3.1. Correlations Between Different Milling, Culinary, and Rice Quality Characteristics
Assessment of the milling quality variables (
Table 1) revealed a wide range of variation among the studied rice varieties, as evidenced by the dispersion measures and intervals corresponding to each attribute. The coefficient of variation (CV) was less than 14% for 83.33% of the quantitative variables analyzed. Notably, the sterility percentage (%S) and disintegration (D) exhibited the highest CV values (71.26% and 149.84%, respectively), effectively distinguishing the ten rice varieties. In contrast, the variables with the lowest variability were husked weight (HW) and polished weight (PW), with CV values of 1.02% and 1.68%, respectively.
As expected, straightforward and positive linear correlations were detected between HW and PW using Pearson’s correlation coefficient (
Table 2). As the HW (g) increased, higher PW values were observed. A strong positive correlation was also observed between the water evaporation (WE) and cooking time (CT). The correlations were the highest and statistically significant (
Table 2).
Negative linear correlations were detected between the polished grain length (PGL), polished grain width (PGW), and %S, with highly significant values of r = −0.87 and r = −0.75, respectively (
Table 2). Additionally, positive correlations were observed between the PGW and %S, with substantial values of r = 0.69.
On the other hand, negative linear correlations were observed between the PGL and PGW, indicating that, as the grain length increases, the width of the polished grains decreases (
Table 2). A negative correlation was also observed between PGL and WE, with r = −0.65.
The correlation indices between the graded weight (GW) and disintegration (D) did not show significant values, possibly due to the high phenotypic variability observed in the analyzed rice materials. In contrast, the amylose content percentage (AC) exhibited a moderate and significant negative correlation with WE, with r = −0.58 (
Table 2). This indicates that a higher WE is associated with a lower AC.
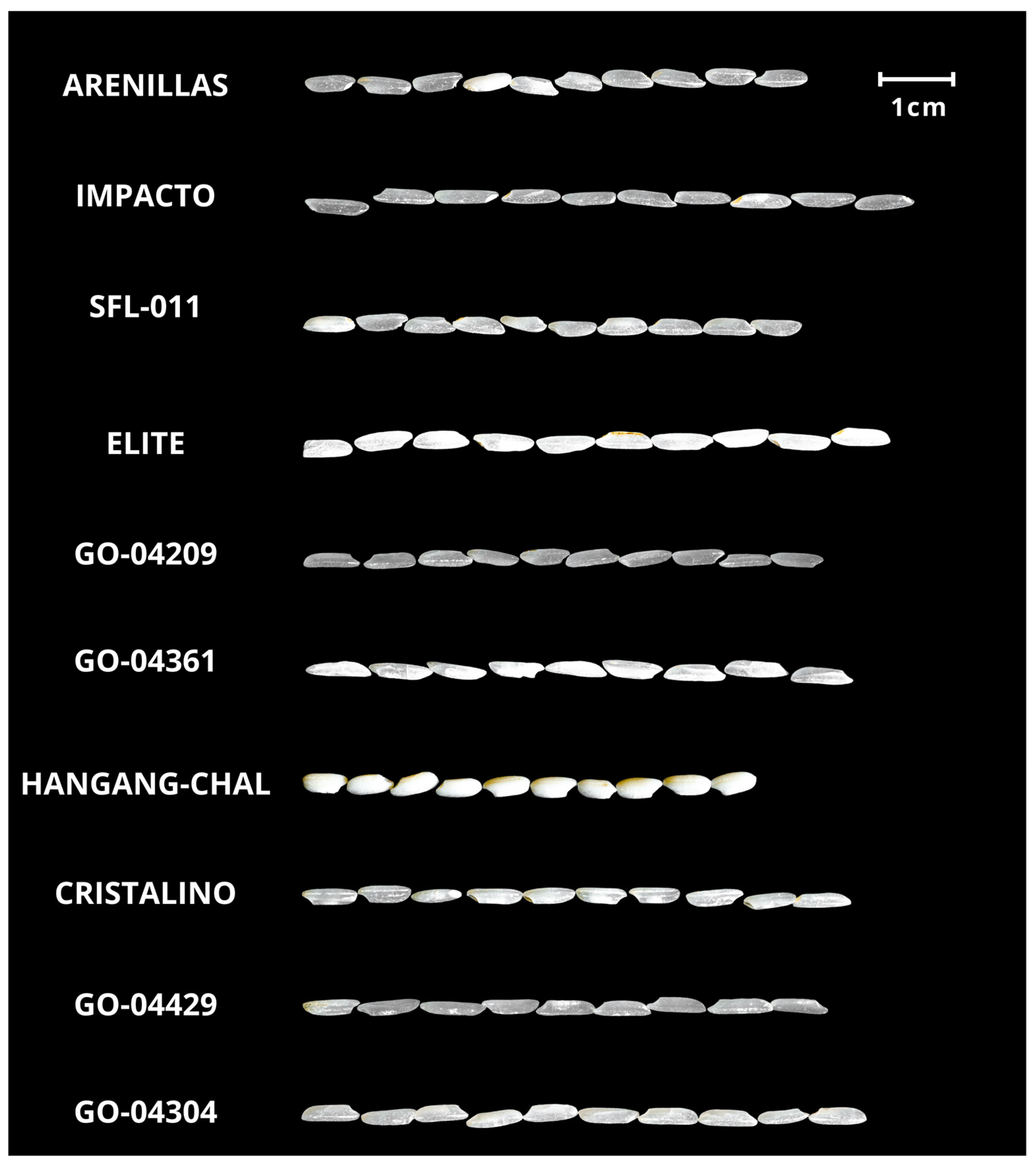
Figure 1 illustrates the variation in the grain size among the different rice varieties and the translucency observed in certain types. Notably, Hangang-Chal (a japonica type variety) stood out for its uniformly white grains and shorter length than other varieties. This highlights the distinctive traits of each cultivar and emphasizes the differences between the indica and japonica varieties, which vary in genetic composition and nutrient content [
33].
3.2. Principal Component Analysis (PCA) of the Spatial Distribution of Groups
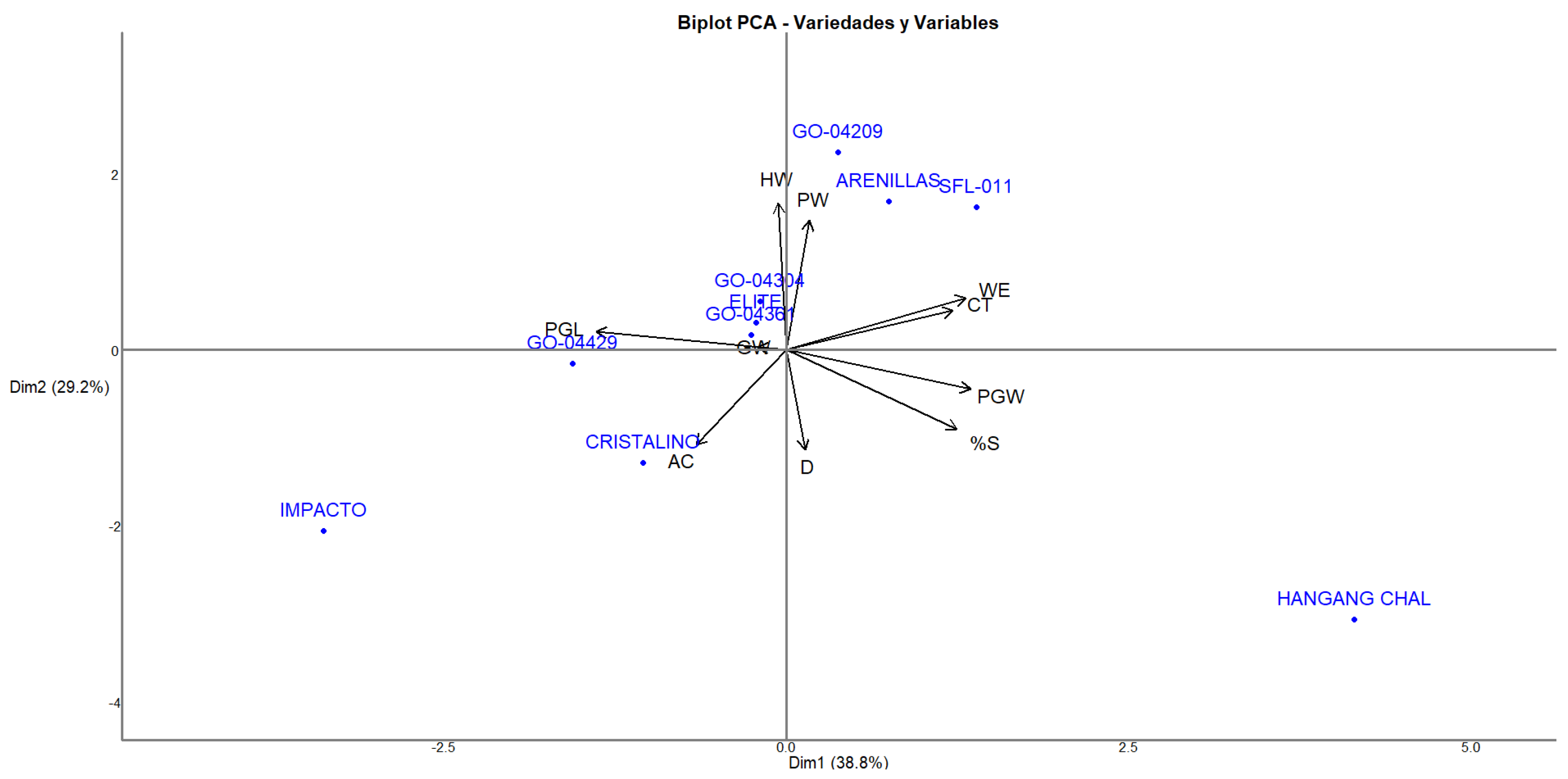
Principal component analysis (PCA) allowed for the identification of the multivariate patterns of variation among the genotypes evaluated, explaining 68.0% of the total variability in the first two dimensions (
Figure 2). The distribution of the genotypes in the different quadrants of the biplot and their relationship with the variables evaluated provides a comprehensive view of their phenotypic profiles of milling and culinary quality.
The distribution observed in the quadrants highlights the usefulness of PCA in identifying extreme or intermediate genotypes. The vector corresponding to amylose content (AC) was oriented toward quadrant II (Dim1 negative and Dim2 positive), indicating that the genotypes projected in that direction had high-amylose content. The vector length suggested a moderate contribution of this variable to genotype differentiation. This was the case for materials such as GO-04209 and GO-04205, which could be considered valuable sources of this trait, and they are relevant for functional properties of the grain and industrial applications. In contrast, the Hangang-Chal genotype in quadrant IV showed a clear separation in the opposite direction, suggesting a low-amylose content, which is a characteristic congruent with its waxy rice profile and is known for its high-amylopectin content.
Principal component analysis (PCA) was performed to reduce the dimensionality of the quantitative variables measured across the ten rice (Oryza sativa L.) varieties. As shown in
Table 3, the first three principal components (PCs) accounted for 80.92% of the total variance, with eigenvalues of 3.88, 2.92, and 1.29, respectively. PC1 alone explained 38.83% of the variation, followed by PC2 (29.16%) and PC3 (12.94%), indicating that these components capture most of the variability present in the dataset. The inclusion of PC4 and PC5 increased the cumulative variance to 95.95%, although their individual contributions were considerably lower. These results justify the use of the first three components for subsequent multivariate analyses, such as biplot interpretation and clustering, as they retain the most informative patterns within the data.
The first component (PC1) was primarily defined by variables related to weight and size, particularly the water evaporation (WE), total classified grain (CT), and reproductive stage, such as sterility percentage (%S). These variables had strong positive loadings on PC1 and are represented by large vectors. Therefore, varieties located in the rightmost region of the biplot, such as Hangang-Chal, were found to be associated with higher values for these descriptors.
The second principal component (PC2) was influenced by grain weight (HW), paddy weight (PW), and, to a lesser extent, polished grain width (PGW). The vectors for these traits pointed upward, indicating that varieties situated higher on the plot, such as Arenillas and SFL-011, exhibited relatively higher values in these variables.
In contrast, Impacto was located at the bottom left of the biplot, opposite the direction of the contributing vectors, suggesting low values across most of the influential variables. Impacto was found to be associated with the polished grain length (PGL) vector, indicating a contrasting phenotypic profile concerning genotypes such as Hangang-Chal.
Similarly, Cristalino appeared in the lower-left quadrant, aligning with negative contributions in both PC1 and PC2. This placement reflects its association with traits such as disintegration (D) and amylose content (AC), which pointed toward the left and slightly downward.
On the other hand, the vector corresponding to the percentage of sterility (%S) was oriented towards quadrant IV, coinciding with the position of Hangang-Chal, which indicated a higher proportion of floral sterility in this genotype. This behavior could be related to its environmental sensitivity or genetic background, influencing the reproductive stability under the conditions evaluated.
The opposite direction between the AC and %S vectors suggested a negative correlation between both variables. A higher amylose content tends to be associated with lower levels of floral sterility. This finding has implications for both the industrial quality and reproductive efficiency of the materials, which are the key aspects of breeding programs.
The clustering pattern observed among the varieties (
Table 4) in the central region, such as GO-04304, GO-04361, GO-04209, and Élite, suggested more balanced or average values across the descriptors, with minimal influence from the extreme values in any single variable. These varieties were aligned near the origin, indicating lower overall variation relative to the main contributing traits, which position them as materials of interest for breeding programs seeking stability.
The PCA effectively distinguished between the rice genotypes based on four significant traits: disintegration, sterility percentage, grain width, and amylose content. PC1 primarily explained the variation associated with the weight- and size-related variables, while PC2 captured the variance driven by the physical grain structure. The PCA biplot confirms that genotypes such as Hangang-Chal and Arenillas occupy opposite regions of the phenotypic space, which is consistent with their distinct performance in the evaluated traits. These findings reinforce using PCA as a robust genotype characterization and selection tool in rice breeding programs [
30,
34].
3.3. Molecular Analysis
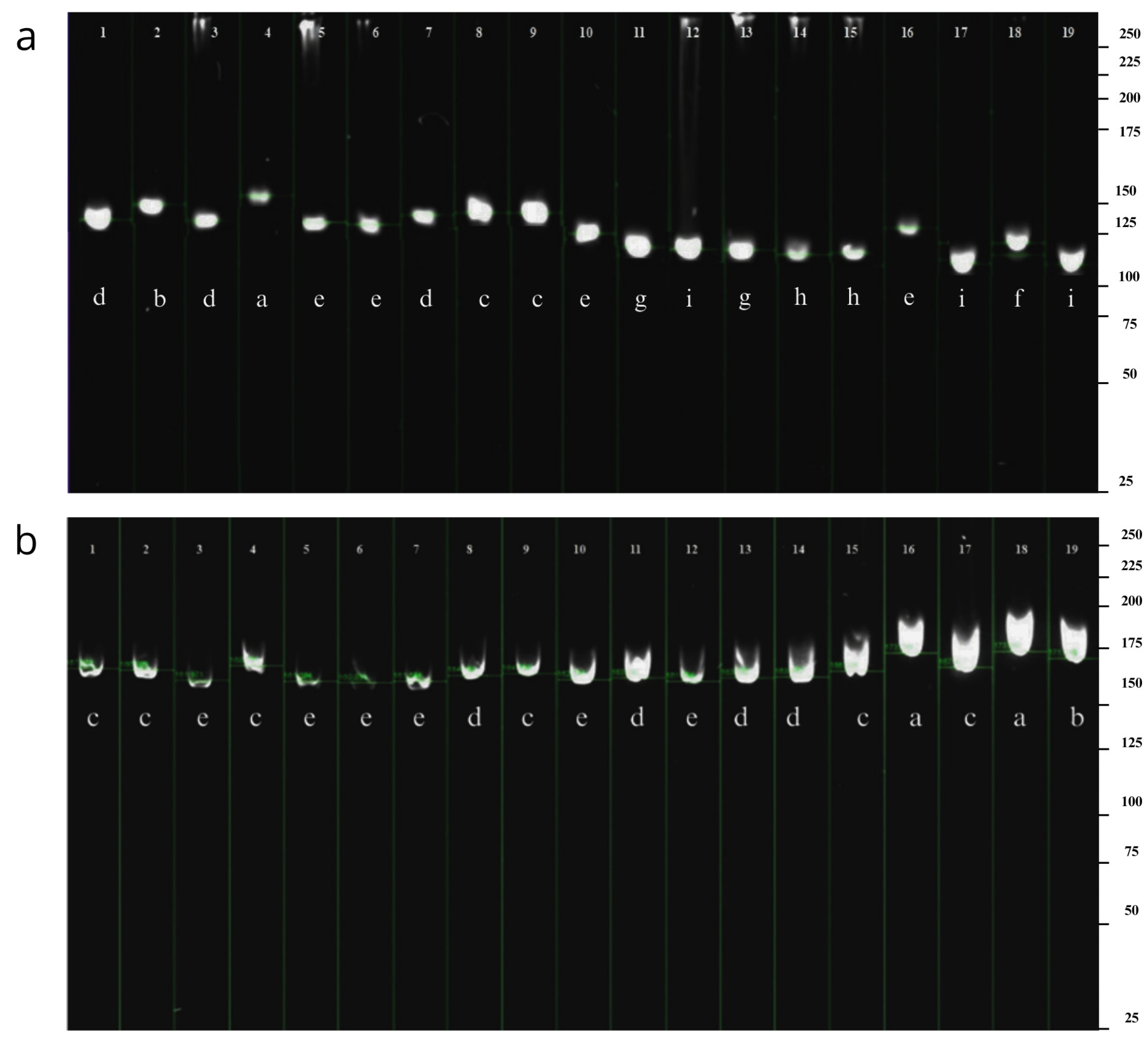
The molecular marker 484-485 exhibited the highest number of alleles, with nine alleles observed and a base pair range of 114–154 bp (
Figure 3). In contrast, the SSR marker 484-W2R displayed fewer waalleles, with five alleles detected on the gel (
Figure 4) and minimum and maximum frequencies ranging from 0.05 to 0.32 (
Table 5).
In the association analysis between the AC in the rice grains and the alleles of each SSR marker, the treatments were grouped into nine alleles for SSR 484-485 and five alleles for SSR 484-W2R using the apparent amylose content percentage as the independent variable in the variance analysis (
Table 6 and
Table 7).
Significant differences were detected between the AC and the molecular marker 484-485 alleles. The coefficient of determination (R
2), explaining 86% of the phenotypic variance in AC percentage, was 0.86 (
Table 6). This result suggests that AC could be considered for potential inclusion in a marker-assisted breeding program using the molecular marker 484-485 to optimize material selection. In contrast, no significant association was observed (
Table 7), between the marker 484-W2R and AC, with an R
2 value of 0.27.
A Tukey multiple comparison test was conducted to determine the correspondence between the 9 alleles of the 484-485 marker (associated with the
waxy gene) and the apparent AC, allowing for the classification of the 19 evaluated materials into their respective AC categories (
Table 8).
Pulut Hitam 9, one of the control varieties, was classified as waxy, while the nursery variety GO-04304 had low-amylose content. Fourteen varieties (73.68%) were classified as having intermediate-amylose percentage (Fedearroz 2000, D-Sativa, GO-04209, GO-04361, Élite, Arenillas, Cristalino, SFL-011, Lemont, GO-04429, Cimarrón, Fedearroz 50, Hangang-Chal, and Impacto). Fonaiap 1, Mahsuri, and Ria were categorized as high-amylose class lines.
They were also associated with alleles 1, 4, 3, 5, 8, and 7 (154, 154, 142, 134, 118, and 110 bp, respectively). Meanwhile, three varieties were classified as high amylose, corresponding to the allele 9 (114 bp).