Comparative Transcriptome Analysis Reveals Genes Associated with Alkaloid Diversity in Javanese Long Pepper (Piper retrofractum) Fruits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. HPLC Analysis
2.3. RNA Extraction, Library Preparation, and Sequencing
2.4. De Novo Transcriptomic Assembly
2.5. Ortholog Identification
2.6. Differential Gene Expression Analysis
2.7. Gene Ontology Enrichment Analysis
2.8. Cluster Analysis
3. Results
3.1. Fruit Ripening Affects the Alkaloid Diversity
3.2. De Novo Transcriptome Construction from P. retrofractum Fruits and Leaves
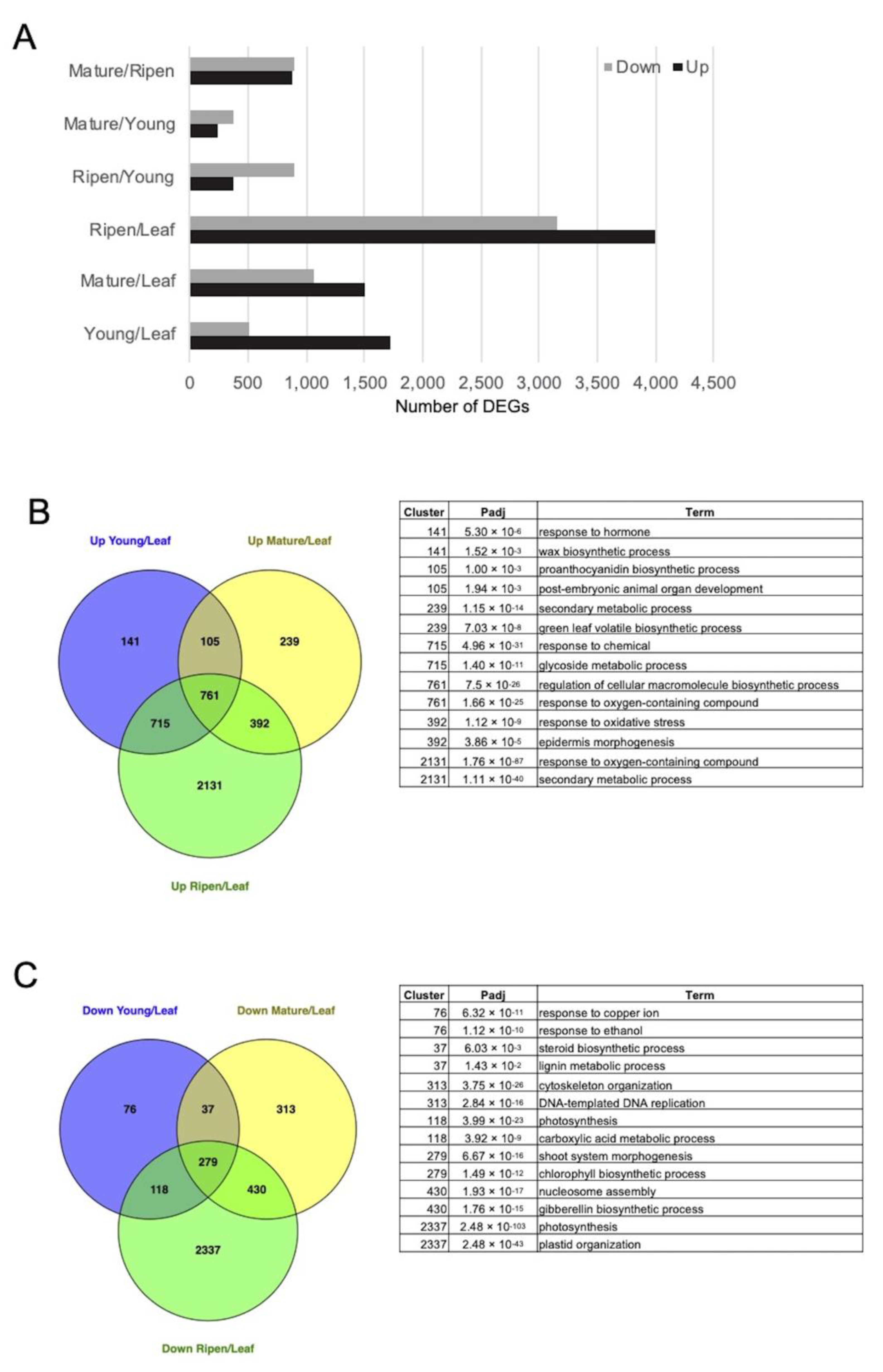
3.3. Transcriptomic Reprogramming during P. retrofractum Fruit Development
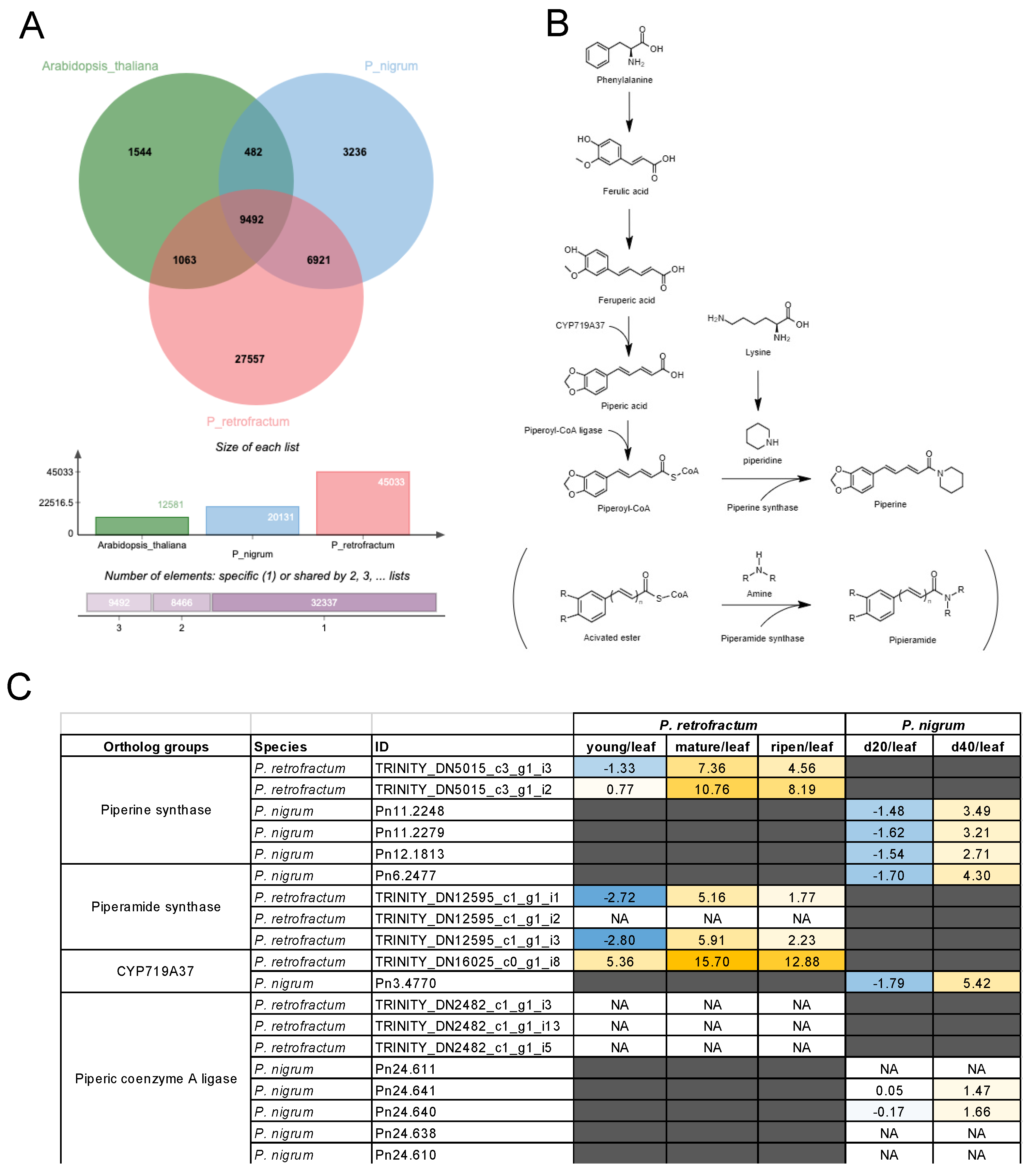
3.4. Cross-Species Transcriptomic Analyses Identified Piperine Biosynthesis Genes in P. retrofractum
3.5. A Group of Non-Piperine Alkaloid Biosynthesis DEGs was Explicitly Upregulated in the Ripened Fruits
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salehi, B.; Zakaria, Z.A.; Gyawali, R.; Ibrahim, S.A.; Rajkovic, J.; Shinwari, Z.K.; Khan, T.; Sharifi-Rad, J.; Ozleyen, A.; Turkdonmez, E.; et al. Piper Species: A Comprehensive Review on Their Phytochemistry, Biological Activities and Applications. Molecules 2019, 24, 1364. [Google Scholar] [CrossRef] [PubMed]
- Jadid, N.; Hidayati, D.; Hartanti, S.R.; Arraniry, B.A.; Rachman, R.Y.; Wikanta, W. Antioxidant activities of different solvent extracts of Piper retrofractum Vahl. using DPPH assay. In Proceedings of the Biodiversity and Biotechnology for Human Welfare, Surabaya, Indonesia, 15 October 2016; p. 020019. [Google Scholar]
- Muharini, R.; Liu, Z.; Lin, W.; Proksch, P. New amides from the fruits of Piper retrofractum. Tetrahedron Lett. 2015, 56, 2521–2525. [Google Scholar] [CrossRef]
- Luyen, B.T.T.; Tai, B.H.; Thao, N.P.; Yang, S.Y.; Cuong, N.M.; Kwon, Y.I.; Jang, H.D.; Kim, Y.H. A new phenylpropanoid and an alkylglycoside from Piper retrofractum leaves with their antioxidant and α-glucosidase inhibitory activity. Bioorganic Med. Chem. Lett. 2014, 24, 4120–4124. [Google Scholar] [CrossRef]
- Kim, K.J.; Lee, M.-S.; Jo, K.; Hwang, J.-K. Piperidine alkaloids from Piper retrofractum Vahl. protect against high-fat diet-induced obesity by regulating lipid metabolism and activating AMP-activated protein kinase. Biochem. Biophys. Res. Commun. 2011, 411, 219–225. [Google Scholar] [CrossRef]
- Tewtrakul, S.; Hase, K.; Kadota, S.; Namba, T.; Komatsu, K.; Tanaka, K. Fruit Oil Composition of Piper chaba Hunt., P. longum L. and P. nigrum L. J. Essent. Oil Res. 2000, 12, 603–608. [Google Scholar] [CrossRef]
- FAO. New Standards to Curb the Global Spread of Plant Pests and Diseases. Available online: https://www.fao.org/news/story/en/item/1187738/icode/ (accessed on 10 August 2023).
- Ratwatthananon, A.; Yooboon, T.; Bullangpoti, V.; Pluempanupat, W. Insecticidal activity of Piper retrofractum fruit extracts and isolated compounds against Spodoptera litura. Agric. Nat. Resour. 2020, 54, 447–452. [Google Scholar] [CrossRef]
- Park, B.-S.; Lee, S.-E.; Choi, W.-S.; Jeong, C.-Y.; Song, C.; Cho, K.-Y. Insecticidal and acaricidal activity of pipernonaline and piperoctadecalidine derived from dried fruits of Piper longum L. Crop Prot. 2002, 21, 249–251. [Google Scholar] [CrossRef]
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Wang, Y.; Hao, R.; Guo, R.; Nong, H.; Qin, Y.; Dong, N. Integrative Analysis of Metabolome and Transcriptome Reveals Molecular Insight into Metabolomic Variations during Hawthorn Fruit Development. Metabolites 2023, 13, 423. [Google Scholar] [CrossRef]
- Ngo, T.H.; Park, J.; Jo, Y.D.; Jin, C.H.; Jung, C.-H.; Nam, B.; Han, A.-R.; Nam, J.-W. Content of Two Major Steroidal Glycoalkaloids in Tomato (Solanum lycopersicum cv. Micro-Tom) Mutant Lines at Different Ripening Stages. Plants 2022, 11, 2895. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhao, X.; Fu, D.; Zhao, Y. Integrated Analysis of Widely Targeted Metabolomics and Transcriptomics Reveals the Effects of Transcription Factor NOR-like1 on Alkaloids, Phenolic Acids, and Flavonoids in Tomato at Different Ripening Stages. Metabolites 2022, 12, 1296. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Salvador, A.; Lana-Costa, J.; Omena-Garcia, R.P.; Batista-Silva, W.; Scossa, F.; Rosado-Souza, L.; Perez-Diaz, J.L.; Menezes-Silva, P.E.; DaMatta, F.M.; Sulpice, R.; et al. Metabolic shifts during fruit development in pungent and non-pungent peppers. Food Chem. 2022, 375, 131850. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Hirose, N.; Ohno, S.; Arakaki, M.; Wada, K. Flavor characteristics and antioxidant capacities of hihatsumodoki (Piper retrofractum Vahl) fresh fruit at three edible maturity stages. J. Food Sci. Technol. 2018, 55, 1295–1305. [Google Scholar] [CrossRef]
- Sirikhachornkit, A.; Suttangkakul, A.; Vuttipongchaikij, S.; Juntawong, P. De novo transcriptome analysis and gene expression profiling of an oleaginous microalga Scenedesmus acutus TISTR8540 during nitrogen deprivation-induced lipid accumulation. Sci. Rep. 2018, 8, 3668. [Google Scholar] [CrossRef]
- Butsayawarapat, P.; Juntawong, P.; Khamsuk, O.; Somta, P. Comparative Transcriptome Analysis of Waterlogging-Sensitive and Tolerant Zombi Pea (Vigna vexillata) Reveals Energy Conservation and Root Plasticity Controlling Waterlogging Tolerance. Plants 2019, 8, 264. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Putri, G.H.; Anders, S.; Pyl, P.T.; Pimanda, J.E.; Zanini, F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 2022, 38, 2943–2945. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Wimalanathan, K.; Lawrence-Dill, C.J. Gene Ontology Meta Annotator for Plants (GOMAP). Plant Methods 2021, 17, 54. [Google Scholar] [CrossRef]
- Horan, K.; Jang, C.; Bailey-Serres, J.; Mittler, R.; Shelton, C.; Harper, J.F.; Zhu, J.-K.; Cushman, J.C.; Gollery, M.; Girke, T. Annotating Genes of Known and Unknown Function by Large-Scale Coexpression Analysis. Plant Physiol. 2008, 147, 41–57. [Google Scholar] [CrossRef]
- Sreeratree, J.; Butsayawarapat, P.; Chaisan, T.; Somta, P.; Juntawong, P. RNA-Seq Reveals Waterlogging-Triggered Root Plasticity in Mungbean Associated with Ethylene and Jasmonic Acid Signal Integrators for Root Regeneration. Plants 2022, 11, 930. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Schnabel, A.; Athmer, B.; Manke, K.; Schumacher, F.; Cotinguiba, F.; Vogt, T. Identification and characterization of piperine synthase from black pepper, Piper nigrum L. Commun. Biol. 2021, 4, 445. [Google Scholar] [CrossRef]
- Hu, L.; Xu, Z.; Wang, M.; Fan, R.; Yuan, D.; Wu, B.; Wu, H.; Qin, X.; Yan, L.; Tan, L.; et al. The chromosome-scale reference genome of black pepper provides insight into piperine biosynthesis. Nat. Commun. 2019, 10, 4702. [Google Scholar] [CrossRef]
- Schnabel, A.; Cotinguiba, F.; Athmer, B.; Yang, C.; Westermann, B.; Schaks, A.; Porzel, A.; Brandt, W.; Schumacher, F.; Vogt, T. A piperic acid CoA ligase produces a putative precursor of piperine, the pungent principle from black pepper fruits. Plant J. 2020, 102, 569–581. [Google Scholar] [CrossRef]
- Schnabel, A.; Cotinguiba, F.; Athmer, B.; Vogt, T. Piper nigrum CYP719A37 Catalyzes the Decisive Methylenedioxy Bridge Formation in Piperine Biosynthesis. Plants 2021, 10, 128. [Google Scholar] [CrossRef] [PubMed]
- Gurung, K.; Manivannan, S. Morphological characterization and secondary metabolites profile of black pepper (Piper nigrum L.) genotypes from Sikkim. J. Spices Aromat. Crops 2020, 29, 98–104. [Google Scholar] [CrossRef]
- Thomas, Q.A.; Ard, R.; Liu, J.; Li, B.; Wang, J.; Pelechano, V.; Marquardt, S. Transcript isoform sequencing reveals widespread promoter-proximal transcriptional termination in Arabidopsis. Nat. Commun. 2020, 11, 2589. [Google Scholar] [CrossRef] [PubMed]
- Osorio, S.; Scossa, F.; Fernie, A. Molecular regulation of fruit ripening. Front. Plant Sci. 2013, 4, 198. [Google Scholar] [CrossRef]
- Moghe, G.; Kruse, L.H.; Petersen, M.; Scossa, F.; Fernie, A.R.; Gaquerel, E.; D’Auria, J.C. BAHD Company: The Ever-Expanding Roles of the BAHD Acyltransferase Gene Family in Plants. Annu. Rev. Plant Biol. 2023, 74, 165–194. [Google Scholar] [CrossRef]
- Chakraborty, P.; Biswas, A.; Dey, S.; Bhattacharjee, T.; Chakrabarty, S. Cytochrome P450 Gene Families: Role in Plant Secondary Metabolites Production and Plant Defense. J. Xenobiot. 2023, 13, 402–423. [Google Scholar] [CrossRef]
- Shojima, S.; Nishizawa, N.-K.; Fushiya, S.; Nozoe, S.; Irifune, T.; Mori, S. Biosynthesis of phytosiderophores. In vitro biosynthesis of 2′-deoxymugineic acid from l-methionine and nicotianamine. Plant Physiol. 1990, 93, 1497–1503. [Google Scholar] [CrossRef]
- Holzer, H.; Schneider, S. Reinigung und charakterisierung einer TPN-abhängigen pyridoxol-dehydrogenase aus bierhefe. Biochim. Biophys. Acta 1961, 48, 71–76. [Google Scholar] [CrossRef]
- Morishige, T.; Tamakoshi, M.; Takemura, T.; Sato, F. Molecular characterization of O-methyltransferases involved in isoquinoline alkaloid biosynthesis in Coptis japonica. Proc. Jpn. Acad. Ser. B 2010, 86, 757–768. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meechuen, M.; Pimsawang, L.; Chaisan, T.; Samipak, S.; Pluempanupat, W.; Juntawong, P. Comparative Transcriptome Analysis Reveals Genes Associated with Alkaloid Diversity in Javanese Long Pepper (Piper retrofractum) Fruits. Int. J. Plant Biol. 2023, 14, 896-909. https://doi.org/10.3390/ijpb14040066
Meechuen M, Pimsawang L, Chaisan T, Samipak S, Pluempanupat W, Juntawong P. Comparative Transcriptome Analysis Reveals Genes Associated with Alkaloid Diversity in Javanese Long Pepper (Piper retrofractum) Fruits. International Journal of Plant Biology. 2023; 14(4):896-909. https://doi.org/10.3390/ijpb14040066
Chicago/Turabian StyleMeechuen, Methat, Lalita Pimsawang, Tanapon Chaisan, Sompid Samipak, Wanchai Pluempanupat, and Piyada Juntawong. 2023. "Comparative Transcriptome Analysis Reveals Genes Associated with Alkaloid Diversity in Javanese Long Pepper (Piper retrofractum) Fruits" International Journal of Plant Biology 14, no. 4: 896-909. https://doi.org/10.3390/ijpb14040066
APA StyleMeechuen, M., Pimsawang, L., Chaisan, T., Samipak, S., Pluempanupat, W., & Juntawong, P. (2023). Comparative Transcriptome Analysis Reveals Genes Associated with Alkaloid Diversity in Javanese Long Pepper (Piper retrofractum) Fruits. International Journal of Plant Biology, 14(4), 896-909. https://doi.org/10.3390/ijpb14040066





