Abstract
Yam (Dioscorea spp.) is an important food security crop with economic, nutritional, and medicinal value. It is a source of carbohydrates for millions of people in tropical and sub-tropical regions of Africa, Asia, South America, the Caribbean, and the South Pacific Islands. Determining the appropriate parents for breeding programs is the most important decision that plant breeders must make to maximize genetic variability and produce excellent recombinant varieties. However, adequate genetic diversity and the population structure of yam accessions in Kenya are not available to guide accurate selection of parents for breeding. In the present study, 25 start-codon-targeted (SCoT) molecular markers were used to determine the genetic diversity and population structure among 20 yam accessions grown in Kenya. A total of 294 fragments were amplified, of which 95% were polymorphic with an average of 11.16 polymorphic fragments per primer. The polymorphic information content (PIC) value and primer resolving power (Rp) of 0.58 and 5.91, respectively, revealed high genetic diversity among the accessions. A dendrogram based on the unweighted pair group method of arithmetic means (UPGMA) grouped the 20 yam accessions into two clusters at 0.61 genetic similarity coefficients. Bayesian structure analysis revealed the existence of three subpopulations and some admixed accessions. Analysis of molecular variance (AMOVA) revealed a variance of 60% within the subpopulations and 40% among the subpopulations. The high degree of genetic diversity in the yam accessions successfully exhibited by SCoT molecular markers may serve as a valuable aid to widen the genetic base in yam breeding programs. The selection and hybridization of parental lines from the different clusters and sub-clusters identified could provide a foundation and could be exploited for yam breeding and variety development.
1. Introduction
Yam (Dioscorea spp.) is an important staple food crop distributed in the sub-tropical and tropical regions of Africa, Asia, South America, the Caribbean, as well as the South Pacific Islands [1]. Globally, yam is ranked fourth after cassava, potato, and sweet potato in terms of production [2]. Its annual production worldwide increased from 8.32 metric tonnes (MT) in 1961 to 60.00 MT in 2019 [2]. In sub-Saharan Africa, over 45 million tonnes of yams are harvested annually, with Nigeria being the world’s leading producer [3]. In Kenya, yam production increased by 47.6% from 1,356,900 tons produced in 2013 to 2,002,800 tons in 2019 [2]. Yams belong to the family Dioscoreaceae comprising more than 644 species, of which only 10 are commonly cultivated for food and have economic importance [4]. The edible starch tubers of the crop are rich in starch, vitamin C, essential minerals, and dietary fibers [5]. In addition to the cultivated species, some uncultivated species have been identified to have medicinal properties, hence serving an important role in the pharmaceutical industry [6].
The cultivation of yams in Kenya has remained traditional and is usually carried out by smallholder farmers. Many cultivars of different Dioscorea species namely the Dioscorea bulbifera, Dioscorea minutifolia, and Dioscorea dumetorum species are mainly grown in the eastern, central, western, and coastal regions of Kenya [7]. Yam has been identified as one of the climate-resilient crops for food and nutrition security, and therefore there is a need for the establishment of breeding programs to develop high-yielding varieties with resistance to pests and diseases as well as quality nutritional traits. However, the lack of adequate information on genetic diversity limits its genetic improvement. It is important to know the genetic diversity of germplasm as a prerequisite for the development of varieties with useful traits. Assessing genetic diversity and relationships in available germplasm is an important task for identifying varied types with useful traits for crop improvement and conservation programs. So far, several studies have been conducted to investigate the genetic diversity of yam germplasm worldwide [8,9,10,11,12,13,14,15]; however, in Kenya, there is limited information on genetic characterization using molecular markers. Thus, characterization of available yam germplasm in terms of their DNA diversity should be prioritized in order to generate information for future conservation strategies, genetic improvement, and variety development.
Assessment of genetic diversity and variety differentiation in yam has been carried out in sub-Saharan Africa through agro-morphological traits and molecular markers [8,9,10,11]. Despite the fact that agro-morphological traits are mostly used in the determination of genetic diversity, they do not demonstrate the true genetic relatedness of the accessions and are strongly influenced by environmental factors [13,14]. Molecular markers have been utilized as powerful tools for the estimating genetic diversity of many plant species with great success and accuracy because they are abundant and unaffected by environment parameters [16]. Various DNA-based markers such as restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), and simple sequent repeats (SSR) have been used in Africa for yam taxonomic classification, genetic diversity, association analysis, and QTL mapping studies [8,9,10,11,12,15]. Start-codon-targeted (SCoT) are based on polymorphisms in the short, highly conserved region of plant genes surrounding the ATG of the start codon [17]. Therefore, it can distinguish genetic variation in a specific gene that link to a specific trait. It is a simple, novel, cost-effective, highly polymorphic, and reproducible molecular marker for which there is no need for prior sequence information. In addition, detection is carried out with agarose-gel-based and therefore, it is simple and relatively cheap to use. The SCoT marker has been used in genetic diversity analysis, phylogenetic relationships, and DNA fingerprinting of economically important food crops and medicinal plants including mango [18], grape [19], orchid [20], durum wheat [21], rose [22], Diospyros [23], Elymus sibiricus [24], Vigna unguiculata [25], Taxus media [26], Dendrobium [27], Chrysanthemum morifolium [28], coconut [29], and Physalis species [30]. The short flanking regions of the ATG start codon is highly conserved across plant species and has led to the widespread use SCoT markers in quantitative trait locus (QTL) mapping, marker-assisted breeding, bulked segregate analysis, and genetic variation [17].
Using SCoT markers to characterize genetic diversity in yam germplasm has not been described in any molecular studies. Therefore, the objective of the current study was to determine the genetic diversity and population structure among yam germplasm cultivated in Kenya. The information generated from this study will be a critical preliminary step towards developing strategies for the selection and breeding of superior varieties of yam with important agronomic traits such as high yield, pest and disease resistance, and nutritional traits.
2. Materials and Methods
2.1. Plant Materials
A total of 20 samples of morphologically distinct yam accessions were collected from major growing regions in Kenya as well as the Genetic Resources Research Institute (GeRRI), Kenya Agricultural Livestock and Research Organization (KALRO) (Table 1). Twelve yam tubers were obtained from central (Muranga, Kirinyaga, and Nyeri counties) and the upper eastern (Meru county) regions while eight young leaf samples were obtained from the GeRRI. Purposive sampling was used to select representative yam collection sites concerning the potential of production. The tubers were placed in a cool box while the leaves were collected in bags containing silica gel and transported to the laboratory at the University of Nairobi for molecular analysis.

Table 1.
Yam accessions collected from Genetic Resources Research Institute (GeRRI) and central and upper eastern regions of Kenya.
2.2. Genomic DNA Extraction
Genomic DNA was isolated from either 0.5 g of leaf samples using the cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle, 1990). The isolated DNA samples were dissolved in 30 μL of nuclease-free water followed by treatment with RNase through the addition of 0.5 mg/mL Ribonuclease A and incubated in a water bath at 37 °C for 30 min. The quality and quantity of the extracted DNA samples were estimated using a UV spectrophotometer and by resolving the DNA samples on 0.8% agarose gel in 1× Tris–Boric–EDTA (TBE) buffer. A final DNA concentration of 50 ng/μL was prepared and stored at −20 °C until use. DNA samples were extracted from three individual plants of the same accession for subsequent SCoT analysis.
2.3. Optimization of Primer Conditions and SCoT-PCR Amplification
The SCoT-PCR analysis was carried out according to protocols established by Collard and Mackill [31] with some minor modifications including optimization of the annealing temperature of the SCoT primers and the duration of PCR thermal cycling conditions. Thirty SCoT primers were tested for their ability to prime to DNA of yam accessions. The primers that either failed to amplify or produced faint bands were excluded from the study. Twenty-five SCoT primers (Supplementary Table S1) that produced consistent amplification and clear banding patterns were used for analysis of the genetic diversity of the twenty yam accessions. PCR reactions were performed in 25 μL volume using 12.5 μL one Taq Quick-Load 2× Master Mix with a standard PCR buffer (New England Biolabs, Hertfordshire, UK), 10 mΜ of the primer, 50 ng of template DNA, and the reaction mixtures were topped up to 25 μL with nuclease-free water. PCR amplifications were performed in a Veriti thermocycler (Bio-Rad, Singapore) with the following thermocycling conditions: initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 1 min, primer annealing at 50 °C for 1 min, and extension at 72 °C for 1.5 min. The amplification process was completed with a 5 min final extension at 72 °C and the PCR products were maintained at 4 °C. The PCR reaction for each SCoT primer was performed at least twice using DNA from two individual plants of the same accession. Where the PCR amplifications and banding patterns were not consistent, a third PCR amplification was carried out. Only clear and reproducible bands were used in the data analysis.
2.4. Visualization of Amplified PCR Products and Data Analysis
The PCR products were resolved by electrophoresis on a 1.5% ethidium-bromide-stained agarose gel in 1× TBE buffer. Electrophoresis was carried out at 70 V for 60 min and PCR products were visualized using a Gel-Doc TM XR+ Imaging System (Bio-Rad, Gmbh, FeldKirchen, Germany). The molecular weights of the PCR products were estimated using a Gene Ruler 1 kb Plus DNA marker (Fischer Thermo Scientific, Waltham, MA, USA).
All PCR-amplified SCoT fragments were detected on gels and scored as binary data for presence (1) or absence (0). Only reproducible and well-defined bands were scored. Polymorphic and monomorphic bands were determined for each SCoT primer. The genetic similarity, Shannon information index, resolving power for each primer, and genetic distances based on Nei’s coefficients between pairs were analyzed using Popgene software, version 3.5 (www.ualberta.ca/fyeh/popgene.pdf, accessed on 8 February 2023).
Polymorphism information content (PIC) per locus was computed using Power Marker (version 3.25). Principal coordinate analysis (PCoA) and analysis of molecular variance (AMOVA) were performed using GENALEX 6.5 software [32] (accessed on 12 September 2022). The distance matrices were generated based on Jaccard’s similarity coefficient. Similarity matrices were subjected to cluster analysis through the unweighted pair group method with arithmetic mean (UPGMA) and a dendrogram was constructed using FigTree software (Version 1.4.2; accessed on 12 October 2022).
The data from the 25 polymorphic SCoT markers were subjected to population structure analysis based on the admixture model clustering method in the software package STRUCTURE 2.3.4 [33]. This model was run by varying the number of assumed population (K) from 1 to 10 (K = group numbers formed according to the STRUCTURE software (Version 2.3.4; accessed on 16 October 2022). A burn-in period of 10,000 and Markov Chain Monte Carlo (MCMC) replications of 100,000 after each burn-in was used. The optimum population (K) which best estimated the structure of the 20 yam accessions was predicted using the Evanno’s method [34] through the online-based software STRUCTURE HARVESTER (Version A.2) [35] (accessed on 16 October 2022). The model was repeated for the K at maximum ΔK with a burn-in period of 100,000 and an MCMC of 100,000 after each burn-in with ten alterations.
3. Results
3.1. SCoT Marker Analysis
Out of the 30 SCoT markers tested for their ability to amplify yam DNA samples, 25 markers which showed polymorphic amplification fingerprint patterns were used to analyze the genetic diversity of the 20 yam accessions. The 25 SCoT markers generated a total of 294 amplified DNA fragments with an average of 12 bands per marker (Table 2). Out of the 294 amplified fragments, 279 (95%) were polymorphic. Out of the 25 SCoT markers, three markers namely SCoT3, SCoT16, and SCoT20 were the most polymorphic with 15, 16, and 20 polymorphic bands, respectively. The lowest numbers of polymorphic bands (3) were amplified using SCoT23 (Table 2). The polymorphic information content (PIC) value of all the SCoT markers ranged from 0.40 (SCoT11) to 0.77 (SCoT6) with an average of 0.58 (Table 2). Nei’s gene diversity varied from 0.25 (SCoT11) to 0.44 (SCoT6) with an average of 0.33, while Shannon information index ranged from 0.37 to 0.63 with an average value of 0.49. The primer resolving power (Rp) was estimated at an average of 5.91. The highest and lowest Rp were 8.70 (for SCoT20) and 3.40 (for ScoT27), respectively (Table 2). A representative amplification profile of the ScoT markers is represented in Supplementary Figure S1.

Table 2.
Total and number of polymorphic bands, gene diversity, resolving power, and Shannon information index per ScoT marker used for the analysis of the 20 yam accessions.
3.2. Genetic Diversity and Cluster Analysis/Grouping of Yam Accessions
The genetic diversity and relationships among the studied yam accessions were determined by Jaccard’s similarity coefficient. The similarity coefficient among the 20 yam accessions ranged from 0.21 to 0.87 with an average of 0.54. The highest similarity coefficient of 0.87 was recorded between accessions E6 and E8 while the lowest value of 0.21 was obtained between E6 and B as well as E4 and B (Supplementary Table S2).
Based on Jaccard’s (J) similarity coefficient and using the UPGMA method, the dendrogram (Figure 1) divided the 20 yam accessions into two major clusters (I and II) at a Jaccard’s similarity coefficient of 0.59. The two major clusters namely I and II composed of 2 and 18 yam accessions, respectively. Cluster I includes two accessions (B and H) from GeRRI–KALRO, while cluster II includes 18 accessions that were divided into sub-clusters, sub-cluster II(a) with 12 yam accessions and sub-cluster II(b) with 6 yam accessions. All the 12 yam accessions (E0, E1, E2, E3, E4, E5, E6, E7, E8, E9, E10, and E11) in sub-cluster II(b) were obtained from Meru, Kirinyaga, Murang’a, and Nyeri coun\ties (central and upper eastern regions in Kenya) while the 6 yam accessions (TDr2579, Makwaka, TDr2436, Amola, TDr00917, and Obiotungu) in sub-cluster II(a) were obtained from GeRRI–KALRO.
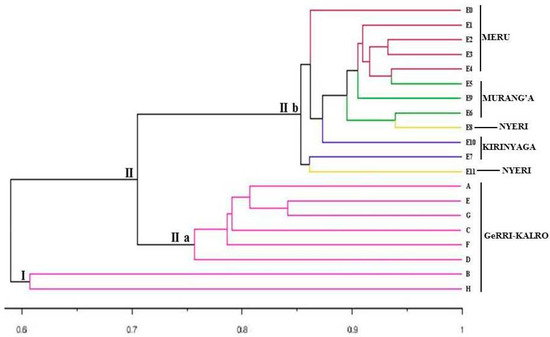
Figure 1.
Dendrogram based on UPGMA showing the relationships among the 20 accessions of yam using SCoT marker data based on Jaccard’s similarity index. Yam accessions represented by similar colors in the dendrogram were obtained from the same region. The letters A–H and E0 to E11 represents the codes of the yam accessions as indicated in Table 1.
Nei’s gene diversity index and Shannon diversity index were calculated and used to evaluate the genetic diversity of yam accessions in the current study. We compared the genetic diversity of yam accessions from four counties in Kenya and the National Genebank of Kenya (GeRRI–KALRO). The estimates of the genetic diversity in each population are summarized in Table 3. Nei’s gene diversity index of the five sampling sites of yam accession ranged from 1.168 to 1.443 and Shannon’s diversity index ranged from 0.116 to 0.386 (Table 3). Within the five accession sources, the GeRRI–KALRO exhibited the highest level of variability (NGDI: 1.443 and SDI: 0.386), whereas accessions from Kirinyaga (NGDI: 1.168 and SDI: 0.116) exhibited the lowest level of variability (Table 3).

Table 3.
Shannon diversity index (SDI) and Nei’s gene diversity index (NGDI) of 20 yam accessions collected from different geographical regions.
3.3. Population Structure Analysis
The maximum peak value of ΔK (83.267) was observed at K = 3 (Figure 2A). Using STRUCTURE software, the Bayesian-model-based clustering analysis grouped the 20 yam accessions into three distinct genetic groups at K = 3, designated here as P1, P2, and P3 (Figure 2B). However, at K = 3, some yam accessions occurred as an admixture group (Figure 2B).
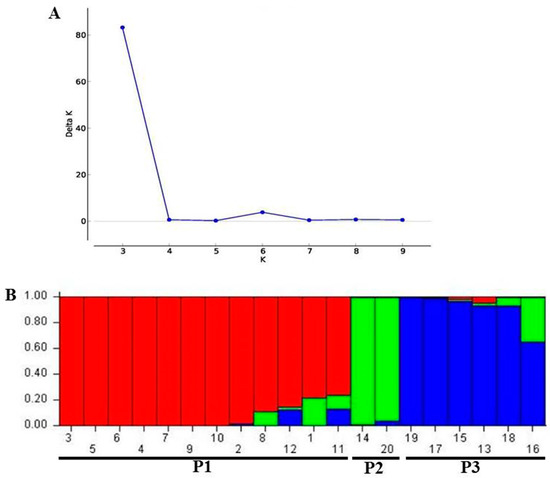
Figure 2.
(A) The estimated membership fraction using LnP(D)-derived delta K (ΔK) with cluster number (K) ranged from 1 to 10 for K = 3. (B) Population structure of 20 yam accessions inferred by STRUCTURE analysis based on SCoT marker data. Each color represents a single genetic group, namely P1, P2, and P3. Each solid bar represents a single accession, while each color represents a genetic group. The numbers 1–20 represents the yam accessions as indicated in Table 1.
3.4. Analysis of Molecular Variance (AMOVA)
Analysis of molecular variance (AMOVA) was used to evaluate the population differentiation among and within the yam accessions. The ScoT markers data revealed 40% of the genetic variation among population and 60% of the variation within the population (Table 4).

Table 4.
Analysis of molecular variance (AMOVA) based on ScoT markers for 20 yam accessions in five populations (Meru, Nyeri, Kirinyaga, Murang’a, and GERRI).
3.5. Principal Coordinates Analysis (PcoA)
The principal coordinate analysis was performed to determine the genetic relations among and within the populations and to analyze the consistency of differentiation among the populations by cluster analysis. The PcoA analysis exhibited a similar classification of the 20 yam accessions compared to the cluster analysis (Figure 3). The percentage of the total variation explained by the first three dimensions of the PcoA axis was 53.21% (first axis = 39.13%, second axis = 8.77%, and third axis = 5.31%). All the accessions from Meru, Murang’a, Kirinyaga, and Nyeri counties (central and upper eastern Kenya regions) were clustered together (Figure 3), similar to the cluster analysis.
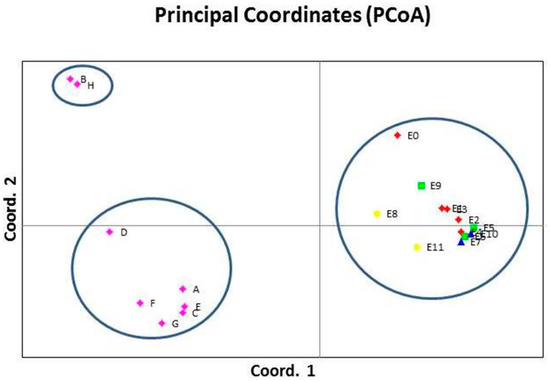
Figure 3.
Bi-plots derived from principal coordinate analysis of 20 yam accessions using ScoT data. The numbers plotted represents individual accessions as listed in Table 1. The colors represent the county/site of collection of the accessions: red = Meru, green = Murang’a, blue = Kirinyaga, yellow = Nyeri, and purple = GeRRI–KALRO.
4. Discussion
The genetic variation in yam (Dioscorea species) should be considered when developing conservation and utilization strategies as well as expediting breeding programs. Molecular markers are efficient and accurate tools to reveal and estimate genetic diversity and to determine the population structure of many plant species [12,13,14,15,36]. There are many gene-based markers that have been developed to aid in the investigation of genetic diversity and population structure analyses in crop plants including SCoT molecular markers. Several studies have indicated that SCoT markers have good capabilities in genetic research due to their ability to reveal polymorphisms in conserved regions and their high reliability as compared with other molecular systems [37,38]. This is the first report on evaluating the genetic diversity and population structure of various yam accessions and establishing phylogenetic relationships among them using SCoT markers. The 25 SCoT markers amplified 294 bands, out of which 279 (95%) were polymorphic and this high polymorphism indicates the informative nature of the SCoT markers used. The high level of polymorphism (95%) demonstrated a relatively high genetic diversity among genotypes of Dioscorea spp. The 95% polymorphisms obtained in this study are higher than the 79.57% polymorphism in indigenous mango (Mangifera indica L.) cultivars previously reported by Gajera et al. [39] and close to 97.26% polymorphism in Crepidium acuminatum reported by Thakur et al. [40]. Similarly, Que et al. [41] revealed 92.85% polymorphism using SCoT markers in determining the genetic diversity of sugarcane accessions within a local sugarcane germplasm collection from China. The SCoT markers used in the current study revealed 95% polymorphism which was comparable to the other markers; for example, Wu et al. [42] assessed the genetic diversity of 21 yam cultivars, including D. opposita, D. alata, D. persimilis, Prain and Purkill, and D. fordii Prain, and Purkill, from seven cultivated populations and found a high level of polymorphism (95.3%) using ISSR markers.
It has been reported that Nei’s gene diversity and Shannon’s information index are important in the study of genetic diversity in plant species [43,44]. In this study, the average Nei’s gene diversity (expected heterozygosity) and Shannon information index were 0.33 and 0.49, respectively, indicating moderate genetic diversity for the yam accessions. The results showed that the germplasm investigated in this study exhibited adequate genetic variation that can be exploited for yam improvement. Wu et al. [42] reported Nei’s gene diversity and a Shannon information index of 0.29 and 0.45, respectively, using ISSR markers in the genetic diversity analysis of 21 cultivated yam (Dioscorea spp.) cultivars in China. The level of heterozygosity could be mainly associated with the breeding system and self-incompatibility in yam [45], as well as the inclusion of genotypes from different localities. A limitation of this study is the use of only 20 yam accessions, which should be addressed by increasing the number of sampling sites in order to fully understand the genetic diversity of yam.
The PIC value represents the informativeness of a marker in detecting polymorphism and is usually used to reveal the differences among crop accessions based on genetic relationships. PIC values of less than 0.25 indicate a low level of polymorphism values between 0.25 and 0.50 indicate intermediate polymorphism, and values more than 0.50 indicate high polymorphism [46,47]. In this study, the PIC values ranged from 0.40 (SCoT11) to 0.77 (SCoT6) with an average of 0.58, which confirms that the SCoT markers used in this study were highly informative and thus possess high discriminatory power based on the description by Botstein et al. [46]. The results revealed high genetic polymorphism among the yam accessions cultivated in Kenya and showed that the accessions are genetically diverse. In addition, these results indicated that the SCoT markers used in this study are very informative and efficient and can be used for species authentication by developing species-specific sequence-characterized amplified region (SCAR) markers. Therefore, in the future, we will develop SCAR markers based on SCoT analysis which could be used in the identification of different Dioscorea species.
The genetic relationships between the tested yam accessions were estimated by calculating the similarity coefficients and the accessions were grouped based on the similarities using the UPGMA method. The similarity coefficient ranged from 0.21 to 0.87 with an average of 0.54, indicating moderate genetic diversity among the 20 yam accessions. The highest similarity coefficient of 0.87 was recorded between accessions E6 and E8 indicating that these accessions are highly genetically similar. The lowest similarity index of 0.21 was obtained between E6 and B as well as between E4 and B, suggesting that these accessions were more genetically diverse and are appropriate for use in the breeding of superior yam varieties. The dendrogram constructed using the UPGMA method separated the 20 yam accessions into two major clusters/groups at the similarity coefficient of 0.2. Cluster I contained accessions from GeRRI–KALRO, while cluster II contained accessions from Meru, Kirinyaga, Murang’a, and Nyeri Counties, as well as GeRRI–KALRO. The SCoT data analysis did not detect any regional groupings associated with the areas of collection of the yam accessions. This indicated that the accessions were genetically distinct from one another and could be used as complementary parents for crossing to obtain a heterotic response. These results will be useful for improving the management of yam as well as for selecting genetically distant parents even from different locations to maximize allele diversity and heterosis in breeding programs.
Based on clustering analysis of the population structure implemented using STRUCTURE software, the best interference of delta K (ΔK) was at K = 3. The Bayesian clustering assists in the identification of population structure and the allocation of individuals or parts of genetic information to several clusters [33]. STRUCTURE software distributes different accessions to different populations depending upon the allele frequencies obtained. In this study, the population structure analysis classified the 20 yam accessions into three populations and an admixture group. The principal coordinate (PCoA) analysis showed similar results to those obtained using STRUCTURE and cluster analysis.
The analysis of molecular variance (AMOVA) revealed high genetic variability among individuals within a population (60%) which indicates that all the samples had a diverse genetic backgrounds and thus a possibility of effective selection from a mini-core collection of the tested population. The presence of high genetic variability within the population signifies the divergence of yam accessions within a single population, indicating that each of the collection sites holds important genetic resources. Similarly, Loko et al. [48] found higher variation within populations than among populations using 41 microsatellite markers to study the genetic diversity and relationships between 64 local varieties of yam. Moreover, Ousmael et al. [10], found a higher proportion of variation within species (63.9%) than among species (36.1%) using ISSR markers to study the genetic diversity of Dioscorea spp. The high genetic variability within populations could be due to evolutionary dynamics/Jaccard similarity and/or admixture in the field of possibly facilitated by pollen flow [10]. Critical information about genetic variation within populations can be helpful in genetic resource conservation, utilization, and establishment in breeding programs [49]. Within-population selection enables identification of genotypes adapted to specific climatic conditions or management practices to achieve higher genetic gains during crop improvement.
The study revealed a considerable genetic diversity among yam germplasm in Kenya. The high degree of genetic diversity in the yam accessions successfully exhibited by SCoT molecular markers may serve as a valuable aid to widen the genetic base of yam breeding programs. The results of genetic diversity and genetic distance are useful for parental selection to improve the breeding value of yam accessions so that crosses between accessions that are farther apart can produce hybrids that are likely to have higher genetic potential than their parents. The present study grouped the accessions into two main genetic clusters with many intermediates and these two genetic clusters would be managed as two evolutionary units; hence, the selection of parents should be based on the wider inter cluster distance. The UPGMA dendrogram and PCoA showed that there was no correlation between the clusters and the geographical origin of the yam accessions. However, the UPGMA dendrogram showed the majority of yam accessions from the same geographic region were found to have a higher genetic similarity compared with samples from different regions. Therefore, crosses between the yam accessions should be made from different geographical regions. These findings provide crucial information about yam accessions in Kenya, which is essential for the conservation of genetic resources and the initiation of germplasm improvement programs.
5. Conclusions
The SCoT markers used in this study are efficient, reliable, and informative in differentiating the studied yam accessions. SCoT markers revealed the presence of wide genetic diversity among the 20 yam accessions from Kenya. The high PIC observed at some of the loci suggested a high level of genetic variability among the yam accessions. The highly polymorphic SCoT markers identified in this study could be used in generating useful molecular descriptors for fingerprinting yam accessions from Kenya. The cluster analysis grouped the accessions into two clusters and sub-clusters irrespective of the collection sites/counties. The wide genetic diversity should be utilized for breeding strategies for the genetic improvement of yam. The information generated in this study is of great interest for the design of future collections and the management of yam germplasm. In addition, some of the accessions used in this study could be a source of desirable genes and can be used by plant breeders to develop varieties with a wide genetic base that can respond better to climate change challenges. The current study used only 20 yam accessions and therefore, there is a need for further research to increase the number of sampling sites in order to fully understand the genetic diversity and population structure of yam.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijpb14010025/s1. Figure S1: Representative agarose gel electrophoresis showing amplification profile: (A) for marker SCoT33 and (B) SCoT35 in yam accessions, where M: 1 kb plus ladder, Lane A–H and E0–E11 denoted different accessions; Table S1: List of SCoT primer sequences used for PCR amplification and genetic diversity of 20 yam accessions; Table S2: Jaccard’s similarity coefficient generated from the binary data.
Author Contributions
Conceptualization, A.A.O., J.L.B., G.O.O. and E.N.N.; methodology, A.A.O., J.L.B., G.O.O. and E.N.N.; validation, J.L.B., G.O.O. and E.N.N.; formal analysis, A.A.O.; investigation, A.A.O.; resources, G.O.O. and E.N.N.; data curation, A.A.O.; writing—original draft preparation, A.A.O.; writing—review and editing, A.A.O., J.L.B., G.O.O. and E.N.N.; supervision, J.L.B. and E.N.N.; project administration, J.L.B., G.O.O. and E.N.N.; funding acquisition, E.N.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genetic data gathered during the current study are included in the manuscript. Other supplementary materials such as the electrophoretic gels of all the SCoT markers are available upon request to the authors.
Acknowledgments
We would like to thank the Centre for Biotechnology and Bioinformatics (CEBIB), University of Nairobi, for allowing us to use their laboratory facilities and reagents for this project.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gebru, H.; Mohammed, A.; Dechassa, N.; Belew, D. Assessment of production practices of smallholder potato (Solanum tuberosum L.) farmers in Wolaita zone, southern Ethiopia. Agric. Food Secur. 2017, 6, 1–11. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Data. 2019. Available online: http://www.fao.org/faostat/en/#home (accessed on 5 September 2022).
- Bassey, E.E. Constraints and prospects of yam production in Nigeria. Eur. J. Phys. Agric. Sci. 2017, 5, 55–64. [Google Scholar]
- Asiedu, R.; Sartie, A. Crops that feed the World 1. Yams for income and food security. Food Secur. 2010, 2, 305–315. [Google Scholar] [CrossRef]
- Polycarp, D.; Afoakwa, E.O.; Budu, A.S.; Otoo, E. Characterization of chemical composition and anti-nutritional factors in seven species within the Ghanaian yam (Dioscorea) germplasm. Int. Food Res. J. 2012, 19, 985–992. [Google Scholar]
- Das, S.U.; Choudhury, M.D.; Mazumder, P.B. In vitro propatation of the genus Dioscorea—A critical review. Asian J. Pharm. Clin. Res. 2013, 6, 2–6. [Google Scholar]
- Maundu, P.M.; Ngugi, G.W.; Kabugi, C.H. Traditional Food Plants of Kenya; National Museums of Kenya: Nairobi, Kenya, 1999. [Google Scholar]
- Muthamia, Z.K.; Morag, F.E.; Nyende, A.B.; Mamati, E.G.; Wanjala, B.W. Estimation of genetic diversity of the Kenyan yam (Dioscorea spp.) using microsatellite markers. Afr. J. Biotechnol. 2013, 12, 5845–5851. [Google Scholar]
- Arnau, G.; Bhattacharjee, R.M.S.; Chair, H.; Malapa, R.; Lebot, V.; Perrier, X.; Petro, D.; Penet, L.; Pavis, C. Understanding the genetic diversity and population structure of yam (Dioscorea alata L.) using microsatellite markers. PLoS ONE 2017, 12, e0174150. [Google Scholar]
- Ousmael, K.M.; Tesfaye, K.; Hailesilassie, T. Genetic diversity assessment of yams (Dioscorea spp.) from Ethiopia using inter simple sequence repeat (ISSR) markers. Afr. J. Biotechnol. 2019, 18, 970–977. [Google Scholar]
- Agre, P.; Asibe, F.; Darkwa, K.; Edemodu, A.; Bauchet, G.; Asiedu, R.; Adebola, P.; Asfaw, A. Phenotypic and molecular assessment of genetic structure and diversity in a panel of winged yam (Dioscorea alata) clones and cultivars. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Tamiru, M.; Becker, H.C.; Maass, B.L. Genetic diversity in yam germplasm from Ethiopia and their relatedness to the main cultivated Dioscorea species sssessed by AFLP markers. Crop Sci. 2007, 47, 1744–1753. [Google Scholar] [CrossRef]
- Tamiru, M.; Heiko, C.; Becker, H.C.; Maass, B.L. Comparative analysis of morphological and farmers’ cognitive diversity in yam landraces (Dioscorea spp.) from Southern Ethiopia. Trop. Agric. Dev. 2011, 55, 28–43. [Google Scholar]
- Mulualem, T.; Mekbib, F.; Shimelis, H.; Gebre, E.; Amelework, B. Genetic diversity of yam (Dioscorea spp.) landrace collections from Ethiopia using simple sequence repeat markers. Aust. J. Crop Sci. 2018, 12, 1223–1230. [Google Scholar] [CrossRef]
- Mignouna, H.D.; Abang, M.M.; Fagbemi, S.A. A comparative assessment of molecular marker assays (AFLP, RAPD and SSR) for white yam (Dioscorea rotundata) germplasm characterization. Ann. Appl. Biol. 2003, 142, 269–276. [Google Scholar] [CrossRef]
- Satya, P.; Karan, M.; Jana, S.; Mitra, S.; Sharma, A.; Karmakar, P.G.; Ray, D.P. Start codon targeted (SCoT) polymorphism reveals genetic diversity in wild and domesticated populations of ramie (Boehmeria nivea L. Gaudich.), a premium textile fiber producing species. Meta Gene 2015, 3, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.C.; Mackill, D.J. Conserved DNA-derived polymorphism (CDDP): A simple and novel method for generating DNA markers in plants. Plant Mol. Biol. Rep. 2009, 27, 558–562. [Google Scholar] [CrossRef]
- Luo, C.; He, X.H.; Chen, H.; Ou, S.J.; Gao, M.P.; Brown, J.S.; Tondo, C.T.; Schnell, R.J. Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochem. Syst. Ecol. 2011, 39, 676–684. [Google Scholar] [CrossRef]
- Guo, D.L.; Zhang, J.Y.; Liu, C.H. Genetic diversity in some grape varieties revealed by SCoT analyses. Mol. Biol. Rep. 2012, 39, 5307–5313. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, P.; Kumaria, S.; Kumar, S.; Tandon, P. Start Codon Targeted (SCoT) marker reveals genetic diversity of Dendrobium nobile Lindl., an endangered medicinal orchid species. Gene 2013, 529, 21–26. [Google Scholar] [CrossRef]
- Etminan, A.; Pour-Aboughadareh, A.; Mohammadi, R.; Ahmadi-Rad, A.; Noori, A.; Mahdavian, Z.; Moradi, Z. Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol. Equip. 2016, 30, 1075–1081. [Google Scholar] [CrossRef]
- Agarwal, A.; Gupta, V.; Haq, S.U.; Jatav, P.K.; Kothari, S.L.; Kachhwaha, S. Assessment of genetic diversity in 29 rose germplasm using SCoT marker. J. King Saud Univ. Sci. 2019, 31, 780–788. [Google Scholar] [CrossRef]
- Deng, L.; Liang, Q.; He, X.; Luo, C.; Chen, H.; Qin, Z. Investigation and analysis of genetic diversity of diospyros germplasms using SCoT molecular markers in Guangxi. PLoS ONE 2015, 10, e0136510. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, W.; Wang, Y.; Zhao, X. Potential of start codon targeted (SCoT) markers to estimate genetic diversity and relationships among Chinese Elymus sibiricus accessions. Molecules 2015, 20, 5987–6001. [Google Scholar] [CrossRef] [PubMed]
- Igwe, D.O.; Afiukwa, C.A.; Ubi, B.E.; Ogbu, K.I.; Ojuederie, O.B.; Ude, G.N. Assessment of genetic diversity in Vigna unguiculata L. (Walp) accessions using inter-simple sequence repeat (ISSR) and start codon targeted (SCoT) polymorphic markers. BMC Genet. 2017, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Jiao, K.; Yu, C.; Guo, H.; Zhu, Y.; Yang, X.; Zhang, S.; Zhang, L.; Feng, S.; Song, Y.; et al. Development of SCoT-based SCAR marker for rapid authentication of Taxus media. Biochem. Genet. 2018, 56, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; He, R.; Yang, S.; Chen, Z.; Jiang, M.; Lu, J.; Wang, H. Start codon targeted (SCoT) and target region amplification polymorphism (TRAP) for evaluating the genetic relationship of Dendrobium species. Gene 2015, 567, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.G.; He, R.F.; Jiang, M.Y.; Lu, J.J.; Shen, X.X.; Liu, J.J.; Wang, Z.A.; Wang, H.Z. Genetic diversity and relationships of medicinal Chrysanthemum morifolium revealed by start codon targeted (SCoT) markers. Sci. Hortic. 2016, 201, 118–123. [Google Scholar] [CrossRef]
- Rajesh, M.K.; Sabana, A.A.; Rachana, K.E.; Rahman, S.; Jerard, B.A.; Karun, A. Genetic relationship and diversity among coconut (Cocos nucifera L.) accessions revealed through SCoT analysis. 3 Biotech 2015, 5, 999–1006. [Google Scholar] [CrossRef]
- Feng, S.; Zhu, Y.; Yu, C.; Jiao, K.; Jiang, M.; Lu, J.; Shen, C.; Ying, Q.; Wang, H. Development of species-specific SCAR markers, based on a SCoT analysis, to authenticate Physalis (Solanaceae) species. Front. Genet. 2018, 9, 192. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Qaderi, A.; Omidi, M.; Pour-Aboughadareh, A.; Poczai, P.; Shaghaghi, J.; Mehrafarin, A.; Nohooji, M.G.; Etminan, A. Molecular diversity and phytochemical variability in the Iranian poppy (Papaver bracteatum Lindl.): A baseline for conservation and utilization in future breeding programmes. Ind. Crops Prod. 2019, 130, 237–247. [Google Scholar] [CrossRef]
- Pour-Aboughadareh, A.; Poczai, P.; Etminan, A.; Jadidi, O.; Kianersi, F.; Shooshtari, L. An analysis of genetic variability and population structure in wheat germplasm using microsatellite and gene-based markers. Plants 2022, 11, 1205. [Google Scholar] [CrossRef]
- Hamidi, H.; Talebi, R.; Keshavarz, F. Comparative efficiency of functional gene-based markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived Polymorphism (CDDP) with ISSR markers for diagnostic fingerprinting in wheat (Triticum aestivum L.). Cereal Res. Commun. 2014, 42, 558–567. [Google Scholar] [CrossRef]
- Gajera, H.P.; Bambharolia, R.P.; Domadiya, R.K.; Patel, S.V.; Golakiya, B.A. Molecular characterization and genetic variability studies associated with fruit quality of indigenous mango (Mangifera indica L.) cultivars. Plant Syst. Evol. 2013, 300, 1011–1020. [Google Scholar] [CrossRef]
- Thakur, J.; Dwivedi, M.D.; Singh, N.; Uniyal, P.L.; Goel, S.; Pandey, A.K. Applicability of Start Codon Targeted (SCoT) and Inter Simple Sequence Repeat (ISSR) markers in assessing genetic diversity in Crepidium acuminatum (D. Don) Szlach. J. Appl. Res. Med. Aromat. Plants 2021, 23, 100310. [Google Scholar] [CrossRef]
- Que, Y.; Pan, Y.; Lu, Y.; Yang, C.; Yang, Y.; Huang, N.; Xu, L. Genetic analysis of diversity within a Chinese local sugarcane germplasm based on Start Codon Targeted Polymorphism. BioMed Res. Int. 2014, 2014, 468375. [Google Scholar] [CrossRef]
- Wu, Z.G.; Li, X.X.; Lin, X.C.; Jiang, W.; Tao, Z.M.; Mantri, N.; Fan, C.Y.; Bao, X.Q. Genetic diversity analysis of yams (Dioscorea spp.) cultivated in China using ISSR and SRAP markers. Genet. Resour. Crop Evol. 2014, 61, 639–650. [Google Scholar] [CrossRef]
- Hamilton, B.M. Population Genetics; Wiley-Blackwell: New York, NY, USA, 2009; p. 234. [Google Scholar]
- Freeland, J.; Kirk, H.; Petersen, S. Genetic analysis of multiple populations. In Molecular Ecology, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2011; pp. 157–165. [Google Scholar]
- Ngailo, S.; Shimelis, H.; Sibiya, J.; Amelework, B.; Mtunda, K. Genetic diversity assessment of Tanzanian sweet potato genotypes using simple sequence repeat markers. S. Afr. J. Bot. 2016, 102, 40–45. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Ge, H.; Liu, Y.; Jiang, M.; Zhang, J.; Han, H.; Chen, H. Analysis of genetic diversity and structure of eggplant populations (Solanum melongena L.) in China using simple sequence repeat marker. Sci. Hortic. 2013, 162, 71–75. [Google Scholar] [CrossRef]
- Loko, Y.L.; Bhattacharjee, R.; Agre, A.P.; Dossou-Aminon, I.; Orobiyi, A.; Djedatin, G.L.; Dansi, A. Genetic diversity and relationship of Guinea yam (Dioscorea cayenensis Lam. D. rotundata Poir. complex) germplasm in Benin (West Africa) using microsatellite markers. Genet. Resour. Crop Evol. 2017, 64, 1205–1219. [Google Scholar] [CrossRef]
- Xiong, F.; Zhong, R.; Han, Z.; Jiang, J.; He, L.; Zhuang, W.; Tang, R. Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol. Biol. Rep. 2011, 38, 3487–3494. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).