Abstract
The plant species Cedrela odorata has been largely exploited in the timber industry due to the high demand for its wood. Therefore, C. odorata has been considered a vulnerable species since 1994, under the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). C. odorata is a key timber species included in the management and conservation plans for the Amazon and Central American rainforests. These plans include the development of genetic and genomic resources to study local populations of the species in Colombia. In this study, two novel chloroplast (cp) genomes were assembled and annotated using the MinION long-read sequencing technology. The new cp genomes were screened for sequence variants (SVs), and a total of 16 SNPs were identified, presumably unique to populations from the Amazon region in Colombia. Comparative genomics with other reported cp genomes from different populations of C. odorata support the hypothesis of intraspecific diversity associated with their place of origin. These cp genome sequences of C. odorata from Colombian individuals represent valuable genomic resources for the species, suitable for identifying novel DNA fingerprinting and barcoding applications.
1. Introduction
Cedrela odorata L. is a widely distributed species from the Meliaceae family that produces valuable and multi-purpose timber. However, due to continuous over-exploitation, it is listed as a vulnerable species on the International Union for Conservation of Nature (IUCN) Red List of Threatened Species in parts of the Americas [1] and as a vulnerable species by the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) under Appendix II (https://cites.org/eng/disc/text.php accessed on 12 December 2022). In Colombia, the cedar timber species C. odorata can be found in different environmental gradients, from tropical dry forest to the pre-montane and tropical moist forests [2]. However, natural populations in the country have been reduced by around 80% due to over-exploitation, habitat fragmentation, and the expansion of the agricultural border [3]. Moreover, illegal trading with false documentation of the species and geographical origin is still used for commercial purposes [4].
Genetic fingerprint using molecular markers and comparative genomics with chloroplast DNA (cpDNA) sequences have been performed on timber species, including C. odorata, to understand the population structure, genetic diversity, phylogenetic structure, and harvesting origin, among others [5,6,7,8,9,10,11]. The cp genome is highly conserved in plants in terms of gene composition and arrangement, uniparentally inherited, and its abundance compared to the nuclear DNA allows for the rapid distinction (i.e., assembly and annotation) of sequences [12]. Cp data could increase the resolution and contribute to the development of new molecular markers to be used as specific barcodes [13,14].
The draft cp genome assemblies of eight individuals of C. odorata collected from Central and South America have been reported as the preliminary resources for the phylogeography of the Cedrela species [15]. Although these draft genomes represent a valuable genomic resource for the Cedrela genus, the assembled sequences are highly fragmented, making their use for comparative genomics with their coding sequences difficult. Nevertheless, Mader et al. [13] identified a set of four SNPs in the chloroplast genome among six individuals of four species of the Meliaceae family, including the reference sequence for the cp genome in C. odorata (GenBank accession MG724915.1). These SNPs could be used as molecular markers to discriminate the species form the Meliaceae family.
Long-read Oxford Nanopore Technology (ONT) has been used in a diverse range of applications, including genome assembly [16], building new reference genomes [17,18]. a few reads longer than 10 kb could be sufficient to cover the entire cp genome, offering great potential for population studies [19,20,21]. ONT is ideal for species with few or null genomic information [22], as is the case for most plant species of the Amazon rainforest, including the populations of C. odorata in Colombia.
In this study, two individuals of C. odorata collected from different populations in the Amazon region of Colombia were selected to sequence their cp genomes, using a cheap and fast portable sequencer MinION as a proof of concept. This resulted in the generation of the first genomic resources of C. odorata obtained from two botanical samples of the Colombian Amazon rainforest. Further comparative genomics showed the presence of unique sequence variants (SVs) and supported the cryptic diversity previously reported for the species in the whole continent. These new genomic resources could be adapted for forensic genomics strategies aimed at combating the illegal timber trade in Colombia.
2. Materials and Methods
2.1. Plant Material and High Molecular Weight (HMW) DNA Extraction
Two C. odorata specimens were collected from the Colombian departments of Caquetá and Putumayo. Plant material is available at the Colombian Amazon Herbarium (COAH) with the ID codes OGP2096 and OGP2143. HMW DNA was extracted from fresh leaves, using the protocol reported for Eucalyptus species by Schalamun et al. [23], with minor modifications. An additional clean-up step with phenol/chlorophorm/Isoamyl alcohol (25:24:1) and isopropyl alcohol was added to ensure DNA quality. The DNA concentration was quantified using the QuibitTMDNA kit (Thermo Fisher Scientific, Eugene, OR, USA), and the quality was estimated with a NanoDropTM (Thermo Scenetific, Waltham, MA, USA) by comparing to the 260/280 and 260/230 nm ratios.
2.2. MinION Sequencing
Libraries were prepared according to the ligation library protocol LibPrep for SQK-LSK109 cells with no DNA fragmentation [24]. Long-read sequencing was carried out on a MinION Mk1C sequencer using MinKNOWTM software, release v20.10 [25]. Data were base-called using Albacore, release v2.0.2 (https://github.com/Albacore/albacore, accessed on 29 September 2022). Quality control (QC) steps were performed according to Wang et al. [20]. Adaptors from long reads were removed using Porechop v0.2.1 (https://github.com/rrwick/Porechop, accessed on 22 July 2024), and bases with quality < 9 were trimmed using Nanofilt v1.2.0 [26]. Reads shorter than 3 kb were discarded.
2.3. Chloroplast Read Extraction and Assembly
The bioinformatics pipeline for chloroplast long-read extraction and assembly for Eucalyptus species was used, with some modifications [20]. The long-read data obtained from the whole genome sequencing strategy, previously described, were filtered for non-chloroplast sources, such as the nuclear and mitochondrial genome and other contaminants. Reads were aligned with the C. odorata reference chloroplast (cp) genome MG724915.1 [13], using BLASR software v5.1 [27]. A duplicated and concatenated reference genome was used to ensure the alignment of long reads spanning the point at which the genome was circularized, according to the strategy used by Wang et al. [20] for circular genomes. The assembly of the filtered reads for the OGP2096 individual was performed using Canu v2.1.1 [28] and Hinge v0.4.1 software [29], with default parameters, and assuming > 40× coverage of the reads against the reference genome. Later, the best performing assembler software was used for the OGP2143 individuals.
2.4. Post-Assembly Polishing and Characterization
The assembled contigs were concatenated using MuMmer v3.23 [30] and polished using Racon and Nanopolish v0.8.1 [31,32] software. Both assemblies were manually inspected for duplicate sequences using Geneious Prime 2020.0.5 (http://www.geneious.com/, accessed on 10 October 2022). Genome annotations and circular gene maps of the cp assemblies were generated using the web-based software GeSeq [33]. The program mVISTA [34] was used to compare both cp assemblies using the LAGAN model and C. odorata cp genome MG724915.1 as a reference. Geneious was also used to perform synonymous codon optimization on the protein-coding genes using a codon table retrieved from the Codon Usage Database (https://www.kazusa.or.jp/codon/, accessed on 7 June 2024). Subsequently, relative synonymous codon usage (RSCU) was calculated for both the assemblies and the reference cp genome. R was used to generate a heatmap based on the RSCU analysis.
2.5. Variant Discovery and Phylogenetic Analysis
A diversity panel of Cedrela species was generated by retrieving the chloroplast genome assemblies of 15 individuals, including the reference genome MG724915.1, additional draft assemblies reported by Finch et al. [15], and the chloroplast genomes of C. odorata generated in this study, as follows: 11 C. odorata, one C. fissilis, C. montana, C angustifolia, and C. saltensis (Table 1). These assemblies were chosen for this analysis due to their proximity to collection sites in Colombia or nearby and represent the largest set of genomic resources for the species. Sequence reads from the COAH specimens (OGP2096 and OGP2143) and draft assemblies were aligned to the C. odorata reference genome using BWA-MEM v. 0.7.17 [35].

Table 1.
Chloroplast genomes of Cedrela genus used for phylogenomic analyses.
Sequence alignments were used as inputs for probabilistic variant calling using Snippy software from the Galaxy bioinformatics toolkit, version [36], and the Genome Analysis Toolkit (GATK) v4.2.4.0 [37], including high-stringency variant filtering for coverage, mapping quality, and variant position. SNP filtering was performed with VCFtools v.0.1.17 [38] to remove insertion/deletion variants, sites with greater than 85% of missing data, and more than two alleles. Manual inspection of the genomic regions with SNPs was performed by comparing their positions with the reference genome MG724915.1, using Geneious v.2020.0.5 software (https://www.geneious.com). Phylogenetic relationships were estimated with 1000 bootstrap Bayesian probabilities using software BEAST (Bayesian Evolutionary Analysis Sampling Trees) v 2.6.1 [39], with default settings. Swietenia mahagoni was used as the outgroup [15]. The resulting phylogenetic trees were visualized with the Interactive Tree of Life (iTOL) v4 web-based software [40].
3. Results
3.1. MinION-ONT Whole Genome Sequencing
A total of 2.6 and 0.99 Gb of raw long-read data were generated for OGP2096 and OGP2143, respectively (Table 2). OGP2096 had 594,819 reads, compared to OGP2143, with 110,484 reads. However, longer reads were observed for OGP2143, with a mean read length of 9.6 kb, compared to OGP2096, with a mean length of 4.3 kb (Table 2, Supplementary Figure S1). The mean quality values for both sequenced individuals were 10 and 9.9, respectively.

Table 2.
Statistics of C. odorata raw sequence data and cp genome assembly.
3.2. Chloroplast Genome Assembly
A total of 1437 reads with a mean read length of ~9 kb from individual OGP2096 and 884 reads with a mean length of ~11 kb from individual OGP2143 resulted from filtering out those reads corresponding to the nuclear and mitochondrial genomes. These subsets of reads were used to generate the chloroplast genome assemblies, with a coverage of 80× and 60×, respectively. The assembly of OGP2096 generated by Canu software was fragmented into three contigs of 102 kb (Contig 4), 55 kb (Contig 2), and 23 kb (Contig 3) in length (Supplementary Figure S2A). Contig 2 contains the large single copy region (LSC) and part of the inverted repeats (IRs) of the chloroplast. Contig 4 contains a part of the IRs and a small single copy (SSC), while Contig 3 contains a part of the IRs. Further polishing of duplicated regions resulted in a unique contig of 138 kb that missed part of the IR and LSC regions (Supplementary Figure S2B).
On the other hand, software Hinge showed accurate performance for long reads for the OGP2096 individual, obtaining an assembly of 190 kb in length, fragmented into two contigs of 70 kb (Contig 1) and 120 kb (Contig 2) (Table 2, Supplementary Figure S2C), and a unique Contig of ~157 kb after post-assembly polishing and concatenation with MuMmer. (Table 2, Supplementary Figure S2D). Considering the better performance of the Hinge assembler, this was used to assemble the OGP2143 cp genome. The resulting assembly was fragmented into two contigs of 40 kb and 142 kb, with a total length of 182 kb. After post-assembly polishing analysis, a single contig of 159 kb was obtained (Table 2, Supplementary Figure S2E,F).
3.3. Genome Annotation
These assemblies were manually inspected and used in the prediction of the gene content and functional annotation of the cp genome. Large and short regions that contain single-copy genes (LSC and SSC) and two inverted repeat (IR) regions, 158.57 kb with a GC content of 37.9% in OGP2096, and 158.59 kb with a GC content of 37.46% in OGP2143, were predicted. A total of 132 protein-coding gene models were predicted and annotated for both cp genomes, similar to the reference sequence (Refseq) MG724915.1 (Table 3). The new cp genomes have three genes annotated as pafI, pafII, and pbfI (assembly factors) of the photosystems I and II (Figure 1). However, these genes are homologues to the assembly factors of photosystem I, ycf3 and ycf4, and pbsN of photosystem II, as reported in the Refseq annotations [13], as shown in Figure S3. Additionally, the SSC intergenic regions are larger in the new cp genome assemblies (5 to 7 kb larger, respectively), compared to the Refseq annotations (Table 3, Figure 1). The cp genome assemblies of OGP2096 and OGP2143 individuals were uploaded into the GenBank under accession numbers OP750006 and OP750007, respectively.

Table 3.
Summary of cp genomic regions in C. odorata.
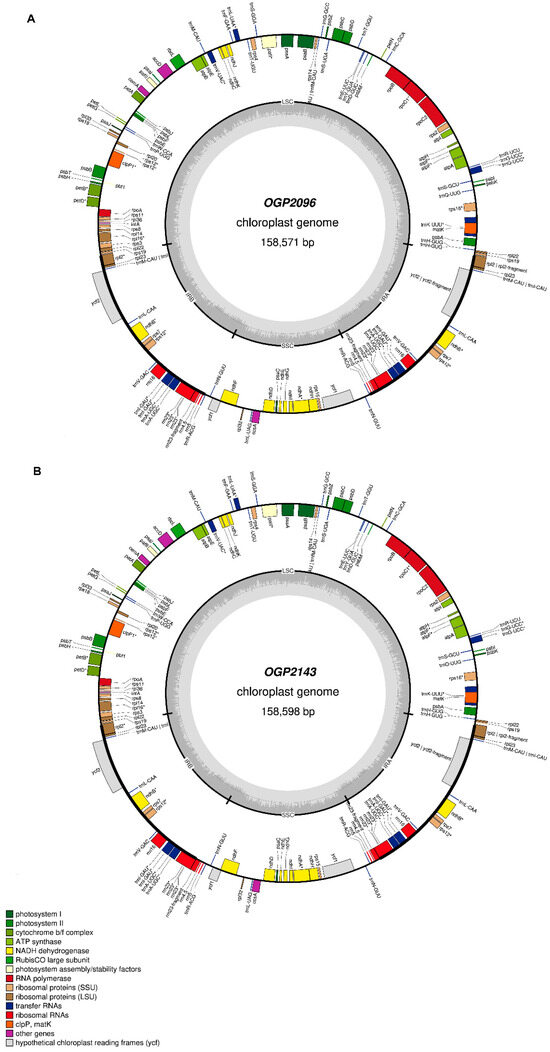
Figure 1.
Graphic representation of the complete genetic map of the chloroplast individuals (A) OGP2096 and (B) OGP2143 sequenced by the long-read MinIon technology. GC content is represented by the inner grey circle. Genes inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise. LSC, large single copy; SSC, small single copy; IR, inverted repeat.
The relative synonymous codon usage frequency was estimated for the protein coding genes of two Cedrela individuals and compared to the RefSeq (Figure 2). Based on the clustering pattern and average RSCU values, two major branches were identified. The smaller branch contains between 22 and 26 codons with an RSCU value higher than one (>1.17) for the OGP2096 and OGP2143 individuals, respectively. Most of the preferred codons (excluding TTG and AGG) ended in A or T for both individuals and the reference, with AGA, encoding arginine (Arg), being strongly preferred (>1.9). The remaining codons, with RSCU values lower than one (34 and 36 codons for OGP2096 and OGP2143 individuals, respectively) are used less frequently than other synonymous codons, while those with values of one are used equally (OGP2096 = 4, OGP2143 = 6) [41]. Based on the RSCU values, the C. odorata cp reference genome and the OGP2096 had the closest values, forming one branch, while the OGP2143 individual formed another branch. In summary, the codon usage patterns were very similar, indicating that the RSCU values for a given codon were comparable across the three individuals evaluated.
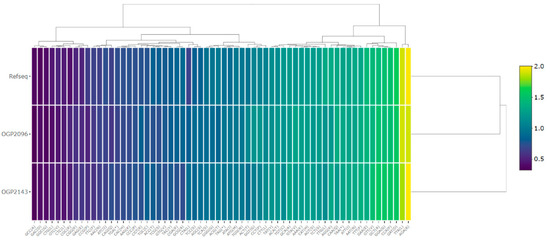
Figure 2.
The RSCU values of two C. odorata individuals and the reference sequence. The RSCU values of 64 codons were used as the basis for tree clustering. The gradient from blue to yellow indicates that the average RSCU value of the codon ranges from low to high.
3.4. Phylogenetics and Structural Variation Analysis
The phylogenetic analysis was performed by aligning the cp genome sequence of the C. odorata individuals OGP2096 and OGP2143 from Colombia, the cp reference genome sequence, and sequences of the Cedrela genus (Table 1), using S. mahagoni as the outgroup. The resulting phylogenetic trees support the monophyletic origin for all taxa of the Cedrela genus, as reported by Finch et al. [15]. The individual OGP2096 grouped in a sister subclade of Cedrela species, closer to the outgrouped C. odorata individual COD202 and other South American individuals (Subclade SA) (Figure 3A). The individual OGP2143 is strongly grouped with the C. monatana individual CEMO50, forming a sister subclade closely related to mostly Central American (Subclade CA) individuals (Figure 3A).
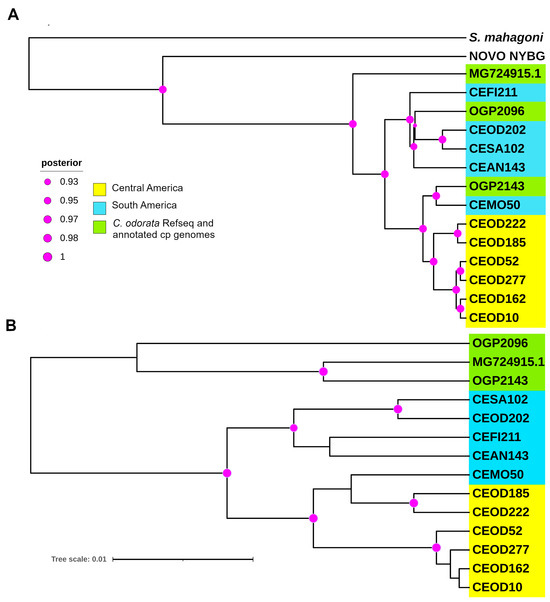
Figure 3.
Phylogenetic analysis using the cp genome of individuals from the Cedrela genus reported by Mader et al. [13], Finch et al. [15], and this study. (A) Tree generated from the alignment the cp genome sequences of all individuals. The analysis shows the grouping clades with significant bootstrap support, differentiating between Central American (yellow colored),South American individuals (light blue colored), and Colombian individuals (green colored). (B) Resulting tree from the alignment of the sequence variants identified with Snippy v4.6.0 of the Galaxy suit. The tree shows similar grouping, according to the place of origin. The phylogenomic analyses were performed using BEAST phylogenetics software, and the visualization was carried out using iTOL software. Green-labeled taxa indicate C. odorata individuals collected in Colombia and deposited in the COAH, and the GenBank accession number of the reference cp genome for C. odorata. The purple circles indicate the bootstrap support as Bayesian posterior probability.
The sequence variants were predicted to identify a group of for genotype studies in local populations of C. odorata from Colombia. A total of 393 deletions, 208 insertions, and 96 multiple variants were identified in the reference cp genome (MG724915.1) compared to the 13 of the 14 cp genomes included in this study (Table 4). Additionally, 2793 sequence variants (SNPs) were identified. Of these, 187 SNPs were unique to each individual compared to the reference. In addition, 131 SNPs were found to segregate in the Cedrela species analyzed (15 with C. montana, 20 with C. saltensis, 64 with C. fissilis, and 32 with C. angustifolia, respectively). A few SNPs were also observed in the genome of C. odorata (e.g., one for CEOD10, CEOD162, and two for OGP2143). The resulting phylogenetic analysis of the Cedrela species based on the sequence variants grouped the Colombian individuals sequenced in this study (OGP2096 and OGP2143) close to the reference genome (Figure 3B), possibly reflecting the nearly complete genome assemblies generated with ONT long reads.

Table 4.
Sequence variants of reference cp genome (MG724915.1) identified in this study.
A total of 16 SVs present only in the OGP2096 and OGP2143 cp genomes were investigated by comparing the corresponding positions in the reference genome. As a result, six SVs were identified as intergenic variants, six as intron variants, and the remaining four were missense variants for specific loci (Table 5).

Table 5.
Potential C. odorata-specific SV for Colombian individuals identified from the chloroplast genome sequencing.
4. Discussion
C. odorata is an important plant species due to its use in the timber industry. However, because of the overexploitation of natural populations of C. odorata and related Cedrela species, the entire Cedrela genus has been declared protected under the CITES convention [42]. Colombia is one of the countries with high biodiversity of Cedrela species, and their wood is primarily used in the timber industry, often illegally. Nevertheless, there is a lack of knowledge about natural populations of C. odorata and the locations where the timber is sourced. Therefore, there is a need to establish suitable policies to limit the exploitation C. odorata’s natural resources.
In this study, the chloroplast genome sequences of two C. odorata individuals were generated using the emerging ONT sequencing technology: (1) as a proof of concept to obtain complete cp sequences as novel genomic resources for the species in Colombia; (2) to use new resources for further phylogenetic analysis, suggesting possible intraspecific diversity (Figure 3) and supporting the hypothesis of differences in local populations, as reported by Finch et al. [15]; and (3) to identify sequence variants that could be used in future tool developments to track illegally collected C. odorata timber.
These are the first cp genomes of C. odorata generated using ONT long-read sequencing technology and the assembly pipeline for chloroplast genomes reported by Wang et al. [20] for the wood forest species Eucalyptus pauciflora. Although the amount of raw long-read data was lower compared to other plant genome sequencing studies [43,44], the number of raw reads (2.6 Gb and ~1 Gb for the individuals OGP2096 and OGP2143, respectively) was sufficient to cover the cp genome up to 40× and generate complete cp genome assemblies for both individuals. These results also showed that the assemblies generated by Hinge with further polishing steps were more accurate than those generated by Canu, as demonstrated for E. pauciflora [20], suggesting that the pipeline developed for assembling chloroplast genomes from long-read sequencing projects could be applied to different plant species.
Plastid sequences of C. odorata can be used to support the geographic identification of derived timber material [15,45,46]. This phylogenetic analysis included a comparison of both individuals, OGP2096 and OGP2143, with a subsample of fragmented cp genomes of C. odorata and other Cedrela species, as reported by Finch et al. [15], and a complete reference genome for the species (Figure 3A). The grouping of these C. odorata individuals, closely related to other Cedrela individuals from Colombia, corroborates the results of Finch et al. [15] and Cavers et al. [45], suggesting that the cp genomes of C. odorata could be used to predict biogeography patterns in C. odorata individuals. Thus, these new chloroplast genomes could be used to identify broad scale local populations of C. odorata in Colombia.
Moreover, recent evidence based on nuclear SNP analysis of different Cedrela species suggests a cryptic diversity in C. odorata, differentiating Mesoamerican from South American individuals [15,46]. In this study, the phylogenetic analysis of the new cp genomes (Figure 3) supports the hypothesis of cryptic diversity for C. odorata, as reported by Finch et al. [46], as OGP2143 groups with the Mesoamerican individuals, while OGP2096 is more related to South American individuals. This hypothesis will be further corroborated in future SNP analyses of local populations of C. odorata from Colombia.
High throughput sequencing technologies allow the identification of SV that can be adapted as molecular markers for plant identification [47,48]. Here, sequence variants between C. odorata individuals were identified, focusing on the unique SVs of OGP2096 and OGP2143 (Table 5). These SVs are located in highly variable genomic regions of the C. odorata chloroplast, compared to other individuals of the Meliaceae family [13]. The intergenic (C/A) and rpl2 intron variants (G/A) in OGP2143 (Table 4) could be included in genotyping studies of C. odorata natural populations, to implement a genomic fingerprint for individuals extracted from the Amazon Region of Colombia.
C. odorata is a threatened timber species in the Americas. Local populations in Colombia are poorly understood and illegally exploited. The new chloroplast genome sequences and their variants reported in this study could be translated into a genomic tool to identify and/or characterize local populations of C. odorata in Colombia. This tool could assist authorities in tracking the origin of illegal timber and combat its exploitation.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijpb15030062/s1, Supplementary Figure S1: ONT quality sequencing analysis of raw data. Read lengths vs. average read from A. OGP2096 and B. OGP2143. Supplementary Figure S2: MuMmer plot of the chloroplast assembly from A. OGP2096 cp genome organization using Canu and B. its concatenated and polished assembly; C. OGP2096 cp genome assembly using Hinge and D. its concatenated and polished assembly; E. OGP2143 cp genome assembly using Hinge and F. its concatenated and polished assembly. Violet and red lines represent the length in kb of each assembled contig. Blue lines represent the genomic regions annotated in the C. odorata reference chloroplast genome (GenBank accession MG724915.1). Supplementary Figure S3: Structure of the OGP2096 and OGP2153 cp genomes constructed using mVISTA, with the C. odorata chloroplast genome (GenBank accession MG724915.1) as the reference.
Author Contributions
Conceptualization, J.S. and N.C.; methodology and formal analysis, G.A.G.-M. and J.S.; writing—original draft preparation, J.S. and G.A.G.-M.; project administration, J.S. and N.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was part of the grant “Investigación en conservación y aprovechamiento sostenible de la diversidad biológica, socioeconómica y cultural de la Amazonia Colombiana”, funded by the Ministry of Environment and Sustainable Development of Colombia.
Data Availability Statement
The sequenced assemblies used in this study can be found in the GenBank database, under accession numbers OP750006 and OP750007.
Acknowledgments
In memoriam to Senior Botanist Dairon Cardenas for his contribution to this study and for his leadership of the Plant Genetic Resources and COAH teams at SINCHI. We thank to Gadys Cardona for her collaboration, establishing the ONT sequencing technology at SINCHI. We are grateful to Ministry of Environment and Sustainable Development of Colombia for the financial support.
Conflicts of Interest
Author Jaime Simbaqueba was employed by the company Nourish Ingredients. Author Gina A. Garzón-Martínez was employed by the company Corporación Colombiana de Investigación Agropecuaria (Agrosavia). The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Mark, J.; Rivers, M.C. Cedrela odorata, Spanish Cedar. IUCN Red List Threatened Species. 2017, p. e.T32292A68080590. Available online: https://www.iucnredlist.org/species/32292/68080590 (accessed on 1 June 2024).
- Cárdenas, D.; Castaño, N.; Tunjuano, S.; Quintero, L. Plan de Manejo Para La Conservación de Abarco, Caoba, Cedro, Palorosa y Canelo de Los Andaquíes; Instituto Amazónico de Investigaciones Científicas—SINCHI, Ed.; Instituto Amazónico de Investigaciones Científicas—SINCHI: Bogota, Colombia, 2015; ISBN 9789588317878. [Google Scholar]
- Franco, N.; Clavijo, C.; Rojas, J.; Talero, C. Plan de Manejo y Conservación Del Cedro (Cedrela odorata L.) Para La Jurisdicción de La Corporación Autónoma Regional de Cundinamarca CAR; Instituto Amazónico de Investigaciones Científicas—SINCHI: Bogotá, Colombia, 2019. [Google Scholar]
- Molinares, C.; Prada, E.; León, E. Condenando el Bosque: Ilegalidad y Falta de Governanza en la Amazonia Colombiana; Environmental Investigation Agency: London, UK, 2019. [Google Scholar]
- Shaw, J.; Shafer, H.L.; Rayne Leonard, O.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA Sequence Utility for the Lowest Phylogenetic and Phylogeographic Inferences in Angiosperms: The Tortoise and the Hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cantino, P.D.; Olmstead, R.G.; Bramley, G.L.C.; Xiang, C.L.; Ma, Z.H.; Tan, Y.H.; Zhang, D.X. A Large-Scale Chloroplast Phylogeny of the Lamiaceae Sheds New Light on Its Subfamilial Classification. Sci. Rep. 2016, 6, 34343. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.J.; Lu, Y.B.; Ye, Q.Q.; Tang, S.Q. Population Genetic Structure and Phylogeography of Camellia flavida (Theaceae) Based on Chloroplast and Nuclear DNA Sequences. Front. Plant Sci. 2017, 8, 718. [Google Scholar] [CrossRef] [PubMed]
- Degen, B.; Fladung, M. Use of DNA-Markers for Tracing Illegal Logging. In Proceedings of the International Workshop “Fingerprinting Methods for the Identification of Timber Origins”, Bonn, Germany, 8–9 October 2007; Volume 321, pp. 6–14. [Google Scholar]
- Cavers, S.; Navarro, C.; Lowe, A.J. Chloroplast DNA Phylogeography Reveals Colonization History of a Neotropical Tree, Cedrela odorata L., in Mesoamerica. Mol. Ecol. 2003, 12, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Ci, X.Q.; Liu, Z.F.; Dormontt, E.E.; Conran, J.G.; Lowe, A.J.; Li, J. Assessing Candidate DNA Barcodes for Chinese and Internationally Traded Timber Species. Mol. Ecol. Resour. 2022, 22, 1478–1492. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Villanueva, K.; de Groot, G.A.; Laros, I.; Bovenschen, J.; Bongers, F.; Zuidema, P.A. Genetic Differences among Cedrela odorata Sites in Bolivia Provide Limited Potential for Fine-Scale Timber Tracing. Tree Genet. Genomes 2019, 15, 33. [Google Scholar] [CrossRef]
- Schroeder, H.; Cronn, R.; Yanbaev, Y.; Jennings, T.; Mader, M.; Degen, B.; Kersten, B. Development of Molecular Markers for Determining Continental Origin of Wood from White Oaks (Quercus L. Sect. Quercus). PLoS ONE 2016, 11, e0158221. [Google Scholar] [CrossRef]
- Mader, M.; Pakull, B.; Blanc-Jolivet, C.; Paulini-Drewes, M.; Bouda, Z.H.N.; Degen, B.; Small, I.; Kersten, B. Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses. Int. J. Mol. Sci. 2018, 19, 701. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; Fazekas, A.J.; et al. A DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Finch, K.N.; Jones, F.A.; Cronn, R.C. Genomic Resources for the Neotropical Tree Genus Cedrela (Meliaceae) and Its Relatives. BMC Genom. 2019, 20, 58. [Google Scholar] [CrossRef]
- Michael, T.P.; Jupe, F.; Bemm, F.; Motley, S.T.; Sandoval, J.P.; Lanz, C.; Loudet, O.; Weigel, D.; Ecker, J.R. High Contiguity Arabidopsis thaliana Genome Assembly with a Single Nanopore Flow Cell. Nat. Commun. 2018, 9, 541. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of Nanopore Sequencing to the Genomics Community. Genome Biol. 2016, 17, 239. [Google Scholar] [CrossRef]
- Scott, A.D.; Zimin, A.V.; Puiu, D.; Workman, R.; Britton, M.; Zaman, S.; Caballero, M.; Read, A.C.; Bogdanove, A.J.; Burns, E.; et al. A Reference Genome Sequence for Giant Sequoia. G3 Genes|Genomes|Genet. 2020, 10, 3907–3919. [Google Scholar] [CrossRef]
- Giordano, F.; Aigrain, L.; Quail, M.A.; Coupland, P.; Bonfield, J.K.; Davies, R.M.; Tischler, G.; Jackson, D.K.; Keane, T.M.; Li, J.; et al. De novo Yeast Genome Assemblies from MinION, PacBio and MiSeq Platforms. Sci. Rep. 2017, 7, 3935. [Google Scholar] [CrossRef]
- Wang, W.; Schalamun, M.; Morales-Suarez, A.; Kainer, D.; Schwessinger, B.; Lanfear, R. Assembly of Chloroplast Genomes with Long- and Short-Read Data: A Comparison of Approaches Using Eucalyptus pauciflora as a Test Case. BMC Genom. 2018, 19, 977. [Google Scholar] [CrossRef]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-Generation Sequencing Technologies: An Overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Schalamun, M.; Nagar, R.; Kainer, D.; Beavan, E.; Eccles, D.; Rathjen, J.P.; Lanfear, R.; Schwessinger, B. Harnessing the MinION: An Example of How to Establish Long-Read Sequencing in a Laboratory Using Challenging Plant Tissue from Eucalyptus Pauciflora. Mol. Ecol. Resour. 2019, 19, 77–89. [Google Scholar] [CrossRef]
- Reiling, S.J.; Chen, S.-H.; Ragoussis, I. McGill Nanopore Ligation LibPrep Protocol SQK-LSK109. Protocolos.io 2020. Available online: https://www.protocols.io/view/mcgill-nanopore-ligation-libprep-protocol-sqk-lsk1-bp2l6b1ezgqe/v1 (accessed on 10 May 2021). [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore Sequencing and Assembly of a Human Genome with Ultra-Long Reads. Nat. Biotechnol. 2018, 36, 338–345. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and Processing Long-Read Sequencing Data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Chaisson, M.J.; Tesler, G. Mapping Single Molecule Sequencing Reads Using Basic Local Alignment with Successive Refinement (BLASR): Application and Theory. BMC Bioinform. 2012, 13, 238. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive k-Mer Weighting and Repeat Separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Kamath, G.M.; Shomorony, I.; Xia, F.; Courtade, T.A.; Tse, D.N. HINGE: Long-Read Assembly Achieves Optimal Repeat Resolution. Genome Res. 2017, 27, 747–756. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and Open Software for Comparing Large Genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef]
- Loman, N.J.; Quick, J.; Simpson, J.T. A Complete Bacterial Genome Assembled de Novo Using Only Nanopore Sequencing Data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and Accurate de Novo Genome Assembly from Long Uncorrected Reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and Accurate Annotation of Organelle Genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. Vista: Visualizing Global DNA Sequence Alignments of Arbitrary Length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows—Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Ech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Van der Auwera, G.; O’Connor, B. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly Media, Inc., Ed.; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2020; ISBN 978-1491975190. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.-H. The Codon Adaptation Index-a Measure of Directional Synonymous Codon Usage Bias, and Its Potential Applications. Nucleic Acids Res. 1986, 14, 4683–4690. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, W.; Zhu, D.; Hong, P.; Zhang, S.; Xiao, S.; Tan, Y.; Chen, X.; Xu, L.; Zong, X.; et al. Chromosome-Scale Genome Assembly of Sweet Cherry (Prunus avium L.) Cv. Tieton Obtained Using Long-Read and Hi-C Sequencing. Hortic. Res. 2020, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Wöhner, T.W.; Emeriewen, O.F.; Wittenberg, A.H.J.; Schneiders, H.; Vrijenhoek, I.; Halász, J.; Hrotkó, K.; Hoff, K.J.; Gabriel, L.; Lempe, J.; et al. The Draft Chromosome-Level Genome Assembly of Tetraploid Ground Cherry (Prunus fruticosa Pall.) from Long Reads. Genomics 2021, 113, 4173–4183. [Google Scholar] [CrossRef]
- Cavers, S.; Telford, A.; Arenal Cruz, F.; Pérez Castañeda, A.J.; Valencia, R.; Navarro, C.; Buonamici, A.; Lowe, A.J.; Vendramin, G.G. Cryptic Species and Phylogeographical Structure in the Tree Cedrela odorata L. throughout the Neotropics. J. Biogeogr. 2013, 40, 732–746. [Google Scholar] [CrossRef]
- Samji, A.; Eashwarlal, K.; Shanmugavel, S.; Kumar, S.; Warrier, R.R. Chloroplast Genome Skimming of a Potential Agroforestry Species Melia Dubia. Cav and Its Comparative Phylogenetic Analysis with Major Meliaceae Members. 3 Biotech 2023, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Finch, K.N.; Jones, F.A.; Cronn, R.C. Cryptic Species Diversity in a Widespread Neotropical Tree Genus: The Case of Cedrela Odorata. Am. J. Bot. 2022, 109, 1622–1640. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and Challenges in Long-Read Sequencing Data Analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Winefield, C.; Bombarely, A.; Prentis, P.; Waterhouse, P. Tools and Strategies for Long-Read Sequencing and De Novo Assembly of Plant Genomes. Trends Plant Sci. 2019, 24, 700–724. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).