Abstract
The tree species Salix caprea shows high adaptability to different habitat conditions and is economically valuable as a woody crop for biomass production. Moreover, S. caprea is dependent on mycorrhizal fungi, which are crucial for its growth and adaptability in different environments. Hence, this study explores the ectomycorrhizal diversity of S. caprea by utilizing the taxonomy (morphotyping and a molecular approach using the ITS and LSU regions) and trait diversity (exploration types) at two test sites in Germany and Poland. In total, 19 ectomycorrhizal (EM) morphotypes of S. caprea were characterized. Seven taxa were identified at the species level (Hebeloma populinum, Cortinarius atrocoerulaeus, Inocybe hirtella, Laccaria cf. ochropurpurea, Tuber maculatum, Cenococcum geophilum, and Phialophora finlandia) and twelve at the genus level (Tomentella spp. 1–8, Hebeloma sp. 1, Inocybe sp. 1, and Tuber spp. 1–2). The EM colonization ranged from 14 to 28% of the fine root tips. At both test sites, the largest portion of the total EM colonization consisted of Thelephoraceae. The exploration types were classified as medium-distance smooth (Tomentella sp. 1–8 and L. ochropurpurea) and medium-distance fringe (C. atrocoerulaeus), while the other taxa were short-distance exploration types, highlighting their potential functional role in the adaptation and growth of S. caprea.
1. Introduction
Ectomycorrhizal (EM) fungi are symbionts associated with the roots of most tree species in boreal and temperate forest ecosystems [1]. Most of these tree species are dependent on EM fungi for their growth and survival [2]. Researchers have shown that EM fungi can positively influence the performance of the tree host by improving soil fertility and seed germination rates, enhancing soil nutrient mobilization (e.g., nitrogen, N) and water uptake, and modulating plant traits to alleviate biotic and abiotic stresses. In exchange, EM fungi receive photosynthetically fixed carbon (C) [3,4,5,6,7]. Mycorrhizal fungi help extend the area of nutrient and water acquisition through the extraradical hyphal network, and they can also aid in the distribution of hydraulic lift water to neighboring plants by forming common mycorrhizal networks, especially during drought [8]. Additionally, many EM fungal partners can efficiently increase carbon sequestration and contribute to soil nutrient mobilization by producing enzymes involved in the hydrolysis of nitrogen and phosphorus compounds from the soil organic matter, and they contribute to the weathering of minerals, e.g., through the release of organic acids [9]. Ectomycorrhizal fungi can alleviate abiotic (e.g., heavy metals) stress through metal exclusion (forming a barrier around plant roots), production of chelating compounds that bind heavy metals (reducing the toxicity), or through metal accumulation in fungal tissue; they can also improve soil structure by forming soil aggregates, reducing the mobility and bioavailability of heavy metals in soil, thereby preventing the metals from reaching the plant roots [10,11]. The EM fungi can also protect plants from biotic stress (e.g., pathogens) through the production of pathogenesis-related biomolecules or they can produce antagonist substances and increase plant fitness resulting from an enhanced nutrient supply [12].
A high diversity of EM fungi is frequently reported in forests; however, the vegetation type and abiotic soil characteristics can either decrease or increase the frequency and diversity of EM formation [13]. Successful EM colonization is problematic at some primary successional forest sites, especially at sites that are degraded or disturbed due to volcanic activity, dunes, mining, clear-cut logging, wildfires, glacier retreat, soil erosion, and the scarcity of ectomycorrhizal vegetation [2].
Willows (Salix species) are well known as pioneer vegetation in disturbed and polluted environments comprising fast-growing shrubs and deciduous trees with high aerial biomass and deep roots [14,15]. They can colonize nutrient-limited sites, such as roadside ditches, abandoned agricultural fields, railroads, old mine tailings, and gravel pits, as well as burned fields or flooded areas where disturbance results in an open community with less competition [14,15]. These species exhibit specific physiological adaptations and are particularly resilient in the face of climate change [16]. Salix spp. are economically valuable as woody crops for biomass production [17]. They are also extensively used for the phytoremediation of trace-element-contaminated environments and to minimize the leaching of pesticides from agricultural soils [18]. Salix caprea (goat willow or pussy willow) is a shrubby tree which is popularly known for its low nutrient requirements and its high tolerance of heavy metals [19]. In general, Salix species are naturally associated with both arbuscular mycorrhizal (AM) and EM fungi. Ectomycorrhizas can colonize more than 80% of the willow’s fine roots, while AM fungi are mainly limited to <5% of the fine roots [20,21,22,23]. Previous studies on the EM fungi of Salix spp. have reported the dominance of Thelephoraceae in different environments, such as contaminated sites [24], S. alba in riparian edge forests [25], and S. viminalis in short rotation coppices [26] and saline soils [27].
Moreover, it is important to understand not only the shifts in EM diversity but also the functional differences in EM taxa based on the development and differentiation of their external mycelium (exploration types) [28]. To understand the functional traits of EM taxa and ecosystem functioning, the classification of EM fungi according to the different exploration types (e.g., short-distance, long-distance, and mat formation), the presence or absence of rhizomorphs, and hyphae type can be relevant [29,30]. The variation in EM exploration types, which differ among EM taxa and differing environments, allows EM fungi to acquire nutrients under unfavorable conditions [31]. In this context, the starting point of field investigation is EM fungal characterization at the species level. This goal can be achieved by combining detailed light microscope-based morphology and anatomical description (morphotyping) [28] with molecular approaches (Sanger sequencing).
Therefore, the present study aims to investigate the abundance and diversity of EM fungi on the fine roots of the dominant shrub species S. caprea (1) and the functional traits (exploration types) of the fungal partners associated with S. caprea species growing at two sites (2); the study also investigates the relationships of the host tree species and soil chemistry with EM composition (3). The recognition of well-adapted EM strains of S. caprea may be an important step before the selection of an EM inoculum for the remediation of poor soils.
2. Materials and Methods
2.1. Test Sites and Sampling
Samples of the fine roots of S. caprea and soil were collected from two test sites: Neuhaus (site 1: 54.277556, 12.298263; 11 m above sea level, northern Germany) and Bydgoszcz (site 2: 53.099306, 18.294809; 36 m above sea level, northern Poland). Site 1 is located close to the dunes of the Baltic Sea (maritime climate) and the neighboring dune vegetation (Ammophila arenaria); site 2 (temperate climate) is situated near agricultural land (without mechanical or chemical human intervention) with surrounding vegetation, including grasses, e.g., perennial ryegrass (Lolium perenne), couch grass (Elymus repens), and dicotyledonous plants, e.g., goosefoot (Chenopodium album), knotgrass (Polygonum aviculare), and common nettle (Urtica dioica).
The vegetation at the two test sites was dominated by 20–30-year-old pioneer individuals of S. caprea. The soil and root samples of S. caprea were taken (20 cm × 20 cm × 20 cm soil cubes) with a sharp knife, c. 20 cm from the stem base of the willow plants. The roots of willows grow highly irregularly with partially concentrated fine root bunches [32]. Bunches of this type were selected for sampling. Five replicates (each from under a different tree; the trees grew 5–6 m apart) were taken randomly from each test site during three sampling seasons in fall 2002, spring 2003, and fall 2003, for a total of fifteen samples per site. The samples were collected in plastic bags and immediately transported at 4 °C to the laboratory for analysis.
2.2. Soil Analysis
The determination of soil pH was performed electrometrically using a glass electrode in 0.01 M CaCl2 with a soil solution ratio of 1:2.5. The C and N contents of the soils were determined using Foss Heraeus CNS—Vario EL (Elementar Analysentechnik GmbH, Langenselbold, Germany). Total phosphorus (Pt) was determined according to the method of Dick and Tabatabai [33]. The P concentration was analyzed using inductively coupled plasma optical emission spectroscopy (ICP-OES, JY 238, HORIBA Jobin Yvon, Lille, France, wavelength 214 nm). The soil texture was determined using Köhn’s method [34].
2.3. Determination of Ectomycorrhizas Colonization
The roots from each sample were carefully separated and washed with distilled water for examination. The root samples of S. caprea at the two study sites (site 1 and site 2) were taken during three seasons: fall_2002, spring_2003, and fall_2003. A total of 15 samples (two sites × three sampling seasons x five replications) from each of the two sites during one sampling season were examined. As the roots of S. caprea grow highly irregularly under natural conditions, the number of root tips varied considerably among the samples taken from different locations and at different times of the year and ranged from 1400 to 5200 root tips (vital and non-vital, mycorrhizal, and non-mycorrhizal). The living roots were identified based on a turgid appearance and the possession of white cortical cells. The EM frequencies associated with each site and each sampling season were calculated (numbers of EM root tips × 100%/total numbers of root tips).
2.4. Morphological and Anatomical Description of the Ectomycorrhizas
The EM types (morphological–anatomical types) were distinguished by the macroscopic characteristics of their fungal mantles, such as color, surface appearance, and the presence of emanating hyphae and hyphal strands, as well as microscopic features such as mantle type [35,36]. The EM types were also classified into exploration types [37]. Based on microscopic identification, two-to-five root tips per EM type were frozen in Eppendorf tubes and stored at −20 °C for molecular analyses.
2.5. Molecular Analysis of the EM Fungi
Analyses of the DNA sequences of the D1/D2 region of the nuclear-encoded large subunit RNA gene (LSU) and the ITS region were used to identify the EM-forming fungi. The DNA was isolated from frozen mycorrhizal samples using the DNAeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. After extraction, fungal DNA was amplified using the universal primers ITS1 or ITS1F and ITS4 or ITS4B [37,38], and some fungal DNA samples were amplified using the primer LSU with the primer pair NL1–NL4 [39]. The PCR reaction was performed according to Haug’s method [40]. The PCR products obtained were purified using the QIAquick protocol (Qiagen, Hilden, Germany). For cycle sequencing, the ABI PRISM Dye-Terminator Cycle Sequencing Ready Reaction Kit was used (Applied Biosystems, Foster City, CA, USA), followed by electrophoresis on an automated sequencer (ABI 373A Stretch, Applied Biosystems, Foster City, CA, USA). The sequencing was carried out according to the manufacturer’s protocol, but the cycle sequencing reaction volumes were reduced by half. Both forward and reverse DNA strands were sequenced.
The sequences were edited using Sequencher TW Version 4.1 (Gene Codes, Ann Arbor, MI, USA). The NCBI BLAST searches with the LSU and ITS sequences were performed on the GenBank [41] and/or UNITE database [42]. For species identification, the ITS region was chosen since the interspecific variability of this region was shown to be fairly high compared to its intraspecific variation [43]. For the identification of mycorrhizal tips formed by Inocybe, we used LSU sequences, since most fungi belonging to this species have less than 3% intraspecific variation in the ITS region of the nuclear ribosomal DNA. Only sequences of EM fungi showing 98% identity over the whole length of the sequence with a known fungus were assumed to be fungal partners of the mycorrhiza. If the identity was 99% or 100%, it was certain that this taxon was involved in the mycorrhiza. The DNA sequences determined for this study were submitted to GenBankwith the accession numbers. The evolutionary history was inferred by using the maximum likelihood method and the Jukes–Cantor model. Initial tree(s) for the heuristic search were obtained automatically by applying the neighbor-join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then by selecting the topology using the superior log likelihood value. Evolutionary analyses were conducted in MEGA X [44].
2.6. Statistical Analysis
Using the Statistica software ver.13.3 (StatSoft, Kraków, Poland), one-way analysis of variance (ANOVA), followed by the Newman–Keuls post hoc test with a significance level of p < 0.05, was carried out to demonstrate the differences between the two test sites during the three sampling seasons in terms of the degree of EM colonization in the fine root tips of S. caprea.
3. Results
The chemical and physical properties of the soil (<2 mm) at the two sites are presented in Table 1. The total P concentration of the soil was below the standards observed in the agricultural soils or short rotation coppices (4.5–6.8 mg/100 g; [45]), which is why these sites were classified as nutrient-deficient. The soil pH in which S. caprea was growing was 6.5 at both test sites.

Table 1.
Chemical and physical properties of soils (0–10 cm depth) at the two test sites: site 1 (Neuhaus, northern Germany) and site 2 (Bydgoszcz, northern Poland). Abbreviations: Corg—organic carbon, Nt—Total nitrogen, Pt—Total phosphorous.
The degree of EM colonization in S. caprea was significantly higher at site 1 during fall (in both the fall sampling seasons) compared to site 2 (Figure 1). The ectomycorrhizal fungal colonization was generally poor in S. caprea during spring at both test sites. The number of different EM morphotypes per site ranged from nine at site 2 to 10 at site 1 (Table 2).
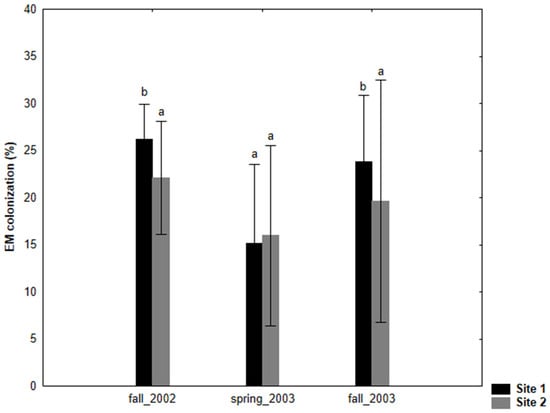
Figure 1.
Ectomycorrhizal (EM) fine root colonization of Salix caprea (% colonized fine root tips, mean value ± SD) at 0–10 cm soil depth grown at two test sites (site 1: NEU and site 2: BYD) during three sampling seasons (fall 2002, spring 2003, fall 2003). The small letters denote significant differences between sampling sites in each year.

Table 2.
The sampling seasons: fall 2002 (1), spring 2003 (2), fall 2003 (3) and description of ectomycorrhizal morphotypes of S. caprea at two test sites: site 1 (NEU) and site 2 (BYD). The nomenclature corresponds to that of Agerer (1991) [35].
Identification of Ectomycorrhizal Morphotypes
A total of 1400 to 5200 root tips (vital and non-vital, mycorrhizal and non-mycorrhizal) of S. caprea from the two sites, site 1 and site 2, taken during three sampling seasons, namely, fall_2002, spring_2003, and fall_2003, were analyzed. Nineteen different morphotypes of S. caprea were identified (Figure 2). These morphotypes mainly comprised the phyla Basidiomycota (14 morphotypes) and Ascomycota (five morphotypes) (Figure 3 and Figure 4). The Basidiomycota group was dominated by the families Thelephoraceae (eight taxa), Hymenogastraceae (two taxa), and Inocybaceae (two taxa), followed by Cortinariaceae and Hydnangiaceae, while Ascomycota was represented by the families Tuberaceae (three taxa), Gloniaceae, and Herpotrichiellaceae.

Figure 2.
Ectomycorrhizal (EM) morphotypes on Salix caprea at the two test sites, site 1 (NEU) and site 2 (BYD). Ectomycorrhizal morphotypes: (1) Tomentella sp. 1 (NEU14), (2) Tomentella sp. 2 (BYD9), (3) Tomentella sp. 3 (NEU1), (4) Tomentella sp. 4 (NEU11), (5) Tomentella sp. 5 (BYD2), (6) Tomentella sp. 6 (BYD3), (7) Tomentella sp. 7 (BYD5), (8) Tomentella sp. 8 (BYD6), (9) Hebeloma populinum (NEU13), (10) Hebeloma sp. (NEU4), (11) Cortinarius atrocoerulaeus (NEU7), (12) Inocybe sp. 1 (NEU10), (13) Inocybe hirtella (BYD1), (14) Laccaria cf. ochropurpurea (NEU3), (15) Tuber sp. 1 (NEU5), (16) Tuber maculatum (BYD4), (17) Tuber sp. 3 (BYD8a), (18) Cenococcum geophilum (NEU9), (19) Phialophora finlandia (NEU2).
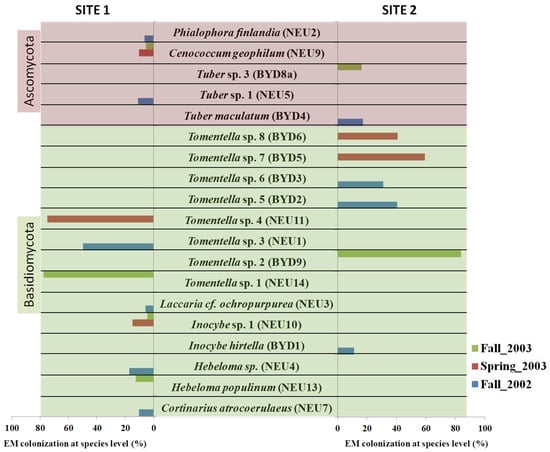
Figure 3.
The colonization percentage of the identified EM morphotypes at the species level at the two test sites: site 1 (Neuhaus) and site 2 (Bydgoszcz) during three sampling seasons (fall 2002, spring 2003, fall 2003).
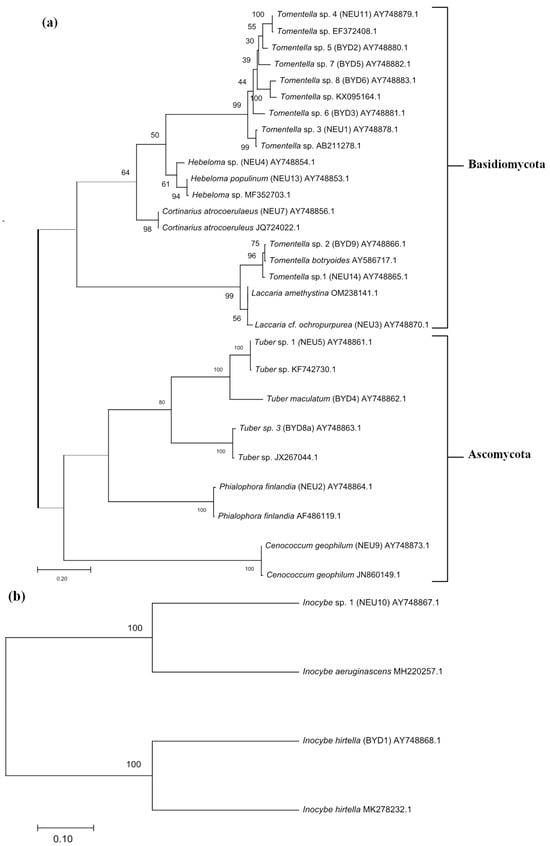
Figure 4.
Phylogenetic tree depicting the different ectomycorrhizal taxa identified from two phyla Ascomycota and Basidiomycota based on ITS (a) and LSU (b) regions of the nuclear ribosomal DNA. The percentage of trees in which the associated taxa clustered together is shown next to the branches.
Seven taxa were identified at the species level: Hebeloma populinum, Cortinarius atrocoerulaeus, Inocybe hirtella, Laccaria cf. ochropurpurea, Tuber maculatum, Cenococcum geophilum, and Phialophora Finlandia (Figure 3 and Figure 4). Twelve were identified at the genus level: Tomentella sp. 1–8, Hebeloma sp. 1, Inocybe sp. 1, and Tuber sp. 1–2 (Table 3 and Figure 3). The EM roots of Inocybe sp. 1 and C. geophilum were sampled at site 1 during two sampling seasons: spring_2003 and fall_2003. The other sixteen morphotypes were found only once. The dominant morphotypes of S. caprea were formed by Thelephoraceae (Table 3 and Figure 3). They were found at both sampling sites and across all sampling seasons (with the exception of site 1 in spring_2003) (Figure 3).

Table 3.
Molecular identification and closest BLAST sequence match of ectomycorrhizas sampled from S. caprea from two sites, site 1 (NEU) and site 2 (BYD), during three sampling seasons (fall 2002, spring 2003, and fall 2003).
Based on the morphotyping of the EM roots, we observed the black, dark brown, or brown color of the hyphal mantle, which was characteristic of all Thelephoraceae morphotypes (Figure 2). Cystidia were observed in three morphotypes of Tomentella sp., and rhizomorphs were found only in one. All these morphotypes were identified at the genus level (Tomentella sp. 1–8) (Table 2). As most Inocybe species have less than 3% intraspecific variation in the ITS region of the nuclear ribosomal DNA [46], we used the LSU sequences of these fungal partners for identification (Table 3 and Figure 4). All Hebeloma species have very similar morphology (Table 3). Their characteristics include a white beige shining mantle and cottony emanating hyphae. The Cortinariaceae had a plectenchymatous mantle. The white and smooth EM fungi of Laccaria cf. ochropurpurea were found at site 1 (Table 2 and Figure 2).
Only five morphotypes were identified among the ascomycetes. The EM fungi of Tuber sp. 1 were found at site 1, while T. maculatum and Tuber sp. 2 were found at site 2 (Table 2). All the EM roots of Tuber sp. were gold-brown with cystidia (Table 2 and Figure 2). Cenococcum geophilum (two sampling seasons: spring_2003 and fall_2003) and P. finlandia (fall_2002) were found only at site 1 (Table 2).
The exploration types of the identified EM fungi were classified as medium-distance smooth (Tomentella sp. 1–8 and Laccaria cf. ochropurpurea) and medium-distance fringe (Cortinarius atrocoerulaeus), while the other taxa were short-distance exploration types with no rhizomorphs (Table 2).
4. Discussion
Combining morphotyping with molecular approaches is a well-accepted method for the characterization of different ectomycorrhizas and the analysis of their community structure [47]. Following this strategy, we highlighted the importance of combining these two methods for the selection and identification of the ectomycorrhizal diversity of Salix caprea, a pioneer tree at two sites.
Species of Betula, Salix, Populus, Alnus, and Sorbus aucuparia L. represent deciduous pioneer tree species in Europe. Pioneer tree species are very common in early successional stages and, due to their rapid regeneration and high seed production, they mitigate the negative consequences associated with disturbed areas, such as soil erosion and the loss of nutrients [48]. It is known that EM fungi may also aid in the establishment and facilitation of early colonizing tree species at disturbed sites. A study by Nara et al. [49] reported that the first plant to colonize the volcanic desert on Mount Fuji, Japan, was an alpine dwarf willow Salix reinii, which is strictly ectomycorrhizal. Similar research at the same site showed that Salix seedlings developed easily only when beside the established Salix shrubs, and no seedling growth was observed in areas without established Salix [50]. Further investigations proved that the growth of successive seedlings was dependent on the EM association of the neighboring EM shrubs, which significantly improved as the early established Salix shrubs facilitated the subsequent establishment of conspecific seedlings by providing EM fungal symbionts [51].
In the present study, a total of 19 different EM morphotypes were characterized in S. caprea from both test sites using an EM morphology examination and ITS/LSU sequencing (Figure 3 and Figure 4). The molecular analysis of EM roots indicated that the EM fungal communities of S. caprea from the sites contained a diverse and broad array of fungi, including basidiomycetes and ascomycetes. Well-known fungal associates of the members of the Salicaceae (Salix and Populus) are, for example, Lactarius controversus, Russula atrorubens, Russula persicina, and Tricholoma cingulatum [24,52]. Many species of Cortinarius [53,54] and Hebeloma [55] also preferentially associate with members of the Salicaceae family. It is striking that only EM fungi formed by Thelephorales (Tomentella spp.) and Agaricales (Inocybe spp.) were observed at both the investigated sites. In addition, Tomentella spp., Inocybe spp., and Cenococcum geophilum were found during both spring and fall sampling seasons. These three EM species can be classified as early colonizers of S. caprea, since they occur in their host in the early stages of growth. The other morphotypes (Hebeloma sp. 1, Hebeloma populinum, Cortinarius atrocoerulaeus, Laccaria cf. ochropurpurea, Tuber sp. 1–2, Tuber maculatum, and Phialophora finlandia) occurred only during the fall sampling season, mostly at site 1, though Tuber species were also found at site 2. There are two explanations for this phenomenon: either the host tree determines the type of EM recruitment, or the spatial heterogeneity may be very high and, consequently, with each sampling, only a small portion of the fungi is cached. Thus, we speculate that further sampling may result in more species at the investigated sites. A report on the underground EM fungal diversity of the pioneer tree S. reinii shows that species of Thelephoraceae, Cortinarius, and Hebeloma were recruited in the later stages of S. reinii growth and that a limited assimilate supply from the plant host inhibited the colonization of some fungi (such as Inocybe lacerna and Scleroderma bovista) and supported the colonization of non-conspicuous sporocarp species such as Sebacina, Thelephoraceae, and C. geophilum [49].
The diversity of EM assemblages also manifests in the functional diversity that is relevant to tree host health and ecosystem functioning [56]. The potential functions of the identified EM fungal isolates are presented in Table 4. Furthermore, the exploration type may also partly explain the dynamics of EM fungi in their host. In this study, Tomentella sp. 1–8 and L. ochropurpurea, which were classified as being of the medium-distance smooth exploration type, had internally undifferentiated rhizomorphs, and the ectomycorrhizal mantles had few or no radiating hyphae close to the root and a smooth appearance. On the other hand, the medium-distance fringe types, i.e., C. atrocoerulaeus, had rhizomorphs with a hairy surface, and they formed fans of emanating hyphae with extended contacts with the soil. The others were identified as short-distance exploration types characterized by the production of abundant, short, non-aggregated hyphae in the vicinity of the root tip; they also produced no rhizomorphs [28]. In the present study, most of the identified EM fungal species were short-distance exploration types, which are known to be present in the fluctuating nutrient pools that are characteristic of disturbed soils [57]. Previously, as Beccarra et al. (2009) [58] and Nara et al. (2003) [49] have suggested, species of Inocybe and Tomentella were well-known as colonizers of EM plants in disturbed or primary habitats.
The most common morphotypes in this study were formed by Tomentella sp., which is a known ‘short-distance explorer’ and is found in forests as well as in disturbed soils [10,28,59]. Most tomentelloid species have a worldwide distribution, with the highest species richness being found in temperate coniferous and broad-leaved forests [60]. The genus Thelephora displays characteristics similar to those of tomentelloid fungi and comprises about 40 species [61]. The ecological characteristics of tomentelloid fungi remain largely unexplored [61,62]. The color of Tomentella sp. morphotypes in this study was always dark (from completely black through to dark brown and brown). Their characteristic dark color results from the incorporation of melanin, a natural dark pigment and a common fungal wall component. Melanized cell walls are not common among EM fungi, except for tomentelloid species [62] and a few other EM fungal taxa. Fungal melanin may act as a barrier between fungal cells and their surrounding environment and protect them against physical, chemical, and biological stress [63]. In addition, the melanized hyphae of the outer mantle layer and the extramatrical mycelium may function as protective barriers between the external environment and the active fungus–plant symbiosis inside the root.
Inocybe sp. is a widely distributed species (mainly generalist fungi) [64]. Ectomycorrhizal fungal species of I. lacera were previously found during the early primary succession of S. reinii [50]. In our study, the morphotypes of Inocybe sp. on S. caprea were characteristic of both sites and occurred across all sampling seasons. Inocybe spp. are abundant in contaminated sites and dry and disturbed habitats and are tolerant to drought conditions [10,11,65]. Another species, Cenococcum geophilum, was observed in both spring and fall of 2003, but only at site 1. This species is cosmopolitan and is well recognized for its extremely wide host and habitant range (e.g., it is found in post-fire soils and colonizes exotic trees) [66,67]. According to Park [56], the early stage of the symbiosis is characterized by white and brown EM fungi, while the black EM roots characterize the most mature stage of EM development. This was confirmed in the present study.
Truffles of the genus Tuber are ascomycetous fungi which form EM with the roots of trees such as Quercus, Populus, and Salix [24,52,68], as well as shrubs such as Cistus and Corylus [69,70]. In the present research, Tuber sp. was isolated from both sites, but was more common at site 2 during fall. Tuber spp. are known to adapt and tolerate extreme conditions [11,24,52].
The genus Cortinarius has a widespread distribution in pioneer trees [53,54]. Many studies have provided reports on different species of Cortinarius found on Salix spp.: C. decipiens and C. tenebricus [71], on S. barratiana—Cortinarius spp. [54], on S. herbacea—C. favrei [72], on S. repens—Cortinarius spp. [73], and on S. reinii—C. alboviolaceus [49]. Similarly, this genus was very common in the present study, but none of the above-mentioned fungal species was identified. The fungi with gold-orange EM roots from site 1 were identified as C. atrocoerulaeus. C. atrocoerulaeus has been previously reported to play a role in assisting plant nutrition [10,74].
Two morphotypes of Hebeloma sp., identified as H. populinum and Hebeloma sp. 1, were found at site 1. Hebeloma species have a broad host range with little ambiguity as the occurrence of these species is naturally associated with various gymnosperms and dicots in the field [75]. Hebeloma populinum, which was found in this study, is also frequently reported as a specialist fungal partner of Populus spp. [10,11].
Laccaria sp. has been reported to form EM associations with numerous tree species, including Pinaceae, Dipterocarpaceae, Fagaceae, Myrtaceae, Tiliaceae, and Salicaceae [76]. An association with S. caprea and L. ochropurpurea was observed at site 1. Hrynkiewicz and colleagues isolated L. ochropurpurea from S. caprea obtained from an ore mining site [24]. The presence of L. ochropurpurea increased pathogen resistance in chestnut plantations (Castanea dentata) [77].
A dark brown morphotype with abundant emanating hyphae, a pseudo-parenchymatous mantle, and fan-like rhizomorphs was found at site 1: it was formed by Phialophora finlandia. Fungus P. finlandia is commonly referred to as Mycelium radicis-atrovirens (MRA) or dark septate endophyte (DSE) [78] and forms ectomycorrhiza and/or ectendomycorrhiza with hardwoods and conifers, respectively [79,80]. Phialophora finlandia has been reported to produce EM morphotypes ranging from light amber, brown, or bicolored to black [79,80], corresponding to the observed morphotype in the present study. This strain can play a role in plant growth promotion and survival in disturbed soils [81] and in the nursery of Picea abies [82].

Table 4.
Potential role of the identified ectomycorrhizal strains reported in the research literature. Abbrev., not defined (ND).
Table 4.
Potential role of the identified ectomycorrhizal strains reported in the research literature. Abbrev., not defined (ND).
| Identified Ectomycorrhiza | Isolated from Pioneer Tree sps. | Generalist or Specialist Fungi | Type of Environment | Potential Function of EM | NCBI Accession No. | Citation |
|---|---|---|---|---|---|---|
| Tomentella sp. | Populus sp. | Generalist | Forest/dry habitat/heavy metal-polluted area | Tolerance to drought or increased concentrations Of heavy metals in the soil | JQ898563 | [10] |
| Salix alba | Generalist | Saline meadow near a soda factory | Adaptation to saline conditions | KP745608 | [59] | |
| Hebeloma populinum | Populus sp. | Specialist | Forest/dry habitat | Tolerance to drought | JQ898545 | [10] |
| Populus tremula | Specialist | Former lead/zinc smelter | Fungal immobilization of heavy metals | EF644130 | [11] | |
| Hebeloma sp. | Populus sp. | Generalist | Heavy metal-polluted area | Tolerance to increased concentrations Of heavy metals in the soil | JQ898546 | [10] |
| Salix caprea | Generalist | Former ore mining area, Heavy metal-polluted soil | Tolerance against toxic metal concentrations in the soil | AY748855 | [24] | |
| Cortinarius atrocoerulaeus | Populus sp. | Generalist | Forest | Plant nutrition | JQ898541 | [10] |
| Quercus sp. | Generalist | Mature mixed forest in the southern Appalachian Mountains | Supplies carbohydrates to seedlings | AY656961 | [74] | |
| Inocybe sp. | Populus sp. | ND | Forest/dry habitat/heavy metal-polluted area | Tolerance to drought or increased concentrations of heavy metals in the soil | JQ898552 | [10] |
| Populus tremula | Generalist | Former lead/zinc smelter | Fungal immobilization of heavy metals | EF644135 | [11] | |
| Inocybe hirtella | Populus sp. | Nd | Forest | Plant nutrition | JQ898549 | [10] |
| Quercus ilex | Nd | Mediterranean forest | Adaptation to climate change | ND | [65] | |
| Laccaria ochropurpurea | Salix caprea | Nd | Former ore mining area, heavy metal-polluted soil | Tolerance against toxic metal concentrations in the soil | AY748870 | [24] |
| Castanea dentata | Nd | Chestnut plantation | Resistance to pathogens | MK077757 | [77] | |
| Tuber sp. | Salix caprea | Nd | Former ore mining area, heavy metal-polluted soil | Tolerance against toxic metal concentrations in the soil | AY748870 | [24] |
| Populus tremula | Generalist | Former lead/zinc smelter | Fungal immobilization of heavy metals | EF644167 | [11] | |
| Tuber maculatum | Populus alba | Nd | Marsh in nature reserve | Adaptation to ecological conditions | HG937633 | [52] |
| Populus nigra, Pinus nigra, Tilia x vulgaris | Generalist | Man-made park, Mediterranean area | Nutritional exchange or competition for the main nutritional resources in fungal communities | ND | [83] | |
| Cenococcum geophilum | Carya ovata | Generalist | Old experimental plantation of exotic trees, temperate mixed forest | Acclimatization of exotic trees outside of their native Range | ND | [67] |
| Quercus sp. | Generalist | Oak savanna/forest, post-fire site | Survival in the extreme environment (after burning) | ND | [66] | |
| Phialophora finlandia | Picea abies | Generalist | Forest nursery | Improvement of the growth and survival of outplanted tree seedlings | DQ508807 | [81] |
| Pinus sylvestris | Generalist | Forest, post-agricultural land (slightly polluted soil) and buffer zone of the copper smelter (heavy metal-polluted soil) | Adaptation to different soil conditions | ND | [83] |
5. Conclusions
In the present study, only the EM symbioses formed by Thelephorales (Tomentella spp.) and Agaricales (Inocybe spp.) were observed at both investigated sites. They can be classified as early colonizers of S. caprea, since they are well known to occur in their host during the early stages of growth. The identified EM taxa were mainly short-distance exploration types, and these taxa typically occurred in the fluctuating nutrient pools that are characteristic of disturbed soils. The EM fungi described in this study were previously reported by other researchers in disturbed environments, e.g., dry and polluted soils, and they have potential functions in the adaptation and growth of S. caprea.
Author Contributions
K.H. participated in all analyses and wrote the first version of the manuscript. BF participated in the preparation of the manuscript. K.H., B.U.F. and J.S. analyzed the results and the statistical output. K.H. and C.B. performed sampling at the locations, selected plant genotypes for the experiment, and provided input for the manuscript. C.B. conducted soil analyses. K.H. designed and managed the field and lab experiments. C.B. was the respondent for the study conceptualization, supervision, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
The work of K. Hrynkiewicz was funded by the European Commission (Marie Curie Host Fellowship HPMD-CT-2001-00078), the Ministry of Scientific Research and Information Technology (Poland, N N305 187337), and DAAD (A/10/05971).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated for this study can be found in the NCBI at https://www.ncbi.nlm.nih.gov (accessed on 5 March 2024) the accession numbers are given in Table 2.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Suz, L.M.; Bidartondo, M.I.; van der Linde, S.; Kuyper, T.W. Ectomycorrhizas and tipping points in forest ecosystems. New Phytol. 2021, 231, 1700–1707. [Google Scholar] [CrossRef] [PubMed]
- Policelli, N.; Horton, T.R.; Hudon, A.T.; Patterson, T.; Bhatnagar, J.M. Back to roots: The role of ectomycorrhizal fungi in boreal and temperate forest restoration. Front. For. Glob. Chang. 2020, 3, 97. [Google Scholar] [CrossRef]
- Karst, J.; Erbilgin, N.; Pec, G.J.; Cigan, P.W.; Najar, A.; Simard, S.W.; Cahill, J.F., Jr. Ectomycorrhizal fungi mediate indirect effects of a bark beetle outbreak on secondary chemistry and establishment of pine seedlings. New Phytol. 2015, 208, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, B.J.; Jones, M.D.; Kranabetter, J.M. Ectomycorrhizae and tree seedling nitrogen nutrition in forest restoration. New For. 2015, 46, 747–771. [Google Scholar] [CrossRef]
- Nara, K. The role of ectomycorrhizal networks in seedling establishment and primary succession. In Mycorrhizal Networks; Springer: Dordrecht, The Netherlands, 2015; pp. 177–201. [Google Scholar] [CrossRef]
- Pickles, B.J.; Simard, S.W. Mycorrhizal networks and forest resilience to drought. In Mycorrhizal Mediation of Soil; Elsevier: Amsterdam, The Netherlands, 2017; pp. 319–339. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M. Mycorrhizal types differ in ecophysiology and alter plant nutrition and soil processes. Biol. Rev. 2019, 94, 1857–1880. [Google Scholar] [CrossRef] [PubMed]
- Egerton-Warburton, L.M.; Querejeta, J.I.; Allen, M.F. Common mycorrhizal networks provide a potential pathway for the transfer of hydraulically lifted water between plants. J. Exp. Bot. 2007, 58, 1473–1483. [Google Scholar] [CrossRef] [PubMed]
- Soudzilovskaia, N.A.; van Bodegom, P.M.; Terrer, C.; Zelfde, M.V.T.; McCallum, I.; Luke McCormack, M.; Fisher, J.B.; Brundrett, M.C.; de Sá, N.C.; Tedersoo, L. Global mycorrhizal plant distribution linked to terrestrial carbon stocks. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Krpata, D.; Peintner, U.; Langer, I.; Fitz, W.J.; Schweiger, P. Ectomycorrhizal communities associated with Populus tremula growing on a heavy metal contaminated site. Mycol Res. 2008, 112, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Karliński, L.; Rudawska, M.; Leski, T. The influence of host genotype and soil conditions on ectomycorrhizal community of poplar clones. Eur. J. Soil Biol. 2013, 58, 51–58. [Google Scholar] [CrossRef]
- Usman, M.; Ho-Plágaro, T.; Frank, H.E.; Calvo-Polanco, M.; Gaillard, I.; Garcia, K.; Zimmermann, S.D. Mycorrhizal symbiosis for better adaptation of trees to abiotic stress caused by climate change in temperate and boreal forests. Front. For. Glob. Chang. 2021, 141, 742392. [Google Scholar] [CrossRef]
- Segnitz, R.M.; Russo, S.E.; Davies, S.J.; Peay, K.G. Ectomycorrhizal fungi drive positive phylogenetic plant–soil feedbacks in a regionally dominant tropical plant family. Ecology 2020, 101, e03083. [Google Scholar] [CrossRef]
- Argus, G.W. Infrageneric classification of Salix (Salicaceae) in the new world. In Systematic Botany Monographs; American Society of Plant Taxonomists: St. Louis, MO, USA, 1997; pp. 1–121. [Google Scholar]
- Weih, M.; Glynn, C.; Baum, C. Willow short-rotation coppice as model system for exploring ecological theory on biodiversity–ecosystem function. Diversity 2019, 11, 125. [Google Scholar] [CrossRef]
- Cannone, N.; Guglielmin, M.; Casiraghi, C.; Malfasi, F. Salix shrub encroachment along a 1000 m elevation gradient triggers a major ecosystem change in the European Alps. Ecography 2022, 2022, e06007. [Google Scholar] [CrossRef]
- Weih, M.; Nordh, N.E.; Manzoni, S.; Hoeber, S. Functional traits ofindividual varieties as determinants of growth and nitrogen use patterns inmixed stands of willow (Salix spp.). For. Ecol. Manag. 2021, 479, 118605. [Google Scholar] [CrossRef]
- Dimitriou, I.; Aronsson, P. Wastewater and sewage sludge application to willows and poplars grown in lysimeters–Plant response and treatment efficiency. Biomass Bioenergy 2011, 35, 161–170. [Google Scholar] [CrossRef]
- Sandil, S.; Gowala, N. Willows: Cost-effective tools for bioremediation of contaminated soils. In Advances in Bioremediation and Phytoremediation for Sustainable Soil Management; Malik, J.A., Ed.; Springer: Cham, Switzerland, 2022; pp. 183–202. [Google Scholar] [CrossRef]
- Trowbridge, J.; Jumpponen, A. Fungal colonization of shrub willow roots at the forefront of a receding glacier. Mycorrhiza 2004, 14, 283–293. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, E.W.; Vries, F.W.; Kuyper, T.W. Mycorrhizal associations of Salix repens L. communities in succession of dune ecosystems. I. Aboveground and below-ground views of ectomycorrhizal fungi in relation to soil chemistry. Can. J. Bot. 2000, 77, 1821–1832. [Google Scholar] [CrossRef]
- Van der Heijden, E.W. Differential benefits of arbuscular mycorrhizal and ectomycorrhizal infection of Salix repens. Mycorrhiza 2001, 10, 185–193. [Google Scholar] [CrossRef]
- Hrynkiewicz, K.; Baum, C.; Leinweber, P. Mycorrhizal community structure, microbial biomass P and phosphatase activities under Salix polaris as influenced by nutrient availability. Eur. J. Soil Biol. 2009, 45, 168–175. [Google Scholar] [CrossRef]
- Hrynkiewicz, K.; Ingeborg, H.; Christel, B. Ectomycorrhizal community structure under willows at former ore mining sites. Eur. J. Soil Biol. 2008, 44, 37–44. [Google Scholar] [CrossRef]
- Parádi, I.; Baar, J. Mycorrhizal fungal diversity in willow forests of different age along the river Waal, The Netherlands. For. Ecol. Manag. 2006, 237, 366–372. [Google Scholar] [CrossRef]
- Hrynkiewicz, K.; Toljander, Y.K.; Baum, C.; Fransson, P.M.A.; Taylor, A.F.S.; Weih, M. Correspondence of ectomycorrhizal diversity and colonisation of willows (Salix spp.) grown in short rotation coppice on arable sites and adjacent natural stands. Mycorrhiza 2012, 22, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Hrynkiewicz, K.; Szymańska, S.; Piernik, A.; Thiem, D. Ectomycorrhizal community structure of Salix and Betula spp. at a saline site in central Poland in relation to the seasons and soil parameters. Water Air Soil Pollut. 2015, 226, 99. [Google Scholar] [CrossRef]
- Agerer, R. Exploration types of ectomycorrhizae. Mycorrhiza 2001, 11, 107–114. [Google Scholar] [CrossRef]
- Regvar, M.; Likar, M.; Piltaver, A.; Kugonič, N.; Smith, J.E. Fungal community structure under goat willows (Salix caprea L.) growing at metal polluted site: The potential of screening in a model phytostabilisation study. Plant Soil 2010, 330, 345–356. [Google Scholar] [CrossRef]
- Kumar, A.; Yadav, A.N.; Mondal, R.; Kour, D.; Subrahmanyam, G.; Shabnam, A.A.; Khan, S.A.; Yadav, K.K.; Sharma, G.K.; Cabral-Pinto, M.; et al. Mycoremediation: A mechanistic understanding of contaminants alleviation from natural environment and future prospect. Chemosphere 2021, 284, 131–325. [Google Scholar] [CrossRef] [PubMed]
- Rúa, M.A. Characterizing Ectomycorrhizal fungal community structure and function of two varieties of Pinus clausa that differ in disturbance history. Forests 2021, 12, 219. [Google Scholar] [CrossRef]
- Kutschera, L.; Lichtenegger, E. Wurzelatlas Mitteleuropäischer Waldbäume und Sträucher; Leopold Stocker Verlag: Graz, Austria, 2002. [Google Scholar]
- Dick, W.A.; Tabatabai, M.A. An alkaline oxidation method for determination of total phosphorus in soils. Soil Sci. Soc. Am. J. 1977, 41, 511–514. [Google Scholar] [CrossRef]
- Schlichting, E.; Blume, H.P. Bodenkundliches Praktikum; Verlag Paul Parey: Berlin, Germany, 1966. [Google Scholar]
- Agerer, R. (Ed.) Colour Atlas of Ectomycorrhizae, 1st–12th ed.; Einhorn, Verlag: Schwäbisch Gmünd, Germany, 1987–2002. [Google Scholar]
- Agerer, R.; Rambold, G. DEEMY, a DELTA-Based Information System for Characterisation and Determination of Ectomycorrhizae; Version 1.1; Institute for Systematic Botany, Section Mycology, University of München: München, Germany, 1998. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Vrålstad, T.; Schumacher, T.; Taylor, A.F. Mycorrhizal synthesis between fungal strains of the Hymenoscyphusericae aggregate and potential ectomycorrhizal and ericoid hosts. New Phytol. 2002, 153, 143–152. [Google Scholar] [CrossRef]
- O’donnell, K. Fusarium and its near relatives. In Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics; Reynolds, D.R., Taylor, J.W., Eds.; CAB International: Wallingford, UK, 1993; pp. 225–233. [Google Scholar]
- Haug, I. Identification of Picea ectomycorrhizae by comparing DNA-sequences. Mycol. Prog. 2002, 1, 167–178. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Larsson, K.H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Pritsch, K.; Boyle, H.; Munch, J.C.; Buscot, F. Characterization and identification of black alder ectomycorrhizas by PCR/RFLP analyses of the rDNA internal transcribed spacer (ITS). New Phytol. 1997, 137, 357–369. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Koczorski, P.; Furtado, B.U.; Gołębiewski, M.; Hulisz, P.; Baum, C.; Weih, M.; Hrynkiewicz, K. The effects of host plant genotype and environmental conditions on fungal community composition and phosphorus solubilization in willow short rotation coppice. Front. Plant Sci. 2021, 5, 647–709. [Google Scholar] [CrossRef] [PubMed]
- Ryberg, M.; Nilsson, R.H.; Kristiansson, E.; Töpel, M.; Jacobsson, S.; Larsson, E. Mining metadata from unidentified ITS sequences in GenBank: A case study in Inocybe (Basidiomycota). BMC Evol. Biol. 2008, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Horton, T.R.; Bruns, T.D. The molecular revolution in ectomycorrhizal ecology: Peeking into the black-box. Mol. Ecol. 2001, 10, 1855–1871. [Google Scholar] [CrossRef] [PubMed]
- Tiebel, K.; Huth, F.; Wagner, S. Soil seed banks of pioneer tree species in European temperate forests: A review. Iforest Biogeo Sci. For. 2018, 11, 48. [Google Scholar] [CrossRef]
- Nara, K.; Nakaya, H.; Wu, B.; Zhou, Z.; Hogetsu, T. Underground primary succession of ectomycorrhizal fungi in a volcanic desert on Mount Fuji. New Phytol. 2003, 159, 743–756. [Google Scholar] [CrossRef]
- Nara, K.; Hogetsu, T. Ectomycorrhizal fungi on established shrubs facilitate subsequent seedling establishment of successional plant species. Ecology 2004, 85, 1700–1707. [Google Scholar] [CrossRef]
- Nara, K. Pioneer dwarf willow may facilitate tree succession by providing late colonizers with compatible ectomycorrhizal fungi in a primary successional volcanic desert. New Phytol. 2006, 171, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Katanić, M.; Grebenc, T.; Orlović, S.; Matavuly, M.; Kovačević, B.; Bajc, M.; Kraigher, H. Ectomycorrhizal fungal community associated with autochthonous white poplar from Serbia. iForest 2015, 9, 330–336. [Google Scholar] [CrossRef]
- Cripps, C.L.; Liimatainen, K.; Niskanen, T.; Dima, B.; Bishop, R.F.; Ammirati, J.F. Intercontinental distributions of species of Cortinarius, subgenus Phlegmacium, associated with Populus in western North America. Botany 2015, 93, 711–721. [Google Scholar] [CrossRef]
- Kokkonen, K. Diversity of boreal small species of Cortinarius subgenus Telamonia with Salix. Karstenia 2020, 58, 60–117. [Google Scholar] [CrossRef]
- Cripps, C.L.; Eberhardt, U.; Schütz, N.; Beker, H.J.; Evenson, V.S.; Horak, E. The genus Hebeloma in the rocky mountain alpine zone. MycoKeys 2019, 46, 1–54. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.W.; Nguyen, N.H.; Stefanski, A.; Han, Y.; Hobbie, S.E.; Montgomery, R.A.; Reich, P.B.; Kennedy, P.G. Ectomycorrhizal fungal response to warming is linked to poor host performance at the boreal-temperate ecotone. Glob. Chang. Biol. 2017, 23, 1598–1609. [Google Scholar] [CrossRef]
- Koide, R.T.; Fernandez, C.; Malcolm, G. Determining place and process: Functional traits of ectomycorrhizal fungi that affect both community structure and ecosystem function. New Phytol. 2014, 201, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Becerra, A.G.; Nouhra, E.R.; Silva, M.P.; McKay, D. Ectomycorrhizae, arbuscular mycorrhizae, and dark-septate fungi on Salix humboldtiana in two riparian populations from central Argentina. Mycoscience 2009, 50, 343–352. [Google Scholar] [CrossRef]
- Jakucs, E.; Kovács, G.M.; Szedlay, G.; Erős-Honti, Z. Morphological and molecular diversity and abundance of tomentelloid ectomycorrhizae in broad-leaved forests of the Hungarian Plain. Mycorrhiza 2005, 15, 459–470. [Google Scholar] [CrossRef]
- Vizzini, A.; Angelini, C.; Losi, C.; Ercole, E. Thelephoradominicana (Basidiomycota, Thelephorales), a new species from the Dominican Republic, and preliminary notes on thelephoroid genera. Phytotaxa 2016, 265, 27–38. [Google Scholar] [CrossRef]
- Kõljalg, U.; Dahlberg, A.; Taylor, A.F.; Larsson, E.; Hallenberg, N.; Stenlid, J.; Larsson, K.H.; Fransson, P.M.; Kårén, O.; Jonsson, L. Diversity and abundance of resupinate thelephoroid fungi as ectomycorrhizal symbionts in Swedish boreal forests. Mol. Ecol. 2000, 9, 1985–1996. [Google Scholar] [CrossRef] [PubMed]
- Toledo, A.V.; Franco, M.E.E.; Lopez, S.M.Y.; Troncozo, M.I.; Saparrat, M.C.N.; Balatti, P.A. Melanins in fungi: Types, localization and putative biological roles. Physiol. Mol. Plant Pathol. 2017, 99, 2–6. [Google Scholar] [CrossRef]
- Matheny, P.B.; Hobbs, A.M.; Esteve-Raventós, F. Genera of Inocybaceae: New skin for the old ceremony. Mycologia 2020, 112, 83–120. [Google Scholar] [CrossRef] [PubMed]
- Richard, F.; Roy, M.; Shahin, O.; Sthultz, C.; Duchemin, M.; Joffre, R.; Selosse, M.A. Ectomycorrhizal communities in a Mediterranean forest ecosystem dominated by Quercus ilex: Seasonal dynamics and response to drought in the surface organic horizon. Ann. For. Sci. 2011, 68, 57–68. [Google Scholar] [CrossRef]
- Dickie, I.A.; Dentinger, B.T.; Avis, P.G.; McLaughlin, D.J.; Reich, P.B. Ectomycorrhizal fungal communities of oak savanna are distinct from forest communities. Mycologia 2009, 101, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Wilgan, R.; Leski, T.; Kujawska, M.; Karliński, L.; Janowski, D.; Rudawska, M. Ectomycorrhizal fungi of exotic Carya ovata in the context of surrounding native forests on Central European sites. Fungal Ecol. 2020, 44, 100908. [Google Scholar] [CrossRef]
- Park, J.Y. A change in color of aging mycorrhizal roots of Tilia americana formed by Cenococcum graniforme. Can. J. Bot. 1970, 48, 1339–1341. [Google Scholar] [CrossRef]
- Garcia-Barreda, S.; Molina-Grau, S.; Reyna, S. Reducing the infectivity and richness of ectomycorrhizal fungi in a calcareous Quercus ilex forest through soil preparations for truffle plantation establishment: A bioassay study. Fungal Biol. 2015, 119, 1137–1143. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Yang, M.; Yan, L.; Kang, Z.; Xiao, Y.; Tang, P.; Ye, L.; Zhang, B.; Zou, J.; et al. Tuber borchii shapes the ectomycorrhizosphere microbial communities of Corylus avellana. Mycobiology 2019, 47, 180–190. [Google Scholar] [CrossRef]
- Louro, R.; Natário, B.; Santos-Silva, C. Morphological characterization of the in vitro mycorrhizae formed between four Terfezia species (Pezizaceae) with Cistus salviifolius and Cistus ladanifer—Towards desert truffles production in acid soils. J. Fungi 2021, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Jumpponen, A.; Trappe, J.M.; Cázares, E. Occurrence of ectomycorrhizal fungi on the forefront of retreating Lyman Glacier (Washington, USA) in relation to time since deglaciation. Mycorrhiza 2002, 12, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Graf, F.; Brunner, I. Natural and synthesized ectomycorrhizas of the alpine dwarf willow Salix herbacea. Mycorrhiza 1996, 6, 227–235. [Google Scholar] [CrossRef]
- Heijden, E.V.D.; Kuyper, T.W. Ecological strategies of ectomycorrhizal fungi of Salix repens: Root manipulation versus root replacement. Oikos 2003, 103, 668–680. [Google Scholar] [CrossRef]
- Walker, J.F.; Miller, O.K., Jr.; Horton, J.L. Hyperdiversity of ectomycorrhizal fungus assemblages on oak seedlings in mixed forests in the southern Appalachian mountains. Mol. Ecol. 2005, 14, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Marmeisse, R.; Gryta, H.; Jargeat, P.; Fraissinet-Tachet, L.; Gay, G.; Debaud, J.C. Hebeloma. In Ectomycorrhizal Fungi Key Genera in Profile; Cairney, J.W.G., Chambers, S.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 89–127. [Google Scholar] [CrossRef]
- Kropp, B.R.; Mueller, G.M. Laccaria. In Ectomycorrhizal Fungi Key Genera in Profile; Cairney, J.W.G., Chambers, S.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 65–88. [Google Scholar] [CrossRef]
- Ritzi, M.V.; Russell, S.D.; Aime, M.C.; McNickle, G.G. First report of ectomycorrhizal fungus Laccaria ochropurpurea, associated with Castanea dentata (American chestnut) roots in a mixed species plantation. Plant Health Prog. 2019, 20, 140–141. [Google Scholar] [CrossRef]
- Jumpponen, A.R.I.; Trappe, J.M. Dark septate endophytes: A review of facultative biotrophic root-colonizing fungi. New Phytol. 1998, 140, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, H.E.; Wang, C.J.K. Mycorrhizal and pathological associations of dematiaceous fungi in roots of 7-month-old tree seedlings. Can. J. For. Res. 1987, 17, 884–899. [Google Scholar] [CrossRef]
- Ruotsalainen, A.L. Dark Septate Endophytes (DSE) in boreal and subarctic forests. In Endophytes of Forest Trees. Forestry Sciences; Pirttilä, A., Frank, A., Eds.; Springer: Cham, Switzerland, 2018; Volume 86, pp. 105–117. [Google Scholar] [CrossRef]
- Trocha, L.K.; Rudawska, M.; Leski, T.; Dabert, M. Genetic diversity of naturally established ectomycorrhizal fungi on Norway spruce seedlings under nursery conditions. Microb. Ecol. 2006, 52, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, M.; Iotti, M.; Oddis, M.; Lalli, G.; Pacioni, G.; Leonardi, P.; Maccherini, S.; Perini, C.; Salerni, E.; Zambonelli, A. Assessment of ectomycorrhizal fungal communities in the natural habitats of Tuber magnatum (Ascomycota, Pezizales). Mycorrhiza 2013, 23, 349–358. [Google Scholar] [CrossRef]
- Rudawska, M.; Leski, T.; Stasińska, M. Species and functional diversity of ectomycorrhizal fungal communities on Scots pine (Pinus sylvestris L.) trees on three different sites. Ann. For. Sci. 2011, 68, 5–15. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).