Survey of Potyviruses, Carlaviruses, and Begomoviruses in Potato Cultivation Centers of West, Central, and East Java Provinces, Indonesia
Abstract
1. Introduction
2. Materials and Methods
2.1. Fields Survey
2.2. Molecular Identification
2.3. Phylogeny and Percentage Identity Analyses
3. Results
3.1. Fields Survey
3.2. Molecular Identification
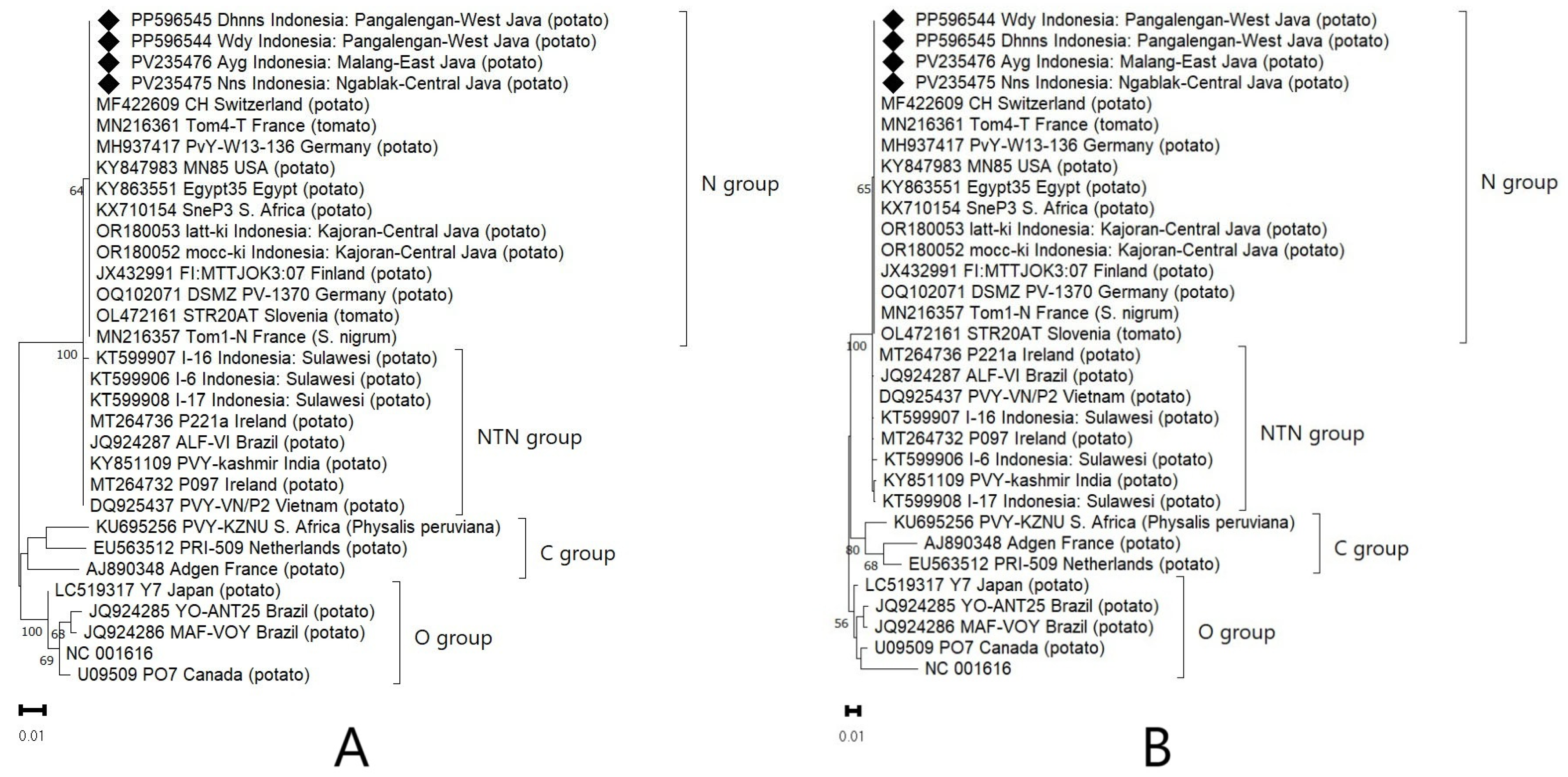
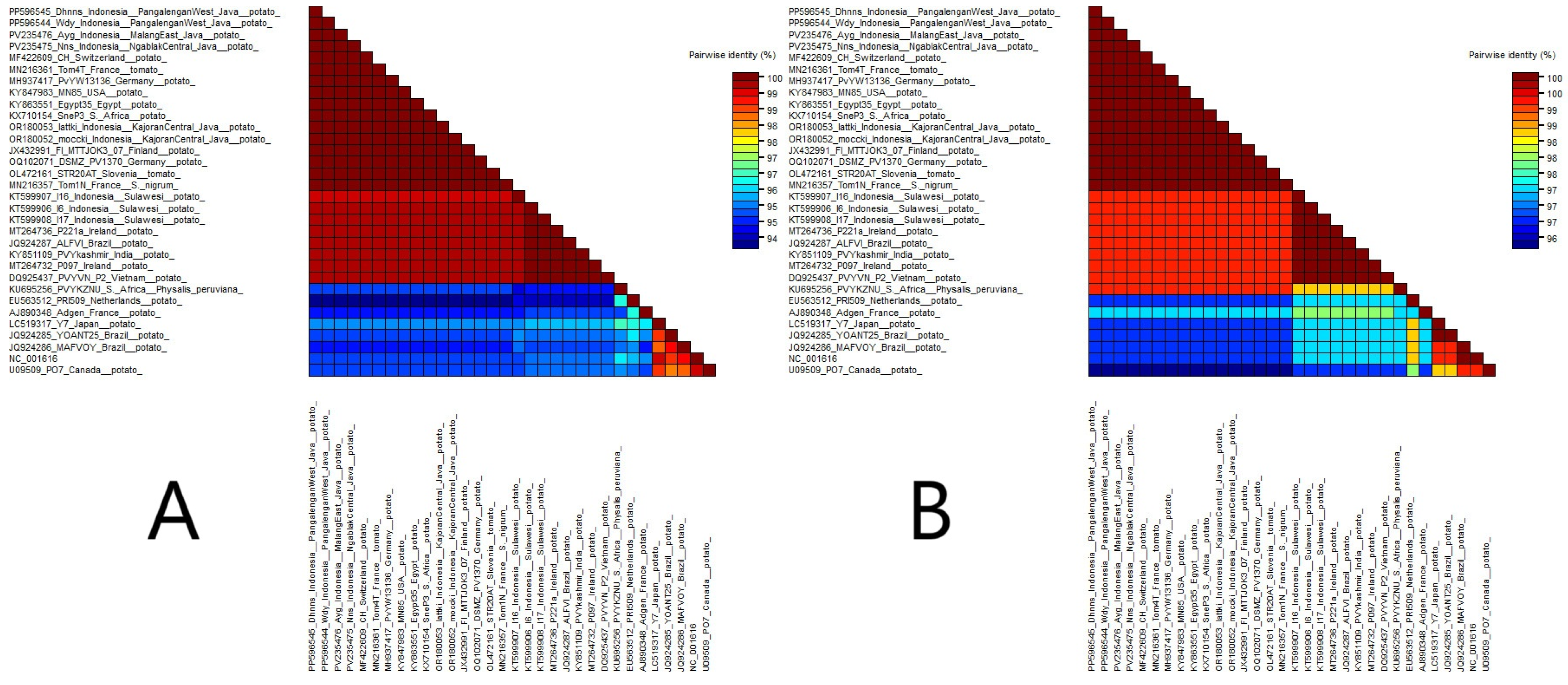
3.3. Phylogeny and Percentage Identity Analyses
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CP | coat protein |
| nt | nucleotide |
| aa | amino acid |
| MEGA | Molecular Evolutionary Genetics Analysis |
| SDT | Sequence Demarcation Tool |
Appendix A
| No. | Accession No. | Isolate Name | Origin | Host | Strain |
|---|---|---|---|---|---|
| 1. | PP596544 | Wdy | Indonesia: West Java | potato | N |
| 2. | PP596545 | Dhnns | Indonesia: West Java | potato | N |
| 3. | PV235476 | Ayg | Indonesia: East Java | potato | N |
| 4. | PV235475 | Nns | Indonesia: Central Java | potato | N |
| 5. | MF422609 | CH | Switzerland | potato | N |
| 6. | MN216361 | Tom4-T | France | tomato | N |
| 7. | MH937417 | PvY-W13-136 | Germany | potato | N |
| 8. | KY847983 | MN85 | USA | potato | N |
| 9. | KY863551 | Egypt35 | Egypt | potato | N |
| 10. | KX710154 | SneP3 | South Africa | potato | N |
| 11. | OR180053 | latt-ki | Indonesia: Central Java | potato | N |
| 12. | OR180052 | mocc-ki | Indonesia: Central Java | potato | N |
| 13. | JX432991 | FI:MTTJOK3:07 | Finland | potato | N |
| 14. | OQ102071 | DSMZ PV-1370 | Germany | potato | N |
| 15. | MN216357 | Tom1-N | France | Solanum nigrum | N |
| 16. | OL472161 | STR20AT | Slovenia | tomato | N |
| 17. | KY851109 | PVY-kashmir | India | potato | NTN |
| 18. | MT264736 | P221a | Ireland | potato | NTN |
| 19. | JQ924287 | ALF-VI | Brazil | potato | NTN |
| 20. | DQ925437 | PVY-VN/P2 | Vietnam | potato | NTN |
| 21. | MT264732 | P097 | Ireland | potato | NTN |
| 22. | KT599906 | I-6 | Indonesia: Sulawesi | potato | NTN |
| 23. | KT599907 | I-16 | Indonesia: Sulawesi | potato | NTN |
| 24. | KT599908 | I-17 | Indonesia: Sulawesi | potato | NTN |
| 25. | JQ924285 | YO-ANT25 | Brazil | potato | O |
| 26. | U09509 | PO7 | Canada | potato | O |
| 27. | JQ924286 | MAF-VOY | Brazil | potato | O |
| 28. | NC 001616 | - | - | - | O |
| 29. | LC519317 | Y7 | Japan | potato | O |
| 30. | KU695256 | PVY-KZNU | South Africa | Physalis peruviana | C |
| 31. | AJ890348 | Adgen | France | potato | C |
| 32. | EU563512 | PRI-509 | Netherlands | potato | C |
References
- Banjarnahor, D.R.V. Faba bean: A promising crop for realizing a healthier potato cropping system in the Dieng highlands. Biota J. Ilm. Ilmu-Ilmu Hayati 2017, 1, 98–102. [Google Scholar] [CrossRef]
- Damayanti, T.A.; Alabi, O.J.; Hidayat, S.H.; Crosslin, J.M.; Naidu, R.A. First report of Potato virus Y in potato in West Java, Indonesia. Plant Dis. 2014, 98, 287. [Google Scholar] [CrossRef]
- Santosa, A.I.; Irbati, A.H.; Dharma, K.S.; Winona, B.; Jaya, R.S.; Andriyani, A.L.; A’yun, C.Q. Potato virus y and shallot latent virus of Kajoran horticultural production center, Magelang Regency, Indonesia: Molecular characterization case study. Caraka Tani J. Sustain. Agric. 2024, 39, 1–9. [Google Scholar] [CrossRef]
- Fuentes, S.; Jones, R.A.C.; Matsuoka, H.; Ohshima, K.; Kreuze, J.; Gibbs, A.J. Potato virus Y; the Andean connection. Virus Evol. 2019, 5, vez037. [Google Scholar] [CrossRef]
- Green, K.J.; Brown, C.J.; Gray, S.M.; Karasev, A.V. Phylogenetic study of recombinant strains of Potato virus Y. Virology 2017, 507, 40–52. [Google Scholar] [CrossRef]
- Scholthof, K.-B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Dupuis, B.; Nkuriyingoma, P.; Ballmer, T. Economic Impact of Potato Virus Y (PVY) in Europe. Potato Res. 2023, 67, 55–72. [Google Scholar] [CrossRef]
- Mastura, F.; Hannum, S.; Elimasni. Deteksi Virus Potato Virus Y (PVY) Pada Tanaman Kentang di Tanah Karo, Sumatera Utara Menggunakan Teknik Molekuler (in Bahasa Indonesia). Bachelor’s Thesis, Universitas Sumatera Utara, Medan, Indonesia, 2018. [Google Scholar]
- Fitriyati, S.S.; Damayanti, T.A.; Mutaqin, K.H. Penyakit Kerdil Pada Kentang di Jawa Tengah: Identifikasi Dan Taksasi Kehilangan Hasilnya (in Bahasa Indonesia). Bachelor’s Thesis, IPB University, Bogor, Indonesia, 2019. Available online: https://repository.ipb.ac.id/handle/123456789/97154 (accessed on 11 May 2025).
- Chikh-Ali, M.; Bosque-Pérez, N.A.; Vander Pol, D.; Sembel, D.; Karasev, A.V. Occurrence and molecular characterization of recombinant Potato virus Y NTN isolates from Sulawesi, Indonesia. Plant Dis. 2016, 100, 269–275. [Google Scholar] [CrossRef]
- Damayanti, T.A.; Kartika, R. Deteksi virus-virus pada kentang di Jawa Barat dengan menggunakan teknik molekuler (in Bahasa Indonesia). J. Hortik. 2015, 25, 171–179. [Google Scholar] [CrossRef]
- Wahyono, A.; Murti, R.H.; Hartono, S.; Nuringtyas, T.R.; Wijonarko, A.; Mulyantoro, M.; Firmansyah, D.; Afifuddin, A.; Purnama, I.C.G. Current status and complexity of three Begomovirus species in pepper plants in lowlands and highlands in Java Island, Indonesia. Viruses 2023, 15, 1278. [Google Scholar] [CrossRef]
- Santosa, A.I.; Somowiyarjo, S. Ageratum yellow vein alphasatellite and tomato leaf curl Java betasatellite association with begomoviruses infecting crops and weeds in Indonesia. Plant Prot. Sc. 2023, 59, 317–324. [Google Scholar] [CrossRef]
- Morales, F.J. History and current distribution of begomoviruses in Latin America. Adv. Virus Res. 2006, 67, 127–162. [Google Scholar] [CrossRef]
- Romay, G.; Chirinos, D.T.; Geraud-Pouey, F.; Torres, M.; Bragard, C. First report of Potato yellow mosaic virus infecting Solanum americanum in Venezuela. New Dis. Rep. 2016, 34, 20. [Google Scholar] [CrossRef]
- Çelik, A.; Santosa, A.I. Impact of coat protein on evolution of ilarviruses. In Current Topics in Membranes, 1st ed.; Andrade, L., Kushmerick, C., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2024; Volume 93, pp. 75–84. [Google Scholar] [CrossRef]
- Revill, P.A.; Ha, C.V.; Porchun, S.C.; Vu, M.T.; Dale, J.L. The complete nucleotide sequence of two distinct geminiviruses infecting cucurbits in Vietnam. Arch. Virol. 2003, 148, 1523–1541. [Google Scholar] [CrossRef]
- Langeveld, S.A.; Dore, J.-M.; Memelink, J.; Derks, A.F.L.M.; van der Vlugt, C.I.M.; Asjes, C.J.; Bol, J.F. Identification of potyviruses using the polymerase chain reaction with degenerate primers. J. Gen. Virol. 1991, 72, 1531–1541. [Google Scholar] [CrossRef]
- Cremer, J.; Campbell, P.; Steele, V.; Persley, D.; Thomas, J.; Harper, S.; Gambley, C. Detection and distribution of viruses infecting garlic crops in Australia. Plants 2021, 10, 1013. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Wardani, T.P.; Hartono, S.; Sulandari, S.; Somowiyarjo, S. Double infections of Rehmannia mosaic virus and Potato virus Y on tobacco plants in Central Java and Special Region of Yogyakarta. J. Perlindungan Tanam. Indones. 2021, 25, 133–140. [Google Scholar] [CrossRef]
- Mehle, N.; Kutnjak, D.; Tušek Žnidarič, M.; Ravnikar, M. First report of Apium Virus Y and Carrot Thin Leaf Virus in parsley in Slovenia. Plant Dis. 2019, 103, 592. [Google Scholar] [CrossRef]
- Kinoti, W.M.; Moran, J.R.; Gambley, C.; Rodoni, B.C.; Constable, F.E. Genome characterization of two Carrot Virus Y isolates from Australia. Microbiol. Resour. Announc. 2020, 9, e00096-20. [Google Scholar] [CrossRef]
- Karanfil, A.; Sarı, M.; Randa-Zelyüt, F.; Santosa, A.I.; Korkmaz, S. First report of Apium virus Y infecting Petroselinum crispum in Turkey. New Dis. Rep. 2022, 45, e12059. [Google Scholar] [CrossRef]
- Randa-Zelyüt, F.; Karanfil, A.; Sarı, M.; Korkmaz, S. First report of natural infection of wild carrot by Watermelon mosaic virus in Turkey. New Dis. Rep. 2022, 46, e12107. [Google Scholar] [CrossRef]
- Pereira-Silva, J.; Boiteux, L.S.; Fonseca, M.E.N.; Reis, L.N.A.; Souza, A.S.; Nery, F.M.B.; Madeira, N.R.; Pereira-Carvalho, R.C. Novel natural hosts of tomato severe rugose virus (ToSRV) in the Fabaceae, Solanaceae, and Oxalidaceae families. J. Plant Dis. Prot. 2022, 129, 425–431. [Google Scholar] [CrossRef]
- Souza-Dias, J.A.C.; Sawazaki, H.E.; Pernambuco-Fo, P.C.A.; Elias, L.M.; Maluf, H. Tomato severe rugose virus: Another begomovirus causing leaf deformation and mosaic symptoms on potato in Brazil. Plant Dis. 2008, 92, 487. [Google Scholar] [CrossRef]
- Onditi, J.; Nyongesa, M.; van der Vlugt, R. Screening for PVYN-Wi resistance in Kenyan potato cultivars. Potato Res. 2021, 64, 469–488. [Google Scholar] [CrossRef]
- Lal, M.; Sharma, S.; Yadav, S.; Kumar, S. Management of late blight of potato. In Potato-From Incas to All Over the World, 1st ed.; Yildiz, M., Ed.; IntechOpen Limited: London, UK, 2018; pp. 83–106. [Google Scholar] [CrossRef]
- Ambarwati, A.D.; Kusmana, K.; Listanto, E. Klon-klon kentang transgenik hasil persilangan terseleksi tahan terhadap penyakit hawar daun Phytophthora infestans tanpa penyemprotan fungisida di empat lapangan uji terbatas (in Bahasa Indonesia). J. Biol. Indones. 2015, 11, 177–186. [Google Scholar]
- Kurniawan, H.; Sulastrini, I.; Suganda, T. Uji ketahanan klon kentang hasil pesilangan Atlantic x Repita terhadap penyakit hawar daun Phytophthora infestans (in Bahasa Indonesia). Agrikultura 2018, 29, 100–104. [Google Scholar] [CrossRef]
- Akbaş, B.; Morca, A.F.; Coşkan, S.; Santosa, A.I.; Çulal-Kılıç, H.; Çelik, A. First complete sequences and genetic variation of plum pox virus T strain in Prunus dulcis and Prunus cerasus. 3 Biotech 2023, 13, 332. [Google Scholar] [CrossRef]
- Coşkan, S.; Morca, A.F.; Akbaş, B.; Çelik, A.; Santosa, A.I. Comprehensive surveillance and population study on plum pox virus in Ankara Province of Turkey. J. Plant Dis. Prot. 2022, 129, 981–991. [Google Scholar] [CrossRef]
- da Silva, W.; Kutnjak, D.; Xu, Y.; Xu, Y.; Giovannoni, J.; Elena, S.F.; Gray, S. Transmission modes affect the population structure of potato virus Y in potato. PLoS Pathog. 2020, 16, e1008608. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santosa, A.I.; Wulandari, R.; Vadilah, M.N.; Sabila, E.; Kusuma, A.F.; Mulyadi, D.; Berlian, I.; Wangi, M.G.P.; Sutejo, A.M.; Çelik, A. Survey of Potyviruses, Carlaviruses, and Begomoviruses in Potato Cultivation Centers of West, Central, and East Java Provinces, Indonesia. Int. J. Plant Biol. 2025, 16, 65. https://doi.org/10.3390/ijpb16020065
Santosa AI, Wulandari R, Vadilah MN, Sabila E, Kusuma AF, Mulyadi D, Berlian I, Wangi MGP, Sutejo AM, Çelik A. Survey of Potyviruses, Carlaviruses, and Begomoviruses in Potato Cultivation Centers of West, Central, and East Java Provinces, Indonesia. International Journal of Plant Biology. 2025; 16(2):65. https://doi.org/10.3390/ijpb16020065
Chicago/Turabian StyleSantosa, Adyatma Irawan, Rossa Wulandari, Meyrin Novia Vadilah, Erlin Sabila, Asista Fatma Kusuma, Dedi Mulyadi, Intan Berlian, Made Getas Pudak Wangi, Ade Mahendra Sutejo, and Ali Çelik. 2025. "Survey of Potyviruses, Carlaviruses, and Begomoviruses in Potato Cultivation Centers of West, Central, and East Java Provinces, Indonesia" International Journal of Plant Biology 16, no. 2: 65. https://doi.org/10.3390/ijpb16020065
APA StyleSantosa, A. I., Wulandari, R., Vadilah, M. N., Sabila, E., Kusuma, A. F., Mulyadi, D., Berlian, I., Wangi, M. G. P., Sutejo, A. M., & Çelik, A. (2025). Survey of Potyviruses, Carlaviruses, and Begomoviruses in Potato Cultivation Centers of West, Central, and East Java Provinces, Indonesia. International Journal of Plant Biology, 16(2), 65. https://doi.org/10.3390/ijpb16020065








