Abstract
Physalis species are used as an indigenous food and medicine in Kenya. However, species identification and an analysis of the health-promoting bioactive compounds and antioxidant properties are lacking. In this study, we report the molecular identification and mineral and phytochemical profiling of wild Physalis accessions. Leaf samples of 10 Physalis accessions were collected and used for species identification using nuclear ITS2 and plastid rbcL barcodes. Ripe fruits were collected from the same accessions and analyzed for mineral, total phenolic, tannin, and flavonoid contents, and antioxidant activities. The Physalis species were discriminated based on the ITS2 barcode and identified as Physalis purpurea. The genetic diversity, distance, and polymorphism of the ITS2 region of Physalis accessions were high due to the high rate of singleton and parsimony mutations. No genetic diversity, distance, or polymorphism was observed based on the rbcL barcode. The mineral content was significantly different (p < 0.05) for calcium, zinc, nickel, copper, and lithium among the Physalis accessions. No significant variation (p > 0.05) was found for phenolic acids or flavonoids, but the tannic acid content varied significantly (p < 0.05). DPPH free radical scavenging varied significantly (p < 0.05) among Physalis accessions. In conclusion, nuclear ITS2 was used to successfully identify the Physalis species of all the accessions as Physalis purpurea. The present study confirmed that Physalis purpurea has a significantly high mineral and phytochemical content and antioxidant activity. The findings from this study can be used to facilitate exploitation of Physalis purpurea in genetic breeding, their application in pharmaceutical, cosmetic, and nutritional value as well as conservation and sustainable use.
1. Introduction
Indigenous varieties adapted to a particular region are ideal resilient crops for climate change adaptation but are neglected and often lost due to the rapid domestication of commercial cultivars. Restoring such plant species would empower local farmers and provide huge economic and nutritional benefits. One such underutilized wild plant species is of the genus Physalis and belongs to the Nightshade (Solanaceae) family [1]. Physalis species are native to the Peruvian and Ecuadorian Andes region of South America; hence, it is referred to as the Peruvian gooseberry [2], although some are also native to Southeast Asia and Eurasia [3]. Studies of Physalis in China have identified five species including Physalis alkekengi, P. angulata, P. pubescens, P. peruviana, and P. minima and two variants of P. alkekengi [4]. Physalis species such as P. philadelphica, P. peruviana, and P. pubescens are grown in various parts of the world [5]. Physalis fruits are exported from several countries including Colombia, Australia, New Zealand, Great Britain, Zimbabwe, Kenya, Egypt, South Africa, Madagascar, and South East Asia [6,7]. The largest producer, consumer, and exporter of Physalis fruits is Colombia [7]. In Kenya, the fruits are seen in vast numbers as wild and self-propagating plants, commonly in farms during and after the harvesting of maize. Currently, there is growing commercial interest in this fruit crop because of its nutritional and pharmacological properties and health benefits. The key steps involved in the proper utilization of this indigenous plant species are accurate identification and authentication.
Taxonomic identification and morphological characterization are the most common methods of plant authentication, although they are limited by environmental or physiological factors and the developmental phase of the plant species [8]. Employing molecular tools such as DNA barcoding could be more successful in species identification. DNA barcoding requires a short universal DNA sequence that exhibits a sufficient level of variation to discriminate species [9,10]. The proposed plant DNA core barcodes from the Consortium for the Barcode of Life (CBOL) Plant Working Group comprise the chloroplast gene large subunit of ribulose bisphosphate carboxylase (rbcL) and matK with the trnH-psbA intergenic sequence and internal transcribed spacer (ITS), and a nuclear gene as the supplement barcode [11]. Chloroplast DNA barcodes such as matK and rbcL have been used in many phylogenetic and plant species identification analyses [12]. The nuclear internal transcribed spacer 2 (ITS2) barcode has also been demonstrated to exhibit 100% species identification and discrimination statistics in plants due to its high intra- and interspecific divergence [13]. Currently, there is lack of information on the genetic characterization of indigenous Physalis plants from the wild in Kenya.
The nutritional and phytochemical profiles of Physalis have been studied to a moderate level in the Andes [14]. The Physalis fruit contains fat and water-soluble vitamins (B-complex, C, A, E, and K), minerals (mainly phosphorus, magnesium, potassium, zinc, and calcium), sugars (e.g., sucrose, glucose, and fructose), fatty acids (palmitate, oleic, and gamma linoleic acid), phytosterols (sitosterol and stigmasterol), and fiber (pectin) [6,15]. The fruit pomace (skin and seed) contains 19.3% oil, 3.1% ash, 17.8% protein, 24.5% carbohydrates, and 28.7% crude fiber [14]. The phytochemicals found in Physalis include withanolides, physalins, carotenoids, phenolics, and flavonoids [15]. A review study states that the most studied phytochemicals are the polyphenolics, which have antioxidant properties and many health-related benefits [16]. The key polyphenolics include flavonoids, stilebenes, phenolic acids, coumarins, and tannins [17]. The Physalis plant’s high content of vitamins, minerals, and antioxidant phytochemicals gives it medicinal properties such as anti-inflammatory activity and this has been specifically determined for Physalis alkekengi species [18]. The characterization of the nutritional and bioactive properties of cultivated and wild fruits of Physalis peruviana growing in the northern Argentinian region identified several types of phytochemicals present in the plant, such as flavonoids and tannins [19]. However, the mineral composition, phytochemical profile, and antioxidant activities of wild fruits of Physalis growing in Kenya have not yet been characterized.
The aim of the current study was to identify and assess the rbcL and ITS2 gene barcodes to discriminate indigenous Physalis accessions collected from the forest. We also evaluated the mineral and phytochemical content, and antioxidant activities of ripe fruits to determine if they are nutrient-rich so that they could be used as sustainable resources for the development of biofortified crops and also promoted as a natural source of antioxidants.
2. Materials and Methods
2.1. Sampling of Plant Material
Leaves and mature fruits of Physalis plants were collected in April 2019 from Sorget forest in Londiani area of Kericho County, Kenya (Figure 1). The area is located at an elevation of 2528 m above sea level and latitude of 0.0684° S and a longitude of 35.5548° E.
Figure 1.
Map showing the location of Physalis sampling in Londiani, Kericho County, Kenya.
The collected Physalis plant samples were identified by the taxonomist Mr. Patrick Mutiso and the samples were preserved in the University of Nairobi herbarium in the Department of Biology (Codes of Voucher Specimens: KP/UON2019/001- KP/UON2019/010). Ten Physalis plants were sampled for their fruits and leaves based on the availability and ripeness of the fruits. The maturity of the fruits was assessed based on the color of the fruit. Ripe fruits had a yellow to orange color. The leaves were used for the molecular identification of the plant species, while the fruits were used for the study of the nutritive value and functional attributes. The collected leaf and fruit samples were wrapped with aluminum foil, kept in an ice box and transferred to the Molecular Biology Laboratory in the Department of Biochemistry, University of Nairobi. The leaf samples were kept at −80 °C prior to genomic DNA extraction. The fruit samples were utilized immediately upon arrival at the laboratory for the extraction of phytochemicals and the determination of the mineral content.
2.2. Molecular Authentication of Physalis Plants
2.2.1. Isolation of Genomic DNA from Leaves of Physalis Accessions
Isolation of genomic DNA from leaves of Physalis accessions was performed using the Cetyl trimethylammonium bromide (CTAB) method [20]. RibonucleaseA (RNase, 0.6 mg/mL) was added to the DNA accessions followed by incubation at 37 °C in a water bath for 30 min to eliminate any contaminating RNA. The integrity of the extracted genomic DNA was verified using 0.8% (w/v) agarose gel stained with ethidium bromide (0.5 µg/mL) and viewed under UV transilluminator in Gel DocTM EZ Imaging System (BioRad, Hercules, CA, USA). DNA was stored at −20 °C until use in the molecular analysis.
2.2.2. Polymerase Chain Reaction (PCR) Amplification and Sequencing
Polymerase chain reaction (PCR) amplification was performed using the DNA barcoding primers rbcL and ITS2 (Table 1). DNA amplification was conducted using a One Taq® Hot start 2× master mix with standard buffer (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s instructions. Amplification was conducted in a Veriti, 96-well Thermal Cycler (Thermo Fischer Scientific, Waltham, MA, USA). Optimization was performed in order to acquire the best conditions for PCR amplification. The annealing temperature for both primers was optimized at the following temperatures: 50 °C, 51 °C, 52 °C, 54 °C, 56 °C, and 58 °C. The best optimum cycling conditions for both primers were used for the PCR amplification of the DNA samples (Table 1). The amplicons were confirmed using 1% agarose gel stained with ethidium bromide (0.5 µg/mL) under a UV transilluminator in the Gel DocTM EZ Imaging System (BioRad, Hercules, CA, USA). Amplicons were cleaned using a gel clean up kit (Applied Biosystems, Thermo Fischer Scientific, Waltham, MA, USA) and sent for sanger sequencing at the University of Nairobi (UoN) Center of Excellence in HIV Medicine (CoEHM) using a 3730 s DNA analyzer (Thermo Fischer, Waltham, MA, USA).

Table 1.
Oligonucleotide primers used for PCR amplification and optimum PCR cycling conditions.
2.3. Sequence and Phylogenetic Analysis
The sequences of each barcode were edited manually in the BioEdit software version 7.2.5.0 [21]. The edited sequences were compared with the available nucleotide sequences in the GenBank database. The sequences were blasted in the NCBI GenBank BLASTn database to determine the sequence homology with other deposited ITS2 and rbcL sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 10 February 2023). The identification of Physalis species was based on the least expected value (E-value), highest query coverage, and similarity percentage. The obtained sequences were also assembled and aligned using the MUSCLE algorithm. Multiple sequence alignment (MSA) of ITS2 and rbcL sequences in this study and the reference sequences retrieved from the NCBI database was performed using the MUSCLE software version 3.8 [22]. This MSA was used in the preparation of a phylogenetic tree. The ITS2 and rbcL sequences were also aligned separately using MUSCLE, and viewed and trimmed on Jalview version 2.11.2.6 to obtain uniform sequence lengths [23,24]. The MSAs performed separately for ITS2 and rbcL sequences were used in the genetic diversity, nucleotide polymorphism, neutrality test, and automatic barcode gap discovery (ABGD) analysis. All MSAs attained were compressed using ESPript 3 (http://espript.ibcp.fr; accessed on 12 February 2023) [25]. The ITS2 and rbcL sequences were submitted to NCBI GenBank through a web-based sequence submission tool and accession numbers were assigned.
Phylogenetic trees were constructed based on the Bayesian inference (BI) method using MrBayes version 3.2.7 (https://nbisweden.github.io/MrBayes/; accessed on 12 February 2023). Statistical analysis was performed using the posterior distribution of the model parameter, which was estimated using the Markov chain Monte Carlo (MCMC) method [26,27,28]. MCMC sampling was performed over 18,000,000 generations at a sampling frequency of 1000 and the first 25% (relburnin = yes burninfrac = 0.25) of samples were discarded when estimating the posterior probabilities of the trees. After 18,000,000 generations, the analysis was stopped when the average standard deviation of the split frequencies was less than 0.01 and tree parameters were summarized. The constructed phylogenetic trees were visualized and modified using the FigTree software version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/; accessed on 12 February 2023).
2.4. Analysis of Genetic Divergence
DNA divergence within Physalis accession populations based on ITS2 and rbcL sequences were determined using the DnaSP software version 6.12.03 [29]. The multiple sequence alignment (MSA) for Physalis accessions based on either the ITS2 or rbcL marker was uploaded into the software and various parameters for the divergence were determined. The number of polymorphic segregating sites (S), nucleotide diversity, and the total number of substitutions were assessed as outlined by the Jukes and Cantor algorithm on DnaSP.
2.5. Determination of Genetic Distance within Physalis Accessions
Intraspecific genetic distances and the overall mean distance of Physalis accessions based on the ITS2 and rbcL sequences were determined using the Kimura 2 parameter (K2P) model with the gamma distribution and a gamma parameter of 0.27 using MEGA version 11.0 [30]. Sequence genetic distance was determined using multiple sequence alignments for Physalis accessions based on ITS2 and rbcL markers.
2.6. Nucleotide Polymorphism and Neutrality Tests
DNA polymorphisms of the ITS2 and rbcL sequences were assessed in all the Physalis accessions. The DNA sequence Polymorphism (DnaSP) software version 6.12.03 was utilized in the DNA polymorphism analysis for ITS2 and rbcL sequences of all Physalis accessions. The DNA polymorphism parameters determined were polymorphic segregating sites, singleton and parsimony informative sites, the nucleotide diversity, and the average number of nucleotide differences.
Tajima’s neutrality test for both ITS2 and rbcL sequences of Physalis accessions were determined to estimate the frequency of mutations among species [31]. The Tajima’s neutrality test determined the Tajima D value among the ITS2 and rbcL sequences of Physalis accessions using the MEGA 11.0 software [32,33]. The analysis involved nine and ten ITS2 and rbcL sequences of Physalis accessions, respectively. The codon positions included were 1st + 2nd + 3rd + noncoding for the rbcL gene sequences. All ambiguous positions were eliminated for each sequence pair (pairwise deletion option) in both the analysis based on ITS2 and rbcL genes. There were a total of 399 and 614 positions in the final dataset for both the ITS2 and rbcL genes, respectively. The MSAs utilized in this analysis were similar those utilized for genetic diversity and DNA polymorphism studies based on ITS2 and rbcL sequences.
2.7. Analysis of DNA Barcoding Gap and Intraspecific Distance
In order to delimit the Physalis species based on their intraspecific divergence within a population, the automatic barcode gap discovery (ABGD) method described by [34] was utilized in this study. Multiple sequence alignments utilized in genetic diversity analysis were also utilized for the ABGD analysis of ITS2 and rbcL sequences of Physalis accessions. The ITS2 and rbcL multiple sequence alignments were separately inputted into the ABGD website (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html; accessed on 17 February 2023) and the distance analysis was performed based on the K80 Kimura measure of distance. The default value for the relative gap width (X) was set at 1.5. Moreover, p values of intraspecific divergence were set at a prior minimum (Pmin) and prior maximum (Pmax) divergence of intraspecific diversity of 0.001 and 0.1, respectively. Default settings were utilized for all other parameters.
2.8. Analysis of Mineral Content in Ripe Fruits
The analysis of macrominerals (calcium (Ca), magnesium (Mg), potassium (K), and sodium (Na)) and trace elements (iron (Fe), zinc (Zn), manganese (Mn), copper (Cu), lithium (Li), and nickel (Ni)) was performed according to the method described by [35]. All the analyses were performed in triplicate. Each fruit sample (1 g) was digested in 5 mL of nitric acid, which was made up to 25 mL using distilled water. The mixture was heated on a hot plate until a third of the volume was left, which was filtered with Whatman filter paper No. 1. The filtrate was appropriately diluted and analyzed for mineral elements using an atomic absorption spectrophotometer (AAS) (Shimadzu, Kyoto, Japan). The wavelengths used for the analysis of each mineral were as follows: Ca—422.42 nm; Zn—213.52 nm; Cu—324.53 nm; Na—588.88 nm; Mg—285.04 nm; Fe—248.23 nm; K—766.74 nm; Ni—231.90 nm; Mn—27.03 nm; Li—670.85 nm. The results were expressed in ppm (1 ppm = 1 mg/L) of a sample of dry weight (DW). All the experiments were carried out three times with different fruit accessions.
2.9. Determination of Phytochemical Content
2.9.1. Estimation of Total Polyphenol Content (TPC)
The estimation of phenol in fruit extracts was assayed using the Folin Ciocalteau method [36]. In a test tube with 2.25 mL of 10% Folin Ciocalteau reagent, 1 mL of filtrate of ethanolic fruit extract was added, mixed thoroughly, and allowed to settle at 23 ± 2 °C for 5 min. To the mixture, 2.25 mL of sodium bicarbonate solution (60 g/L) was added, vortexed, and incubated at 23 ± 2 °C for 90 min. The absorbance was measured at 725 nm using a spectrophotometer (Shimadzu, Kyoto, Japan). A blank was prepared using the same method, but the fruit extract was replaced with sterile distilled water. A standard curve was used to determine the phenol content using gallic acid. All the experiments were carried out three times with different fruit accessions. The TPC was assessed as mg/mL of gallic acid equivalents per gram of fruit extract.
2.9.2. Estimation of Total Tannin Content (TTC)
The content of tannins in the fruit samples was assayed using the Folin Ciocalteau method [36]. In a test tube, 0.25 mL of 10% Folin Ciocalteau reagent was added to 0.1 mL of ethanolic fruit extracts and vortexed. The mixture was allowed to settle at 23 ± 2 °C for 5 min, and then 1.25 mL of sodium hydroxide was added and the mixture was incubated at room temperature for 40 min. Absorbance was measured at 725 nm using a spectrophotometer. A blank was prepared using the same method, but the fruit sample was replaced with sterile distilled water. A standard curve was then prepared for the estimation of the tannin content from Physalis fruits using tannic acid. All the experiments were carried out three times with different fruit accessions. The TTC was assessed as mg/mL of tannic acid equivalents per gram of fruit extract.
2.9.3. Estimation of Total Flavonoid Content (TFC)
The determination of the flavonoid content in the fruit extracts was performed using the aluminum chloride (AlCl3) colorimetric method [37]. In a test tube, 1 mL of ethanolic Physalis fruit extract was added to 4 mL of water and 0.3 mL of 5% sodium nitrate solution. The content was mixed thoroughly followed by the addition of 0.6 mL of aluminum chloride. The mixture was mixed thoroughly and incubated for 6 min at 23 ± 2 °C before the addition of 2 mL sodium hydroxide. A precipitate was formed on mixing, which was centrifuged and the absorbance of the supernatant was measured at 510 nm. A blank was prepared using the same method, but the fruit extract was replaced with sterile distilled water. A standard curve was prepared for the estimation of the flavonoid content using rutin. All the experiments were carried out three times with different fruit accessions. The TFC was assessed as mg/mL of rutin equivalents per gram of fruit extract.
2.10. Estimation of Antioxidant Activity
The antioxidant activity was measured using 2, 2-diphenyl-2-picrylhydrazyl (DPPH) and hydrogen peroxide radical scavenging in vitro assays. The DPPH radical scavenging (RS) assay was performed based on the radical degradation method described by [38] with some modifications. The sample extracts (0.5 mL) were mixed with 0.1 mM DPPH radical solution (0.3 mL) prepared in an ethanol solution. Color change from deep violet to light yellow was observed and the absorbance was measured at 517 nm using a UV-Vis spectrophotometer after 100 min of reaction in the dark. The blank was prepared using 3.3 mL of ethanol and 0.5 mL of the sample. A control was prepared using 3.5 mL of absolute ethanol and 0.3 mL of the DPPH radical solution and the absorbance was measured. The percentage inhibition of the DPPH radical of the sample extract relative to the control was used to determine the antioxidant capacity using the equation:
The hydrogen peroxide scavenging assay was performed using a method determined by [39] with a few modifications. In a test tube, 0.5 mL of the sample extract was mixed with 4 mL of 4 mM hydrogen peroxide solution prepared in 0.1 M phosphate buffer (pH 7.4). The mixture was incubated for 10 min at room temperature and the absorbance was measured at 230 nm using a UV-Vis spectrophotometer. A blank was prepared using phosphate buffer and sample extract. A control was prepared using phosphate buffer and hydrogen peroxide and the absorbance was measured. The hydrogen peroxide radical scavenging (HRS) activity percentage was calculated using the equation:
2.11. Statistical Analysis
The results for the antioxidant activity, and mineral and phytochemical content of the Physalis accessions obtained were reported as mean ± standard deviation (SD) using SPSS version 20 [40]. All the measurements were performed in triplicate. One-way analysis of variance (ANOVA) was performed using the statistical software SPSS version 20 [40]. The means from all analyses were separated by Tukey’s HDS multiple comparisons test at α = 0.05. A regression and correlation analysis was also performed to determine the impact of polyphenols on the antioxidant activity. Regression was analyzed using the statistical software SPSS version 20 [40] while correlation studies were analyzed using Microsoft excel version 2016.
3. Results
3.1. Amplification and Sequencing Success Rate
The PCR amplification results in both ITS2 and rbcL regions achieved success rates of 100% (Table 2). The lengths of the ITS2 and rbcL sequences were in the range of 301–663 bp and 520–733 bp, respectively. The average lengths of ITS2 and rbcL sequences were 561 bp and 616 bp, respectively. The GC contents of the ITS2 and rbcL sequences were in the ranges of 60–65.2% and 42.7–43.9%, respectively. The average GC content of ITS2 sequences was 61.1%, which was significantly higher than that of the rbcL sequences (43.1%). The sequencing success rate was 99% and 100% for ITS2 and rbcL sequences, respectively (Table 2).

Table 2.
Efficiency of PCR amplification and sequencing for Physalis accessions based on ITS2 and rbcL DNA barcode regions.
3.2. Species Discrimination Based on BLASTn Analysis
According to the BLASTn analysis of ITS-2 sequences, seven of the nine Physalis accessions were identified as Physalis purpurea, and one was identified as Physalis peruviana and one as Physalis aff philadelphica (Supplementary Table S1). The BLASTn analysis of rbcL sequences identified all the Physalis accessions as Physalis minima (Supplementary Table S1). The percentage identity for the nine Physalis accessions based on ITS2 sequences ranged from 86.00 to 94.4%. The percentage identity for 10 Physalis accessions based on rbcL sequences ranged from 99.86 to 100%. Eight out of the ten rbcL sequences of Physalis accessions gave a 100% sequence similarity with Physalis minima (NC_048515.1 from the GenBank). Two accessions (OQ507154.1 and OQ507156.1) were also identified as Physalis minima (NC_048515.1 from the GenBank) with a similarity identity of 99.59 and 99.86%, respectively (Supplementary Table S1).
3.3. Multiple Sequence Alignment
The multiple sequence alignment of combined ITS2 and rbcL gene sequences and retrieved sequences from the BLASTn analysis based on MUSCLE had a sequence length of 730 bp. The MSA is presented in Supplementary Figure S1. A high rate of substitution mutations was noted for the ITS2 sequences, while very few substitutions, deletions, and insertion mutations were noted for the rbcL sequences.
The MSA of ITS2 sequences of the nine Physalis accessions had a sequence length of 399 bp (Supplementary Figure S2). There was a high rate of substitution and deletion mutation within this MSA. A deletion point mutation was observed at position 10 of the MSA whereby, in Physalis accessions OQ372026.1 and OQ372023.1, the nucleotide thymine was deleted while all other accessions contained a thymine at this position. Another deletion point mutation was noted at position 46 of the MSA whereby guanine was deleted for the Physalis accessions OQ372023.1, OQ372026.1, OQ372027.1, and OQ372029.1. These Physalis accessions also had a deletion macrolesion mutation of eight nucleotides from position 179 to 187. Both transition and transversion point mutations were also identified in the alignment. At position 107 of this MSA, there was a transition point mutation for the Physalis accession OQ372023.1, whereby guanine replaced adenine. A transversion point mutation was observed at position 119 for the Physalis accession OQ372023.1 whereby guanine replaced the thymine found on all other sequences. Insertion mutations were not identified on this MSA. The multiple alignment of rbcL sequences of 10 Physalis accessions had a sequence length of 614 bp (Supplementary Figure S3). There were no deletions, insertions or substitution mutations noted in this MSA. All Physalis accessions had similar sequences with the key difference being the length of the sequences.
3.4. Physalis Species Identification Based on Phylogenetic Analysis
The phylogram tree was constructed based on Bayesian inference and the combination of ITS2 and rbcL sequences. Based on rbcL sequences, all the Physalis accessions were clustered together as indicated by colour red in Figure 2. The rbcL sequences did not form clades with their reference sequences on the phylogram and there was no species discrimination. The species names (Physalis minima) of the accessions indicated in the phylogram were based on the BLASTn analysis. Based on the ITS2 gene sequences, the Physalis accessions were identified with a percentage posterior probability of 100 as Physalis purpurea (Figure 2) and the sequences were deposited in GenBank with accession numbers OQ372021.1–OQ372029.1.
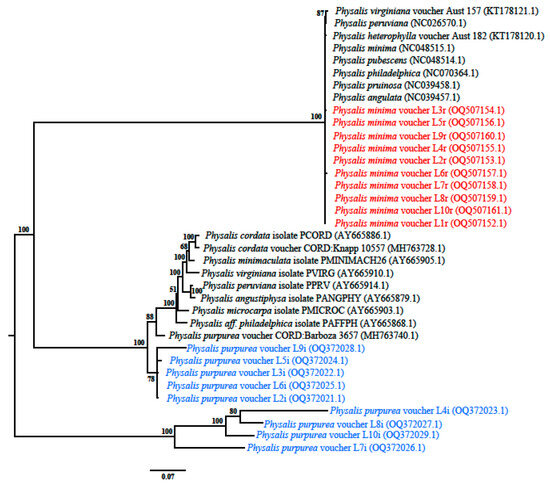
Figure 2.
Phylogenetic tree from MrBayes for the Physalis accessions based on ITS2 and rbcL gene sequences. Different colors are used to represent Physalis accessions (experimental) as the variant species and reference sequences used in this study. Black represents the two clusters and sequences retrieved from GenBank for both ITS2 and rbcL genes. Blue represents Physalis purpurea sequences and red represents Physalis minima sequences. The values above the branches represent the percentage posterior probability statistic from the Mr. Bayes phylogram.
3.5. Intraspecific Divergence of Physalis Accessions
DNA divergence was determined based on the number of polymorphic (segregating) sites (S), nucleotide diversity, and the total number of substitutions. Based on ITS2 gene sequences, nucleotide diversity, the total number of nucleotide substitutions, and the number of polymorphic (segregating) sites were 0.27629, 134, and 124, respectively. There was no nucleotide diversity (0), nucleotide substitutions (0.00000), or polymorphic segregating sites (0) among the Physalis accessions based on rbcL gene sequences.
3.6. Genetic Distance within Physalis Accessions
The genetic distance within Physalis accessions based on ITS2 and rbcL gene sequences was assessed. The overall average genetic distance among the Physalis accessions based on ITS2 gene sequences was 1.67 ± 0.77, while there was no genetic distance based on rbcL gene sequences. The mean average genetic distance within (intraspecific) Physalis species was 1.67 ± 0.84 and 0 based on ITS2 and rbcL gene sequences, respectively.
3.7. Nucleotide Polymorphism and Genetic Diversity of Physalis Accessions
There were 124 segregating sites identified with ITS2 gene sequences, while no segregating sites were recorded for the rbcL gene sequences (Table 3). The lack of segregating polymorphic sites within a population is an indication that all plants within this population are identical. The high number of polymorphic segregating sites for the ITS2 Physalis gene sequences is an indication that this gene is highly diverse among the Physalis accessions and has undergone differentiation. There were 21 singleton and 103 parsimony sites identified among the segregating sites of Physalis accessions based on the ITS2 gene sequences.

Table 3.
DNA polymorphism of Physalis accessions based on ITS2 and rbcL markers.
3.8. Tajima’s Neutrality Test
Tajima’s neutrality test was performed for both the ITS2 and rbcL gene sequences to assess the selection and nucleotide diversity of the Physalis accessions. The number of segregating sites (S) for ITS2 and rbcL were 180 and 0, respectively. The Tajima values of the Physalis accessions based on the ITS2 and rbcL gene sequences were 0.779171 and 0, respectively. The Tajima D of the ITS2 sequences indicated a negative selection pressure in the Physalis population. The nucleotide diversity of the ITS2 and rbcL sequences based on Tajima’s test was 0.190894 and 0, respectively, indicating a variation in the ITS2 barcode region of Physalis accessions.
3.9. Genetic Differences and Barcoding Gap Analysis
Automatic barcode gap discovery (ABGD) results generated by the K80 Kimura measure of distance (K2P) based on the ITS2 gene sequences for Physalis accessions were used to assess the presence of a barcoding gap. The rbcL gene sequences for Physalis accessions were not able to provide results on barcoding as there was no variation in the sequences at an intraspecific and interspecific level. Based on the ITS2 gene sequences, all pairwise distances were ranked by increasing the distance values from 0.02 to 0.70 and three barcoding gaps were detected (Figure 3). The first and smallest barcode gap was observed between distances of 0.15 (15%) and 0.18 (18%) (Figure 3). The second and largest barcode gap was evident between the distances of 0.22 (22%) and 0.44 (44%) (Figure 3). The third barcode gap was found between the distances of 0.59 (59%) and 0.70 (70%).
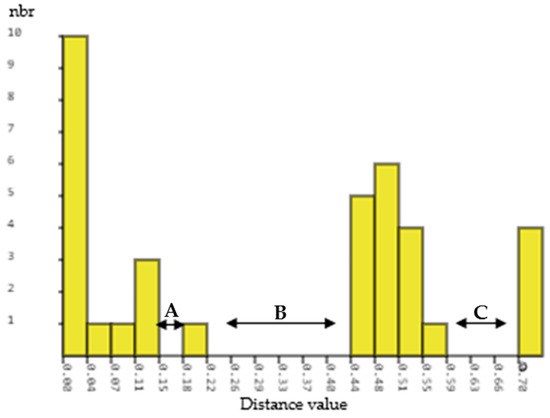
Figure 3.
A histogram indicating the hypothetical distribution of pairwise differences of ITS2 gene sequences for nine Physalis accessions. Low divergence is presumably intraspecific divergence, whereas higher divergence indicates interspecific divergence. The abbreviation nbr on the y-axis of the histogram stands for the number of pairwise comparisons. A, B, and C represent barcode gaps.
3.10. Mineral Analysis
The mineral content of Physalis accessions was determined for macro- and microminerals (Table 4 and Table 5, Supplementary Figure S4). The highest macromineral content among the Physalis accessions was noted for potassium at a mean of 527.778 ± 260.526, while the lowest was the magnesium content at a mean of 33.911 ± 29.942 (Table 4). Sodium and calcium contents were at moderate levels in Physalis accessions with means of 377.46 ± 147.193 and 128.121 ± 20.976, respectively (Table 4).

Table 4.
Macromineral content of fruits of Physalis accessions collected from the same environmental conditions. Data are expressed as mean ± SD of three independent accessions. CV: coefficient of variation; n = 3; means followed by single letters in a column differ significantly at a 5% level of significance.

Table 5.
Trace element content in fruits of Physalis accessions.
The micromineral content of Physalis accessions was also determined (Table 5). The highest micromineral content was noted for zinc at a mean of 24.364 ± 12.572 (Table 5). The lowest micromineral content was noted for lithium at a mean of 0.047 ± 0.024 (Table 5). A moderate micromineral content was noted for iron, manganese, copper, and nickel (Table 5).
3.11. Phytochemical Content and In Vitro Antioxidant Activity
The ripened fruits of Physalis accessions were assessed for phytochemical (total phenolic acid, tannic acid, and flavonoid) content and antioxidant activities (Table 6 and Supplementary Figures S5 and S6). The phytochemical contents and antioxidant activities did not change significantly (p > 0.05) among the Physalis accessions (Table 6). The TPC of different Physalis accessions did not change considerably, ranging from 0.024 ± 0.025 to 0.092 ± 0.053 mg GAE/g DW (Table 6). Physalis accessions L1 and L5 recorded significantly higher TTC compared with the other eight accessions (Table 6). The TFC of the different Physalis accessions did not change significantly and ranged from 0.058 ± 0.034 to 0.152 ± 0.089 mg Rutin/g DW. Two different chemical assays (DPPH and HRS) were performed to assess the antioxidant activity of Physalis accessions (Table 6 and Supplementary Figure S6). The DPPH and HRS values obtained for the antioxidant property did not show significant differences (p > 0.05) among the different Physalis accessions (Table 6). The DPPH and HRS among the different Physalis accessions ranged from 29.846 ± 13.537 to 97.344 ± 2.263% and 8.696 ± 7.609 to 64.131 ± 9.962%, respectively (Table 6).

Table 6.
Phytochemical content and radical scavenging activities of different indigenous Physalis accessions collected from the same environmental conditions.
3.12. Correlation Analysis between Phytochemical Contents and Antioxidant Activities
Correlation and regression studies were performed to assess the effect of each phytochemical compound (phenolic acid, tannins, and flavonoids) on the radical scavenging activity of Physalis accessions (Table 7). DPPH radical scavenging was largely facilitated by the phenolic acid content, with a correction of r = 0.327, while flavonoids and tannins had a negative correlation (Table 7). Hydrogen peroxide radical scavenging was largely facilitated by phenolic acids and flavonoids at a correlation value of r = 0.3599 and 0.2877, while tannins had a negative correlation (Table 7). The regression analysis showed that all phytochemicals (phenolic acids, tannins, and flavonoids) had a linear relationship to the radical scavenging of DPPH and hydrogen peroxide radicals (Table 7).

Table 7.
Correlation and regression analysis of phytochemical content and radical scavenging activity of different Physalis accessions from the same environmental conditions.
4. Discussion
DNA barcoding is relatively fast in terms of species identification and discrimination [41]. DNA barcodes such as rbcL, matK, psbA-tnrH, and ITS2 are very efficient in identifying unknown plant species, with ITS2 being among the best DNA barcodes for species identification and discrimination [13,41]. Based on BLASTn and the phylogenetic analysis of ITS2 and rbcL gene sequences, the Physalis accessions used in the current study were all confirmed to belong to the genus Physalis.
The results of this study showed that the used barcode regions have different abilities of species discrimination and identification. ITS2 was proposed as a core barcode for seed plants by the Consortium for the Barcode of Life (CBOL) Plant Working Group [42]. It was clear that the Physalis accessions used in the current study were identified as Physalis purpurea based on ITS2 barcode. The phylogenetic tree was able to discriminate the Physalis accessions and had reliable clades with a posterior probability of 80%. Species discrimination based on the rbcL gene was not possible as there was no formation of clades on the Bayesian inference phylogenetic tree. The high conservation of the rbcL barcode gene in Physalis accessions makes it a less ideal candidate for DNA barcoding when compared to other barcode genes such as ITS2 [43]. The ability of ITS2 to emerge as a better barcode than rbcL is clearly supported in other studies on Physalis and other plants [2,13,41].
The rbcL barcode gene of Physalis accessions under study did not display genetic distance, diversity, or polymorphism. This is an indication that there were no genetic differences between the Physalis accessions used. Therefore, the rbcL barcode is highly conserved in some species of Physalis, which has also been reported in other studies [44,45]. The ITS2 barcode gene showed genetic variation among the Physalis accessions and a high nucleotide diversity of 0.27629 was observed. This has also been reported in the DNA barcoding and identification of Solanaceae plants [43]. The genetic distance observed among the Physalis accessions in this study based on ITS2 concurs with the results of previous studies where the ITS2 barcode region was reported to have a high genetic variation due to its high rate of mutation [46]. A high intraspecies variation was observed based on the ITS2 sequences, which supports the successful identification of Physalis accessions based on the ITS2 barcode region. The Tajima D value of Physalis accessions based on the ITS2 gene was 0.779171, an indication that the gene had a low level of low-frequency mutations and a balancing selection within the population [47]. Three barcode gaps were identified for the ITS2 gene sequences among Physalis accessions based on the ABGD method. The presence of barcode gaps is crucial for species delimitation and forms the basis for plant species identification and discrimination [48].
Commercial interest in plants of the genus Physalis has been rising worldwide due to its nutritional value, edible fruits, and the current and potential medicinal uses [2]. The Physalis accessions were investigated for mineral content and the analysis revealed that P. purpurea fruits are rich in potassium, sodium, calcium, and magnesium, which concurs with reports from other studies on the wild edible fruits of Physalis [49]. Based on these findings, this fruit can be used as an alternative for the daily intake of minerals, which are essential for human health. Potassium is important to the function of the cardiovascular system in humans [50]. Potassium/sodium balance is fundamental for the transmission of electrical impulses in the heart [51]. Magnesium is an important mineral in protein synthesis, oxidative phosphorylation, the regulation of body temperature and muscle contractions, and it is a cofactor for many enzymes [52]. Calcium is also another important element required for blood clotting, growth, bone formation, cell metabolism, and heart function [53,54]. The Physalis fruits also contain trace elements including zinc, iron, copper, nickel, lithium, and manganese. Low levels of iron and nickel in Physalis have also been reported in other studies [49,55]. Zinc is important in the catalytic activity of enzymes, cellular signaling, and facilitates the modification of the structures of DNA and RNA, proteins, and cellular membranes [56]. Iron is an essential vitamin that is required in small amounts for DNA synthesis, oxygen transportation, and the electron transport chain [57]. Copper is also an essential element that is required in small amounts for facilitating lung elasticity, neurovascularization, the metabolism of iron, adequate growth, energy metabolism, reactive oxygen species detoxification, and cardiovascular integrity [58,59]. The functions of nickel in animals are not well known [60]. However, in plants and bacteria it is required as a cofactor for the enzymes involved in growth and germination in plants [60]. Lithium is an essential element in the physiological regulation of mood [61]. Manganese is required as a cofactor for enzymes that perform cholesterol, carbohydrate, and protein metabolism [62]. The presence of minerals in plant accessions can be linked to their medicinal properties as well [63]. Our analyses show the potential of Physalis purpurea as an excellent mineral supplement in nutraceuticals. Though little attention is given to this wild fruit, our findings indicate a richness of nutrients and its potential application as a nutritional supplement.
The Physalis accessions were rich in phytochemicals such as phenols, tannins, and flavonoids, which have also been identified in the genus in other previous studies [64]. The levels of these different types of phytochemicals were different in the Physalis accessions, an indication of the existence of different cultivars within Physalis purpurea. Variation in phytochemical secondary metabolites in plants, such as phenolic acid and flavonoids, has been linked to environmental stress during growth and development [65]. Ecologically limiting factors like lighting, carbon dioxide, soil salinity, temperature, and soil fertility can affect the biochemical and physiological responses of plants and their secondary metabolite production [66]. Abiotic stressors lead to fluctuations in the chemical constituents of plants, selectively altering the content of secondary metabolites such as phytochemicals [65]. Soil salinity, an abiotic stressor for plants, has been shown to cause the accumulation of secondary metabolites such as flavonoids in plants as a response to nutritional imbalance, decreased photosynthesis, and the uptake of nutrients [67]. Flavonoid accumulation when plants are under stress due to the increased salinity of soils provides a curative effect for affected plants [68]. The tannin and flavonoid contents were the highest among the phytochemicals identified in the Physalis accessions studied. Previous studies observed the highest phytochemical content to be phenols followed by tannins and the lowest being flavonoids [69,70]. The concentration of phenols might have varied in this study as compared to others due to the geographical variations, environmental/abiotic stressors, the method of extraction of phenols, the sugars present, and the carotenoid and ascorbic acid contents [65,71,72]. The presence of phenols has been associated with antioxidant properties and, therefore, the ability to scavenge for free reactive oxygen species is facilitated by phytochemicals [69,73]. According to the radical scavenging activity assays conducted, the fruit extracts of Physalis accessions scavenged free radicals such as DPPH and hydrogen peroxide due to the presence of phytochemicals. Phenolic and flavonoid contents in plants are important for their antioxidant properties, which allow them to scavenge reactive free radicals by donating hydrogen atoms to the free radicals [74].
Correlation studies of the polyphenol content and the ability of polyphenols to promote radical scavenging have shown a linear relationship between phenolic and flavonoid contents in relation to radical scavenging capacity [75]. In this study, the correlation analysis showed a positive correlation between the phenolic content and the DPPH radical scavenging activity. However, a negative correlation was observed for the tannin and flavonoid contents (r = −0.1150) and the DPPH radical scavenging activity. This is an indication that the phenolic content of P. purpurea contributed more towards the DPPH radical scavenging activity than tannins and flavonoids. This concurs with similar studies on the role of phenolic content from the genus Physalis in DPPH radical scavenging [76]. Phenol and flavonoid contents showed a moderate correlation to the hydrogen peroxide scavenging capability. This concurs with other studies showing that phenol and flavonoid contents contribute towards hydrogen peroxide scavenging activity [72]. The presence of tannins in the Physalis accessions did not show any DPPH radical and hydrogen peroxide scavenging activities. However, tannic acid in other plant studies has been shown to have radical scavenging activity against DPPH radicals and hydrogen peroxide [76,77].
5. Conclusions
The Physalis accessions used in the current study were identified as Physalis purpurea based on the ITS2 barcode region. The high genetic variation among the Physalis accessions based on the ITS2 sequences allowed for the clear identification of Physalis species as Physalis purpurea. There was no genetic variation among the rbcL sequences of Physalis, an indication that the gene is relatively conserved. The study confirmed that the fruits of Physalis purpurea contained a high content of minerals, including calcium, sodium, magnesium, and potassium. The fruits were also rich in phenolic acids, tannins, and flavonoids, and exhibited antioxidant properties. The phenolic compounds and flavonoids were the major contributors to the radical scavenging activity of the Physalis purpurea fruits. Therefore, the underutilized Physalis purpurea can be used as an excellent source of antioxidants for the management of oxidative stress-induced human diseases.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijpb14040073/s1. Table S1: BLASTn analysis results for Physalis accessions based on ITS2 and rbcL barcode regions; Figure S1: Multiple sequence alignment for Physalis accessions ITS2 and rbcL gene sequences and their reference sequences based on BLASTn analysis; Figure S2: Multiple sequence alignment for Physalis accessions based on ITS2 gene only; Figure S3: Multiple sequence alignment of Physalis accessions based on rbcL gene only; Figure S4: Heat map representation of the mineral content of Physalis accessions; Figure S5: Heat map for the polyphenol content distribution of Physalis accessions; Figure S6: Heat map for the radical scavenging activity of Physalis accessions.
Author Contributions
Conceptualization, K.P., K.M., E.K.M., J.M.W. and E.N.N.; methodology, K.P. and E.N.N.; software, K.P.; validation, K.M., E.K.M., J.M.W. and E.N.N.; formal analysis, K.P.; investigation, K.P.; resources, K.M. and E.N.N.; data curation, K.P.; writing—original draft preparation, K.P.; writing—review and editing, K.M., E.K.M., J.M.W. and E.N.N.; supervision, K.M., E.K.M., J.M.W. and E.N.N.; funding acquisition, K.M. and E.N.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The nucleotide sequences of ITS2 and rbcL regions for Physalis accessions used in this study were deposited in Genebank through online submission portal and were assigned the following accession numbers: OQ372021.1–OQ372029.1 for ITS2 and OQ507152.1 to OQ507161.1 for rbcL.
Acknowledgments
We would like to appreciate the Department of Biochemistry and Center for Biotechnology and Bioinformatics (CEBIB), University of Nairobi where the research was carried out. The results reported in this study are part of the PhD work for the first author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Afroz, M.; Akter, S.; Ahmed, A.; Rouf, R.; Shilpi, J.A.; Tiralongo, E.; Sarker, S.D.; Göransson, U.; Uddin, S.J. Ethnobotany and antimicrobial peptides from plants of the solanaceae family: An update and future prospects. Front. Pharmacol. 2020, 11, 565. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jiang, M.; Shi, Y.; Jiao, K.; Shen, C.; Lu, J.; Ying, Q.; Wang, H. Application of the ribosomal DNA ITS2 region of Physalis (Solanaceae): DNA barcoding and phylogenetic study. Front. Plant Sci. 2016, 7, 1047. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Hu, X.; Yang, J.; Yang, W. Identification of single-copy orthologous genes between Physalis and Solanum lycopersicum and analysis of genetic diversity in Physalis using molecular markers. PLoS ONE 2012, 7, e50164. [Google Scholar] [CrossRef] [PubMed]
- Chinese Academy of Sciences. Flora of China; Science Press: Beijing, China, 1978; Volume 67, p. 50. [Google Scholar]
- Sang-Ngern, M.; Youn, U.J.; Park, E.J.; Kondratyuk, T.P.; Simmons, C.J.; Wall, M.M.; Ruf, M.; Lorch, S.E.; Leong, E.; Pezzuto, J.M.; et al. Withanolides derived from Physalis peruviana (Poha) with potential anti-inflammatory activity. Bioorganic Med. Chem. Lett. 2016, 26, 2755–2759. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, M.F.; Mörsel, J.T. Oil goldenberry (Physalis peruviana L.). J. Agric. Food Chem. 2003, 51, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Deng, G.F.; Xu, X.R.; Wu, S.; Li, S.; Li, H.B. Chemical components and bioactivities of Cape gooseberry (Physalis peruviana). Int. J. Food Nutr. Saf. 2013, 3, 15–24. [Google Scholar]
- Menzel, M.Y. The cytotaxonomy and genetics of Physalis. Proc. Am. Philos. Soc. 1951, 95, 132–183. Available online: http://www.jstor.org/stable/3143331 (accessed on 12 December 2022).
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA barcoding as a molecular tool to track down mislabeling and food piracy. Diversity 2015, 8, 2. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Kang, Y.; Deng, Z.; Zang, R.; Long, W. DNA barcoding analysis and phylogenetic relationships of tree species in tropical cloud forests. Sci. Rep. 2017, 7, 12564. [Google Scholar] [CrossRef]
- Feng, S.; Jiao, K.; Zhu, Y.; Wang, H.; Jiang, M.; Wang, H. Molecular identification of species of Physalis (Solanaceae) using a candidate DNA barcode: The chloroplast psbA–trnH intergenic region. Genome 2018, 61, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.L.; Feng, S.J.; Tian, J.Y.; Wei, A.Z.; Yang, T.X. Internal transcribed spacer 2 (ITS 2) barcodes: A useful tool for identifying Chinese Zanthoxylum. Appl. Plant Sci. 2018, 6, e01157. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, M.F.; Moersel, J.T. Impact of enzymatic treatment on chemical composition, physicochemical properties and radical scavenging activity of goldenberry (Physalis peruviana L.) juice. J. Sci. Food Agric. 2007, 87, 452–460. [Google Scholar] [CrossRef]
- Puente, L.A.; Pinto-Muñoz, C.A.; Castro, E.S.; Cortés, M. Physalis peruviana Linnaeus, the multiple properties of a highly functional fruit: A review. Food Res. Int. 2011, 44, 1733–1740. [Google Scholar] [CrossRef]
- Somani, S.J.; Modi, K.P.; Majumdar, A.S.; Sadarani, B.N. Phytochemicals and their potential usefulness in inflammatory bowel disease. Phytother. Res. 2015, 29, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Bayir, A.G.; Kiziltan, H.S.; Kocyigit, A. Plant family, carvacrol, and putative protection in gastric cancer. In Dietary Interventions in Gastrointestinal Diseases; Academic Press: Cambridge, MA, USA, 2019; pp. 3–18. [Google Scholar] [CrossRef]
- Hong, J.M.; Kwon, O.K.; Shin, I.S.; Song, H.H.; Shin, N.R.; Jeon, C.M.; Oh, S.R.; Han, S.B.; Ahn, K.S. Anti-inflammatory activities of Physalis alkekengi var. franchetii extract through the inhibition of MMP-9 and AP-1 activation. Immunobiology 2015, 220, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bazalar Pereda, M.S.; Nazareno, M.A.; Viturro, C.I. Nutritional and antioxidant properties of Physalis peruviana L. fruits from the Argentinean Northern Andean region. Plant Foods Hum. Nutr. 2019, 74, 68–75. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Troshin, P.V.; Procter, J.B.; Barton, G.J. Java bioinformatics analysis web services for multiple sequence alignment—JABAWS: MSA. Bioinformatics 2011, 14, 2001–2002. [Google Scholar] [CrossRef] [PubMed]
- Troshin, P.V.; Procter, J.B.; Sherstnev, A.; Barton, D.L.; Madeira, F.; Barton, G.J. JABAWS 2.2 distributed web services for Bioinformatics: Protein disorder, conservation and RNA secondary structure. Bioinformatics 2018, 34, 1939–1940. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Kartavtsev, Y.P. Divergence at Cyt-b and Co-1 mtDNA genes on different taxonomic levels and genetics of speciation in animals. Mitochondrial DNA 2011, 22, 55–65. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; ISBN 9780195135855. [Google Scholar]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G.J. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Hernández, O.M.; Fraga, J.M.; Jiménez, A.I.; Jimenez, F.; Arias, J.J. Characterization of honey from the Canary Islands: Determination of the mineral content by atomic absorption spectrophotometry. Food Chem. 2005, 93, 449–458. [Google Scholar] [CrossRef]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocalteu reagent. Meth. Enzymol. 1999, 299, 152–178. [Google Scholar] [CrossRef]
- Chang, C.C.; Yang, M.H.; Wen, H.M.; Chern, J.C. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J. Food Drug Anal. 2002, 10, 3. [Google Scholar] [CrossRef][Green Version]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C.L. Use of a free radical method to evaluate antioxidant activity. LWT—Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Ruch, R.J.; Cheng, S.J.; Klaunig, J.E. Prevention of cytotoxicity and inhibition of intercellular communication by antioxidant catechins isolated from Chinese green tea. Carcinogenesis 1989, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- IBM SPSS. SPSS for Windows (Version 20); IBM: Chicago, IL, USA, 2010. [Google Scholar]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- China Plant BOL Group 1; Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef]
- Ralte, L.; Singh, Y.T. Use of rbcL and ITS2 for DNA barcoding and identification of Solanaceae plants in hilly state of Mizoram, India. Res. Crops 2021, 22, 616–623. [Google Scholar] [CrossRef]
- Xu, J.; Zang, F.; Wu, Q.; Wang, Y.; Wang, B.; Huang, P.; Zang, D.; Ma, Y.; Zheng, Y. Analysis of the genetic diversity and molecular phylogeography of the endangered wild rose (Rosa rugosa) in China based on chloroplast genes. Glob. Ecol. Conserv. 2021, 28, e01653. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, D.H.; Choi, B.H. Phylogeography and genetic diversity of East Asian Neolitsea sericea (Lauraceae) based on variations in chloroplast DNA sequences. J. Plant Res. 2013, 126, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.H.; Chai, L.C.; Chin, C.F. Efficacy of DNA barcode internal transcribed spacer 2 (ITS 2) in phylogenetic study of Alpinia species from Peninsular Malaysia. Physiol. Mol. Biol. Plants 2020, 26, 1889–1896. [Google Scholar] [CrossRef] [PubMed]
- Korneliussen, T.S.; Moltke, I.; Albrechtsen, A.; Nielse, R. Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinform. 2013, 14, 289. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.A.; Cruickshank, R.H. The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 2013, 13, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Musinguzi, E.; Kikafunda, J.K.; Kiremire, B.T. Promoting indigenous wild edible fruits to complement roots and tuber crops in alleviating vitamin A deficiency in Uganda. In Proceedings of the 13th ISTRC Symposium, Arusha, Tanzania, 10–14 November 2007; pp. 763–769. [Google Scholar]
- Sica, D.A.; Struthers, A.D.; Cushman, W.C.; Wood, M.; Banas, J.S., Jr.; Epstein, M. Importance of potassium in cardiovascular disease. J. Clin. Hypertens. 2002, 4, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Kowey, P.R. The role of potassium. In Women’s Health and Menopause: New Strategies—Improved Quality of Life; Springer: Berlin/Heidelberg, Germany, 2002; pp. 151–157. [Google Scholar]
- Jahnen-Dechent, W.; Ketteler, M. Magnesium basics. Clin. Kidney J. 2012, 5, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.C.; Taylor, C.L.; Yaktine, A.L.; Valle, H.B. Overview of calcium—Dietary reference intakes for calcium and vitamin D—NCBI bookshelf. Pediatrics 2012, 130, 10–542. [Google Scholar] [CrossRef]
- Berchtold, M.W.; Brinkmeier, H.; Muntener, M. Calcium ion in skeletal muscle: Its crucial role for muscle function, plasticity, and disease. Physiol. Rev. 2000, 80, 1215–1265. [Google Scholar] [CrossRef]
- Erkaya, T.; Dağdemir, E.; Şengül, M. Influence of Cape gooseberry (Physalis peruviana L.) addition on the chemical and sensory characteristics and mineral concentrations of ice cream. Food Res. Int. 2012, 45, 331–335. [Google Scholar] [CrossRef]
- Brown, K.H.; Wuehler, S.E.; Peerson, J.M. The importance of zinc in human nutrition and estimation of the global prevalence of zinc deficiency. Food Nutr. Bull. 2001, 22, 113–125. [Google Scholar] [CrossRef]
- Abbaspour, N.; Hurrell, R.; Kelishadi, R. Review on iron and its importance for human health. J. Res. Med. Sci. 2014, 19, 164. [Google Scholar]
- National Research Council. Copper in Drinking Water; National Research Council: Ottawa, ON, Canada, 2000; ISBN 0-309-06939-4. [Google Scholar]
- Ruiz, L.M.; Libedinsky, A.; Elorza, A.A. Role of copper on mitochondrial function and metabolism. Front. Mol. Biosci. 2021, 8, 711227. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G.; Carocci, A.; Lauria, G.; Sinicropi, M.S.; Catalano, A. Nickel: Human health and environmental toxicology. Int. J. Environ. Res. Public Health 2020, 17, 679. [Google Scholar] [CrossRef] [PubMed]
- Demling, J.H.; Eglau, M.C.; Autenrieth, T. On the physiological function oflithium from a psychiatric view point. Med. Hypotheses 2001, 57, 506–509. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc; Institute of Medicine/Food and Nutrition Board, National Academy Press: Washington, DC, USA, 2001. [Google Scholar] [CrossRef]
- Okwu, D.E. Phytochemicals, vitamins and mineral contents of two Nigerian medicinal plants. Int. J. Mol. Med. Adv. Sci. 2005, 1, 375–381. [Google Scholar]
- Sathyadevi, M.; Subramanian, S. Extraction, isolation and characterization of bioactive flavonoids from the fruits of Physalis peruviana Linn extract. Asian. J. Pharm. Clin. Res. 2015, 8, 152–157. [Google Scholar]
- Pant, P.; Pandey, S.; Dall’Acqua, S. The influence of environmental conditions on secondary metabolites in medicinal plants: A literature review. Chem. Biodivers. 2021, 18, e2100345. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. Effect of salinity stress on growth and physiology of medicinal plants. In Medicinal Plants and Environmental Challenges; Springer: Berlin/Heidelberg, Germany, 2017; pp. 177–188. [Google Scholar] [CrossRef]
- Gengmao, Z.; Yu, H.; Xing, S.; Shihui, L.; Quanmei, S.; Changhai, W. Salinity stress increases secondary metabolites and enzyme activity in safflower. Ind. Crops Prod. 2015, 64, 175–181. [Google Scholar] [CrossRef]
- El-Beltagi, H.S.; Mohamed, H.I.; Safwat, G.; Gamal, M.; Megahed, B.M. Chemical composition and biological activity of Physalis peruviana L. Gesunde Pflanzen 2019, 71, 113–122. [Google Scholar] [CrossRef]
- Kasali, F.M.; Tusiimire, J.; Kadima, J.N.; Tolo, C.U.; Weisheit, A.; Agaba, A.G. Ethnotherapeutic uses and phytochemical composition of Physalis peruviana L.: An overview. Sci. World J. 2021, 2021, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Oszmiański, J.; Lachowicz, S.; Gorzelany, J.; Matłok, N. The effect of different maturity stages on phytochemical composition and antioxidant capacity of cranberry cultivars. Eur. Food Res. Technol. 2018, 244, 705–719. [Google Scholar] [CrossRef]
- Aryal, S.; Baniya, M.K.; Danekhu, K.; Kunwar, P.; Gurung, R.; Koirala, N. Total phenolic content, flavonoid content and antioxidant potential of wild vegetables from Western Nepal. Plants 2019, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Ghosh, G.; Das, D.; Nayak, S. Phytochemical investigation and in vitro antioxidant activity of an indigenous medicinal plant Alpinia nigra BL Burtt. Asian Pac. J. Trop. Biomed. 2013, 3, 871–876. [Google Scholar] [CrossRef]
- Amarowicz, R.; Pegg, R.B.; Rahimi-Moghaddam, P.; Barl, B.; Weil, J.A. Free-radical scavenging capacity and antioxidant activity of selected plant species from the Canadian prairies. Food Chem. 2004, 84, 551–562. [Google Scholar] [CrossRef]
- Shrestha, P.M.; Dhillion, S.S. Diversity and traditional knowledge concerning wild food species in a locally managed forest in Nepal. Agrofor. Syst. 2006, 66, 55–63. [Google Scholar] [CrossRef]
- Karpagasundari, C.; Kulothungan, S. Free radical scavenging activity of Physalis minima Linn. leaf extract (PMLE). J. Med. Plants Stud. 2014, 2, 59–64. [Google Scholar]
- Gülçin, İ.; Huyut, Z.; Elmastaş, M.; Aboul-Enein, H.Y. Radical scavenging and antioxidant activity of tannic acid. Arab. J. Chem. 2010, 3, 43–53. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).