.T. Genetic Alterations in the P53 Gene in the Blast Crisis of Chronic Myelogenous Leukemia: Analysis by Polymerase Chain Reaction Based Techniques. Leukemia 1993, 7, 593–600. [Google Scholar]
- Ahmadi, S.E.; Rahimian, E.; Rahimi, S.; Zarandi, B.; Bahraini, M.; Soleymani, M.; Safdari, S.M.; Shabannezhad, A.; Jaafari, N.; Safa, M. From Regulation to Deregulation of P53 in Hematologic Malignancies: Implications for Diagnosis, Prognosis and Therapy. Biomark. Res. 2024, 12, 137. [Google Scholar] [CrossRef]
- Sobczyńska-Konefał, A.; Jasek, M.; Karabon, L.; Jaskuła, E. Insights into Genetic Aberrations and Signalling Pathway Interactions in Chronic Lymphocytic Leukemia: From Pathogenesis to Treatment Strategies. Biomark. Res. 2024, 12, 162. [Google Scholar] [CrossRef]
- Zenz, T.; Häbe, S.; Denzel, T.; Mohr, J.; Winkler, D.; Bühler, A.; Sarno, A.; Groner, S.; Mertens, D.; Busch, R.; et al. Detailed Analysis of P53 Pathway Defects in Fludarabine-Refractory Chronic Lymphocytic Leukemia (CLL): Dissecting the Contribution of 17p Deletion, TP53 Mutation, P53-P21 Dysfunction, and miR34a in a Prospective Clinical Trial. Blood 2009, 114, 2589–2597. [Google Scholar] [CrossRef] [PubMed]
- Dufies, M.; Jacquel, A.; Belhacene, N.; Robert, G.; Cluzeau, T.; Luciano, F.; Cassuto, J.P.; Raynaud, S.; Auberger, P. Mechanisms of AXL Overexpression and Function in Imatinib-Resistant Chronic Myeloid Leukemia Cells. Oncotarget 2011, 2, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Sharma, A.; Patne, K.; Tabasum, S.; Suryavanshi, J.; Rawat, L.; Machaalani, M.; Eid, M.; Singh, R.P.; Choueiri, T.K.; et al. AXL Signaling in Cancer: From Molecular Insights to Targeted Therapies. Signal Transduct. Target. Ther. 2025, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Ben-Batalla, I.; Erdmann, R.; Jørgensen, H.; Mitchell, R.; Ernst, T.; von Amsberg, G.; Schafhausen, P.; Velthaus, J.L.; Rankin, S.; Clark, R.E.; et al. Axl Blockade by BGB324 Inhibits BCR-ABL Tyrosine Kinase Inhibitor–Sensitive and -Resistant Chronic Myeloid Leukemia. Clin. Cancer Res. 2017, 23, 2289–2300. [Google Scholar] [CrossRef]
- Neubauer, A.; Burchert, A.; Maiwald, C.; Gruss, H.-J.; Serke, S.; Huhn, D.; Wittig, B.; Liu, E. Recent Progress On the Role of Axl, A Receptor Tyrosine Kinase, in Malignant Transformation of Myeloid Leukemias. Leuk. Lymphoma 1997, 25, 91–96. [Google Scholar] [CrossRef]
- Deininger, M.W.N.; Goldman, J.M.; Melo, J.V. The Molecular Biology of Chronic Myeloid Leukemia. Blood 2000, 96, 3343–3356. [Google Scholar] [CrossRef]
- Morgan, M.A.; Dolp, O.; Reuter, C.W. Cell-Cycle-Dependent Activation of Mitogen-Activated Protein Kinase Kinase (MEK-1/2) in Myeloid Leukemia Cell Lines and Induction of Growth Inhibition and Apoptosis by Inhibitors of RAS Signaling. Blood 2001, 97, 1823–1834. [Google Scholar] [CrossRef][Green Version]
- Shang, Z.-C.; Sun, B.-Z.; Chen, Z.-N.; Wang, W.; Feng, Q.; Wang, S.; Zhang, T. Effects of mitogen-activated protein kinase(MAPK) in cell signal transduction of chronic myelogenous leukemia cell line K562. Ai Zheng 2003, 22, 140–142. [Google Scholar]
- Reuter, C.W.M.; Morgan, M.A.; Bergmann, L. Effect of Mutationally Activated Ras on the Ras to MAP Kinase Signaling Pathway and Growth Inhibition of Myeloid Leukemia Cells by Inhibitors of the MAP Kinase Cascade. In Acute Leukemias VIII; Büchner, T., Hiddemann, W., Wörmann, B., Schellong, G., Ritter, J., Creutzig, U., Eds.; Haematology and Blood Transfusion/Hämatologie und Bluttransfusion; Springer: Berlin/Heidelberg, Germany, 2001; Volume 40, pp. 100–108. ISBN 978-3-642-62109-3. [Google Scholar]
- LeMaistre, A.; Lee, M.; Talpaz, M.; Kantarjian, H.; Freireich, E.; Deisseroth, A.; Trujillo, J.; Stass, S. Ras Oncogene Mutations Are Rare Late Stage Events in Chronic Myelogenous Leukemia. Blood 1989, 73, 889–891. [Google Scholar] [CrossRef]
- Li, N.; Chen, M.; Yin, C.C. Advances in Molecular Evaluation of Myeloproliferative Neoplasms. Semin. Diagn. Pathol. 2023, 40, 187–194. [Google Scholar] [CrossRef]
- Parikh, C.; Subrahmanyam, R.; Ren, R. Oncogenic NRAS, KRAS, and HRAS Exhibit Different Leukemogenic Potentials in Mice. Cancer Res. 2007, 67, 7139–7146. [Google Scholar] [CrossRef] [PubMed]
- Tyner, J.W.; Erickson, H.; Deininger, M.W.N.; Willis, S.G.; Eide, C.A.; Levine, R.L.; Heinrich, M.C.; Gattermann, N.; Gilliland, D.G.; Druker, B.J.; et al. High-Throughput Sequencing Screen Reveals Novel, Transforming RAS Mutations in Myeloid Leukemia Patients. Blood 2009, 113, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Basu, S.; Shah, S.; Bhattacharyya, D.; Gupta, P.P.; Acharjee, M.; Roychoudhury, S.; Nath, S. Deep Sequencing Reveals the Spectrum of BCR-ABL1 Mutations upon Front-Line Therapy Resistance in Chronic Myeloid Leukemia: An Eastern-Indian Cohort Study. Cancer Treat. Res. Commun. 2022, 33, 100635. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Moon, J.H.; Ahn, J.-S.; Kim, Y.-K.; Lee, S.-S.; Ahn, S.-Y.; Jung, S.-H.; Yang, D.-H.; Lee, J.-J.; Choi, S.H.; et al. Next-Generation Sequencing–Based Posttransplant Monitoring of Acute Myeloid Leukemia Identifies Patients at High Risk of Relapse. Blood 2018, 132, 1604–1613. [Google Scholar] [CrossRef]
- Langabeer, S.E.; Haslam, K.; Kelly, J.; Quinn, J.; Morrell, R.; Conneally, E. Targeted Next-Generation Sequencing Identifies Clinically Relevant Mutations in Patients with Chronic Neutrophilic Leukemia at Diagnosis and Blast Crisis. Clin. Transl. Oncol. 2018, 20, 420–423. [Google Scholar] [CrossRef]
- Endrullat, C.; Glökler, J.; Franke, P.; Frohme, M. Standardization and Quality Management in Next-Generation Sequencing. Appl. Transl. Genom. 2016, 10, 2–9. [Google Scholar] [CrossRef]
- Federici, G.; Soddu, S. Variants of Uncertain Significance in the Era of High-Throughput Genome Sequencing: A Lesson from Breast and Ovary Cancers. J. Exp. Clin. Cancer Res. 2020, 39, 46. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to Accessing Data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef]
- Khotskaya, Y.B.; Mills, G.B.; Mills Shaw, K.R. Next-Generation Sequencing and Result Interpretation in Clinical Oncology: Challenges of Personalized Cancer Therapy. Annu. Rev. Med. 2017, 68, 113–125. [Google Scholar] [CrossRef]
- Zhong, J.; Stucky, A.; Sun, F.; Huang, J. PB1800: DEEPDECON: A DEEP-LEARNING METHOD FOR DETECTING MINIMAL RESIDUAL DISEASE OF ACUTE MYELOID LEUKEMIA. Hemasphere 2022, 6, 1680–1681. [Google Scholar] [CrossRef]
- Luh, F.; Yen, Y. FDA Guidance for next Generation Sequencing-Based Testing: Balancing Regulation and Innovation in Precision Medicine. NPJ Genom. Med. 2018, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves NGS-Based Test for Patients With ALL or MM|ASH Clinical News|American Society of Hematology. Available online: https://multiplemyeloma.org.nz/trial-represents-bold-move-for-treatment-of-newly-diagnosed-myeloma-2-2-2-2/ (accessed on 23 March 2025).
- Evans, B.J. The Limits of FDA’s Authority to Regulate Clinical Research Involving High-Throughput DNA Sequencing. Food Drug Law J. 2015, 70, 259–287. [Google Scholar] [PubMed]
- CMS Finalizes Coverage of Next Generation Sequencing Tests, Ensuring Enhanced Access for Cancer Patients|CMS. Available online: https://www.cms.gov/newsroom/press-releases/cms-finalizes-coverage-next-generation-sequencing-tests-ensuring-enhanced-access-cancer-patients (accessed on 23 March 2025).
- Hedblom, A.H.; Pruneri, G.; Quagliata, L.; Costa, J.L.; Dumanois, R.; Rolando, C.; Saunders, R. Cancer Patient Management: Current Use of next-Generation Sequencing in the EU TOP4. J Cancer Policy 2023, 35, 100376. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and Management of AML in Adults: 2022 Recommendations from an International Expert Panel on Behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Béné, M.C.; Grimwade, D.; Haferlach, C.; Haferlach, T.; Zini, G.; European LeukemiaNet. Leukemia Diagnosis: Today and Tomorrow. Eur. J. Haematol. 2015, 95, 365–373. [Google Scholar] [CrossRef]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing–Based Oncology Panels. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef]
- Horgan, D.; Curigliano, G.; Rieß, O.; Hofman, P.; Büttner, R.; Conte, P.; Cufer, T.; Gallagher, W.M.; Georges, N.; Kerr, K.; et al. Identifying the Steps Required to Effectively Implement Next-Generation Sequencing in Oncology at a National Level in Europe. JPM 2022, 12, 72. [Google Scholar] [CrossRef]
- Kaminski, A.; Szamreta, E.A.; Shah, R.; Ning, N.; Aggarwal, J.; Hussain, A.; Adeboyeje, G. Barriers to Next-Generation Sequencing despite Increased Utilization: U.S. Physician Survey Results. JCO 2021, 39, e18754. [Google Scholar] [CrossRef]
- Dhawale, T.; Nardi, V.; Wang, C.; Bard, A.; Essigmann, N.; Leonard, D.; Graubert, T.; Fathi, A.T. Impact of Ultra Rapid Molecular Profiling on Treatment Delays and Healthcare Utilization of Patients Hospitalized for Acute Leukemia. Blood 2024, 144, 5058. [Google Scholar] [CrossRef]
- Llop, M.; Sargas, C.; Barragán, E. The Role of Next-Generation Sequencing in Acute Myeloid Leukemia. Curr. Opin. Oncol. 2022, 34, 723–728. [Google Scholar] [CrossRef]
- Shah, N.P.; Bhatia, R.; Altman, J.K.; Amaya, M.; Begna, K.H.; Berman, E.; Chan, O.; Clements, J.; Collins, R.H.; Curtin, P.T.; et al. Chronic Myeloid Leukemia, Version 2.2024, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2024, 22, 43–69. [Google Scholar] [CrossRef]
- Said, B.; Gilles, S.; Weisdorf, D.; Rashidi, A. The Role of Bone Marrow Morphology in the Diagnosis of Relapsed Acute Myeloid Leukemia. Leukemia 2018, 32, 1052. [Google Scholar] [CrossRef]
- Szamreta, E.A.; Kaminski, A.; Shah, R.; Ning, N.; Aggarwal, J.; Hussain, A.; Adeboyeje, G. Survey Study of Barriers to Evidence-Based Decision-Making in Oncology Care Using next-Generation Sequencing. JCO 2021, 39, e18757. [Google Scholar] [CrossRef]
- Akhoundova, D.; Rubin, M.A. Clinical Application of Advanced Multi-Omics Tumor Profiling: Shaping Precision Oncology of the Future. Cancer Cell 2022, 40, 920–938. [Google Scholar] [CrossRef] [PubMed]
- Ha, K.C.; Lalonde, E.; Li, L.; Cavallone, L.; Natrajan, R.; Lambros, M.B.; Mitsopoulos, C.; Hakas, J.; Kozarewa, I.; Fenwick, K.; et al. Identification of Gene Fusion Transcripts by Transcriptome Sequencing in BRCA1-Mutated Breast Cancers and Cell Lines. BMC Med. Genom. 2011, 4, 75. [Google Scholar] [CrossRef]
- Lilljebjörn, H.; Orsmark-Pietras, C.; Mitelman, F.; Hagström-Andersson, A.; Fioretos, T. Transcriptomics Paving the Way for Improved Diagnostics and Precision Medicine of Acute Leukemia. Semin. Cancer Biol. 2022, 84, 40–49. [Google Scholar] [CrossRef]
- Hou, S.; Liu, J.; Zhu, Y. Multi-Omics Advances for Molecular Characterization, Precision Medicine, and Prognostic Implications in Leukemia. Cell Investig. 2025, 1, 100007. [Google Scholar] [CrossRef]
- Casado, P.; Cutillas, P.R. Proteomic Characterization of Acute Myeloid Leukemia for Precision Medicine. Mol. Cell. Proteom. 2023, 22, 100517. [Google Scholar] [CrossRef]
- Casado, P.; Wilkes, E.H.; Miraki-Moud, F.; Hadi, M.M.; Rio-Machin, A.; Rajeeve, V.; Pike, R.; Iqbal, S.; Marfa, S.; Lea, N.; et al. Proteomic and Genomic Integration Identifies Kinase and Differentiation Determinants of Kinase Inhibitor Sensitivity in Leukemia Cells. Leukemia 2018, 32, 1818–1822. [Google Scholar] [CrossRef]
- Bao, Y.; Qiao, J.; Gong, W.; Zhang, R.; Zhou, Y.; Xie, Y.; Xie, Y.; He, J.; Yin, T. Spatial Metabolomics Highlights Metabolic Reprogramming in Acute Myeloid Leukemia Mice through Creatine Pathway. Acta Pharm. Sin. B 2024, 14, 4461–4477. [Google Scholar] [CrossRef]
- Shafiei, F.S.; Abroun, S.; Vahdat, S.; Rafiee, M. Omics Approaches: Role in Acute Myeloid Leukemia Biomarker Discovery and Therapy. Cancer Genet. 2025, 292–293, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhao, H.; Wang, R.; Chen, Y.; Ouyang, X.; Li, W.; Sun, Y.; Peng, A. Cancer Epigenetics: From Laboratory Studies and Clinical Trials to Precision Medicine. Cell Death Discov. 2024, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Metzeler, K.H.; Walker, A.; Geyer, S.; Garzon, R.; Klisovic, R.B.; Bloomfield, C.D.; Blum, W.; Marcucci, G. DNMT3A Mutations and Response to the Hypomethylating Agent Decitabine in Acute Myeloid Leukemia. Leukemia 2012, 26, 1106–1107. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Shen, K.; Xiao, M. TET2 Mutation in Acute Myeloid Leukemia: Biology, Clinical Significance, and Therapeutic Insights. Clin. Epigenet. 2024, 16, 155. [Google Scholar] [CrossRef]
- Wang, T.; Cui, S.; Lyu, C.; Wang, Z.; Li, Z.; Han, C.; Liu, W.; Wang, Y.; Xu, R. Molecular Precision Medicine: Multi-Omics-Based Stratification Model for Acute Myeloid Leukemia. Heliyon 2024, 10, e36155. [Google Scholar] [CrossRef]
- Kosvyra, A.; Karadimitris, A.; Papaioannou, Μ.; Chouvarda, I. Machine Learning and Integrative Multi-Omics Network Analysis for Survival Prediction in Acute Myeloid Leukemia. Comput. Biol. Med. 2024, 178, 108735. [Google Scholar] [CrossRef]
- Mohr, A.E.; Ortega-Santos, C.P.; Whisner, C.M.; Klein-Seetharaman, J.; Jasbi, P. Navigating Challenges and Opportunities in Multi-Omics Integration for Personalized Healthcare. Biomedicines 2024, 12, 1496. [Google Scholar] [CrossRef]
- Tsagiopoulou, M.; Gut, I.G. Machine Learning and Multi-Omics Data in Chronic Lymphocytic Leukemia: The Future of Precision Medicine? Front. Genet. 2024, 14, 1304661. [Google Scholar] [CrossRef]
- Abbasi, E.Y.; Deng, Z.; Ali, Q.; Khan, A.; Shaikh, A.; Reshan, M.S.A.; Sulaiman, A.; Alshahrani, H. A Machine Learning and Deep Learning-Based Integrated Multi-Omics Technique for Leukemia Prediction. Heliyon 2024, 10, e25369. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Flach, J.; Shumilov, E.; Wiedemann, G.; Porret, N.; Shakhanova, I.; Bürki, S.; Legros, M.; Joncourt, R.; Pabst, T.; Bacher, U. Clinical Potential of Introducing Next-Generation Sequencing in Patients at Relapse of Acute Myeloid Leukemia. Hematol. Oncol. 2020, 38, 425–431. [Google Scholar] [CrossRef]
- Chanan-Khan, A.A.; Challagulla, S.; Kaushiva, A.; Miller, V.; Wang, Y.; Smith, H.; Yang, K. Impact of Real-World Treatment Sequencing Patterns on Time to Next Treatment among Patients with Chronic Lymphocytic Leukemia in the United States. Blood 2023, 142, 5143. [Google Scholar] [CrossRef]
- McGowan, P.F.; Hyter, S.D.; Cui, W.; Plummer, R.M.; Godwin, A.K.; Zhang, D. Comparison of Flow Cytometry and Next-Generation Sequencing in Minimal Residual Disease Monitoring of Acute Myeloid Leukemia: One Institute’s Practical Clinical Experience. Int. J. Lab. Hematol. 2022, 44, 118–126. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.; Jiang, M.; Hou, H.; Gao, Y.; Li, Y.; Wang, F.; Wang, J.; Peng, K.; Liu, Y.-X. Nanopore Sequencing: Flourishing in Its Teenage Years. J. Genet. Genom. 2024, 51, 1361–1374. [Google Scholar] [CrossRef]
- Wagner, U.; Wong, C.; Camenisch, U.; Zimmermann, K.; Rechsteiner, M.; Valtcheva, N.; Theocharides, A.; Widmer, C.C.; Manz, M.G.; Moch, H.; et al. Comprehensive Validation of Diagnostic Next-Generation Sequencing Panels for Acute Myeloid Leukemia Patients. J. Mol. Diagn. 2022, 24, 935–954. [Google Scholar] [CrossRef]
- Doostparast Torshizi, A.; Wang, K. Next-Generation Sequencing in Drug Development: Target Identification and Genetically Stratified Clinical Trials. Drug Discov. Today 2018, 23, 1776–1783. [Google Scholar] [CrossRef]
- Hux, A.; Lewis, A.; Sachwitz, D.; Gregory, T. Clinical Utility of Next-Generation Sequencing in Precision Oncology. JAAPA 2019, 32, 35–39. [Google Scholar] [CrossRef]
- Duan, X.-P.; Qin, B.-D.; Jiao, X.-D.; Liu, K.; Wang, Z.; Zang, Y.-S. New Clinical Trial Design in Precision Medicine: Discovery, Development and Direction. Signal Transduct. Target. Ther. 2024, 9, 57. [Google Scholar] [CrossRef]
- Venugopal, S.; Watts, J. The Future Paradigm of HMA + VEN or Targeted Inhibitor Approaches: Sequencing or Triplet Combinations in AML Therapy. Hematology 2023, 2023, 192–197. [Google Scholar] [CrossRef]
- Dhawan, D. Clinical Next-Generation Sequencing. In Progress and Challenges in Precision Medicine; Elsevier: Amsterdam, The Netherlands, 2017; pp. 35–54. ISBN 978-0-12-809411-2. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).