Abstract
In every organism, the expression of genes is regulated in response to the changes in the surrounding environment. The study of epigenetics in plants is essential in view of the improvement of agricultural productivity. Epigenetic modifications can enhance crops’ yield and stress tolerance without making any alteration within their genomic sequences. The routes of epigenetic modifications include processes such as methylation of DNA, modifications of histone proteins, chromatin remodeling, and non-coding RNA-mediated regulation of genes. Genome-wide epigenetic profiles, coined as the epigenome, of several plants have been identified in recent years. In the scope of this review, we are going to discuss progress made in the field of plant epigenomics under the limelight of stress tolerance, especially saline conditions.
1. Introduction
Introducing the scope of this review, the most pertinent question is what is epigenetics? Epigenetics is defined as any kind of heritable phenotypic deviation which involves changes in the expression of the gene(s) of an organism [1,2]. These deviations are essentially brought in by alterations in chromosomal structure but not by a change in DNA sequence, i.e., chromosomal mutations. These deviations affect the working of the genome. They pass on from one generation to the next generation. Epigenetic modifications are influenced by a variety of external factors and the cell’s innate developmental package [1]. The study of epigenetics elucidates how newly formed cells that possess identical genomes go through the process of differentiation to give rise to various cell types and how such cells maintain a particular physio-morphology.
Epigenetic changes occur via 1. methylation of DNA stretches 2. modifications of the nucleosomal histone proteins and 3. RNA interference [1]. These three modes have been thoroughly studied over the decades. Inheritance of epigenetic changes also contributes to the evolutionary process of adaptation and acclimation. It occurs in two ways, firstly through the inducible epigenetic marks (epigenetic modifications induced by stress) that enable transgenerational plasticity and secondly through non-inducible epigenetic marks (stable epigenetic modifications non-induced by stress) which are resultants of selection pressure. DNA methylation pattern, chromatin remodeling as changes in the core histone proteins, or small RNAs (sRNAs) ushers phenotypic diversities and this eventually pays off towards a quick evolutionary adaptive process. Epigenetic modification alone is not a clue or signage of epigenetic inheritance. The changes that matter for evolution, whether they are epigenetic or genetic, are those that get passed onto the next generation through the germline. Epigenetic modification that occurs in a single generation is a part of phenotypic plasticity. Transgenerational inheritance give rise to epialleles. Epialleles are forms of a gene that do not vary in their nucleotide sequence but still show variation in their epigenetic marks [1]. Abiotic stresses such as salinity generate transgenerational epigenetic inheritance and in some cases this response has been studied at the genomic level.
2. Methods for Detecting Changes in Epigenome
Under the scope of this review, for understanding epigenetic changes and their role in salinity stress, it’s necessary to understand how these epigenetic modifications are assessed. In the recent decade, numerous tools have been developed for detecting methylation imprints [3]. Important methods have been discussed here. Whole Genome Bisulfite Sequencing (WGBS) is the utmost used method for the identification of 5-methylcytosine (5mC) pattern at a resolution of a single nucleotide [4]. This method can recognize each of the methylated sites and measure the degree of methylation at a specific region in the genome. The differential conversion of unmethylated cytosine into uracil is the basis of this method. Unmethylated cytosine residues get converted into uracil, on the other hand, methylated cytosines remain unchanged. Further, the evaluation of methylated cytosines is done by various sequencing methods. High-Resolution Melt Analysis/High-resolution melt (HRM) study is another simple and cost-effective method. In this method, DNA is exposed to sodium bisulfite for conversion of unmethylated cytosine residue into uracil followed by amplification through PCR. The PCR amplicon is subjected to melt temperature analysis. In the final step, the degree of methylation between known standard DNA samples and unknown samples is compared to interpret the results [5]. The melting profile from resultant PCR products is suggestive of a DNA methylation pattern. HRM has been highly successful in providing results where single nucleotide polymorphism (SNP) is detected and scanning for mutation is carried out. It is a precise method which can also identify species [5].
Enzymatic digestion of DNA by methylation-sensitive restriction endonucleases followed by PCR amplification is the basis of Chop-PCR [6]. This method can detect methylated cytosine residue at the restriction sites of such endonucleases as methylated cytosine shelters DNA from being digested, and then intact DNA can be amplified through PCR [6].Enzymatic digestion takes place in the case of unmethylated cleavage sites and consequent target sequence cannot be amplified. Methylation Sensitive Amplification Polymorphism (MSAP) sequencing is another rigorously applied technique for assessing methylated CCGG sites as studied in various plants [7]. MSAP sequencing uses isoschizomers with altered sensitivity to methylation patterns. Enzymes such as HpaII and MspI are used in this method [8]. For extraction of methylated DNA from genomic DNA via immunoprecipitation, very specific antibodies are used in the techniques such as DNA immunoprecipitation or MeDIP-Seq (Methylated DNA immunoprecipitation sequencing) and 6mA-IP (N6-methyladenine Immunoprecipitation) [9]. In these techniques, DNA needs to be extracted and fragmented using sonication or other methods. Antibodies that can identify methylated DNA fragments are used to mark off methylated regions. DNA microarrays or DNA sequencing tools are used to identify these sheared methylated DNA fragments. Interaction between DNA and proteins can be studied with coimmunoprecipitation (ChIP) as well. ChIP requires antibodies that are highly specific for DNA-binding proteins. Further analysis of DNA is done for the identification of putative target sequences bound by protein. ChIP is beneficial for finding sections of the genome associated with histone variations [9]. Other refined ChIP-based modes of epigenetic detection involve ultra-low-input native ChIP-seq which works efficiently with very sparse cell or tissue mass [10], though not chalked out in plant systems well. In this technique, an ultra-low-input micrococcal nuclease-based native ChIP (ULI-NChIP) is used followed by a well-established sequencing method to generate genome-wide histone mark profiles [10]. Another helpful tool for studying epigenetic modifications is an enzymatic method that involves chromatin endogenous cleavage (ChEC), with the aid of fusion protein comprising a chromatin protein of interest and MNase, which degrades unprotected DNA in presence of calcium [11]. This method has been heavily worked out in yeast but not in plants.
These techniques have made the way easier for the study of methylation patterns. Methods to analyze the tightly packaged chromatin material which is not easy to get through, have also been developed. Native chromatin can be studied using assays that probe on the basis of nuclease accessibility. Nucleases can chop nucleic acids into their monomeric constituents. Though, nucleases fail to degrade inaccessible chromatin material. Kits have been developed which can test chromatin accessibility. EpiQ™ chromatin analysis kit (Biorad, USA) is one such tool. Real-time quantitative PCR is done in this method to determine the accessibility of the chromatin material. This kit analyses the accessibility of the DNA by using the nuclease treatment, followed by the qPCR step. Nuclease cannot digest heterochromatin region. So, after treatment with nuclease, such regions remain intact. Thus, the DNA of heterochromatin remains available for qPCR. The trick of EpiQ analysis lies here, heterochromatin region generates a minute shift in melt curves of samples (PCR products) that are subjected to nuclease treatment and those which are not given nuclease treatment. The euchromatin regions, on the other hand, are accessible to the nuclease, thus the nuclease cut these regions and the digested DNA becomes unapproachable for subsequent qPCR analysis. Thus, euchromatin regions render a larger change in the quantification cycle amongst the nuclease-treated and untreated samples. Two other methods including digital DNase and DNase Seq work similarly with digestion by the nuclease to generate shorter stretches of DNA of the accessible genomic regions. These shorter fragments are extracted and analyzed based on sequencing results, to provide insightful details on genomic stretches which are accessible to digestion by the nuclease enzyme. This practice can then be used to relate structural DNA differences amid samples on genome-wide scales. Spatial variations in the genomic constituent in the nucleus influence gene expression. Chromatin looping causes changes in gene positioning by placing varying chromosome positions in close proximity to each other. This in turn brings in changes in gene transcription and regulation. Chromosome conformation capture or 3C assay is a tool that works with chromosome looping. This assay studies the 3-dimensional conformation of chromatin materials, by inspecting the arrangement of chromatin looping. Firstly, the crosslinking of the chromatin is carried out, followed by lysis of the cells and digestion by restriction enzymes. Specific primers are designed based on the genes to be studied in the loops in close proximity [12]. The modified version of 3C is Circular Chromosome Conformation Capture (4C) and Chromosome Conformation Capture Carbon Copy (5C). 4C identifies the interactions between genetic loci in a genome-wide manner [12] and 5C further offers a high-throughput analysis of genomic interactions through deep sequencing in order to distinguish novel parts of the DNA which are in the vicinity of each other [13].
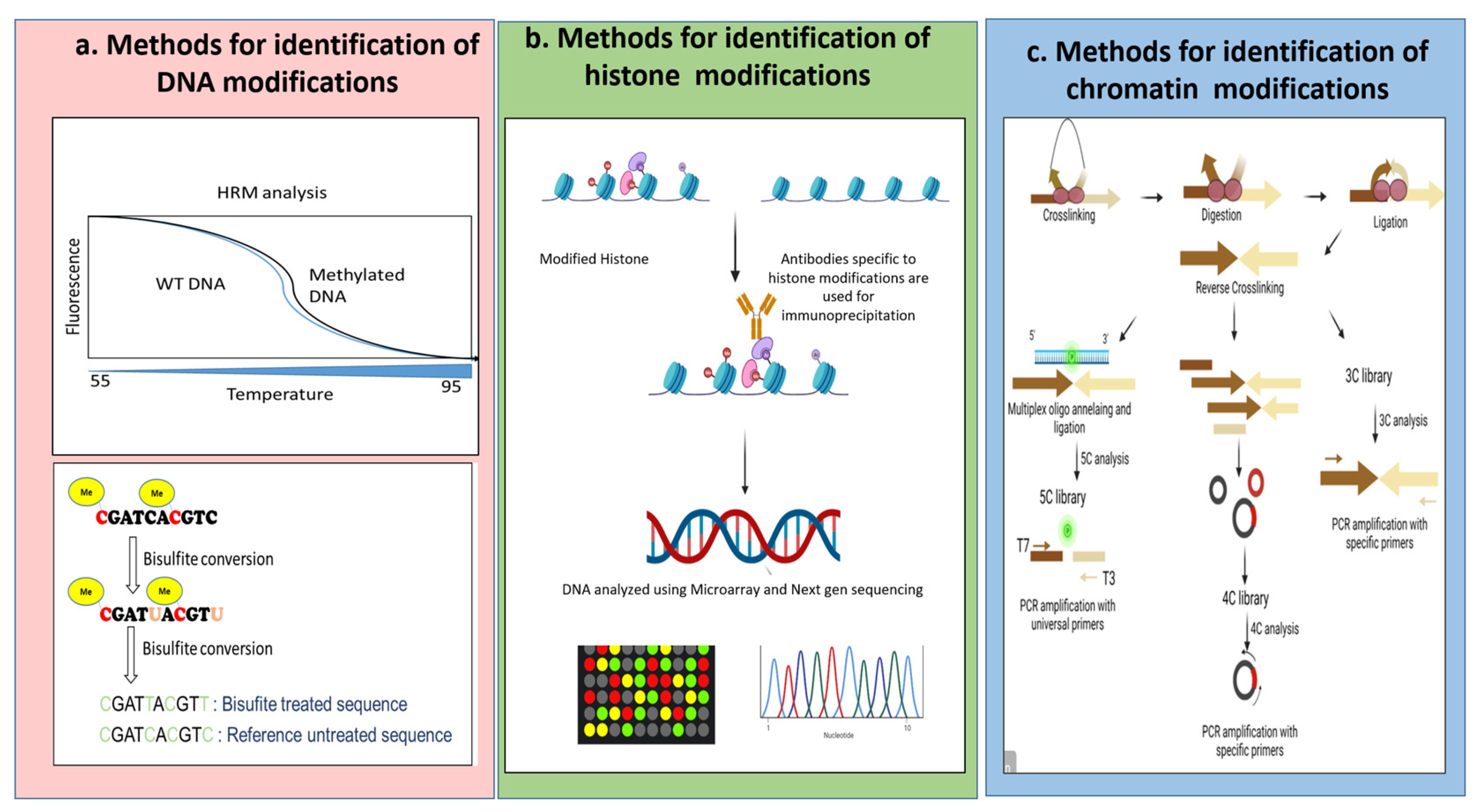
Other than this, with the advancement of deep sequencing methods, expression analysis of a plethora of miRNAs [14] has been done. With the upsurge in modified versions of sequencing platforms and availability of precise databases, identification of putative genes and authentication of sRNAs and further assigning their target mRNAs has become easier. Successively, sRNAs are getting enormous importance in relation to stress resistance in plants. Reports have highlighted the role of DNA methylation and miRNAs in transcriptional and post-transcriptional regulations of abiotic stress tolerance [15]. In Figure 1 we have added the schematic representation of some of the most used techniques for epigenetic detection.
Figure 1.
Schematic representation of some of the methods to identify epigenetic modifications. (a) Describes the methods for identification where DNA modification occurs. It can be analyzed via HRM analysis or the most prevalent technique of Bisulphite sequencing. (b) Describes the methods for identification where a histone modification occurs. It can be performed using methods of immunoprecipitation followed by sequencing and microarray. (c) Describes the methods for identification where chromatin modification occurs. It can be done by a series of steps of crosslinking, digestion, and ligation followed by the generation of libraries and sequencing using universal or specific primers following the 3C, 4C and 5C techniques. Image developed using Biorender.com (accessed on 7 September 2022).
3. Salinity-Induced Epigenetic Changes in Plants
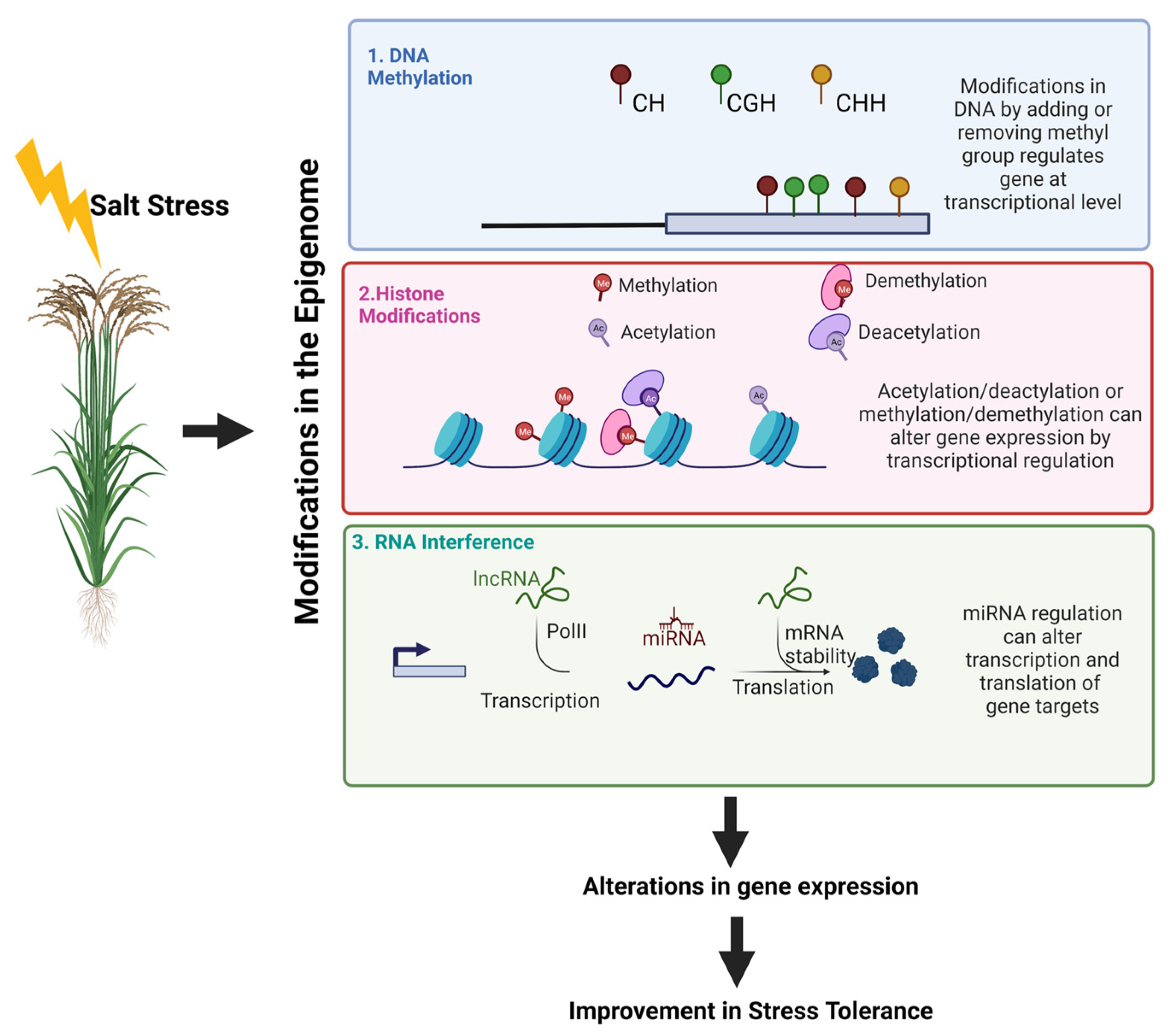
A vast expanse of cultivable land is affected by a larger extent of sodic and saline conditions, not allowing a proper growth of crops and production per capita of land. It has become important for scientists to understand the molecular mechanism prevailing over every stress, in order to handle them and meet the need of feeding the world population. In such a scenario, epigenetic solutions growing from the modifications of core histone proteins such as methylation, acetylation, and phosphorylation have been a boon in disguise to influence salt-induced stress responses in plants [16]. Biotechnological improvement has aided scientists to elucidate the underlying epigenetic mechanism that governs flowering, germination, fruit ripening, vernalization of fruiting and photoperiodism, etc. It has also helped in the identification of the configurational changes that the genome undergoes during cell differentiation. Studies have been done regarding the processes of natural selection and other adaptive responses of plants to their environment by assessing the heritable epigenetic marks of a plant [17]. Through studies on several abiotic stress responses, it was vividly observed that some abiotic stresses trigger somatic memory that lasts for some longer durations through mitotic cell divisions [18]. This kind of memory quickly resets to baseline levels when normalcy can thrive in. On the other hand, some memories affect chromatin in cells that undergo meiosis. They are heritable and thus have the potential to pass on from parent plants to the stress-free offsprings of next-generation. This is known as a transgenerational epigenetic inheritance [19]. This memory may offer descendants the tenacity to be a better fit in adaptability [19]. Our present understanding regarding the roles of epigenetics in response to plants towards salinity stresses is growing. Vast areas of study remain to be worked on to bring on and transfer the benefits of these changes to the field plants. A summary of how epigenetics and salinity are related in terms to bring in modifications is given in Figure 2.
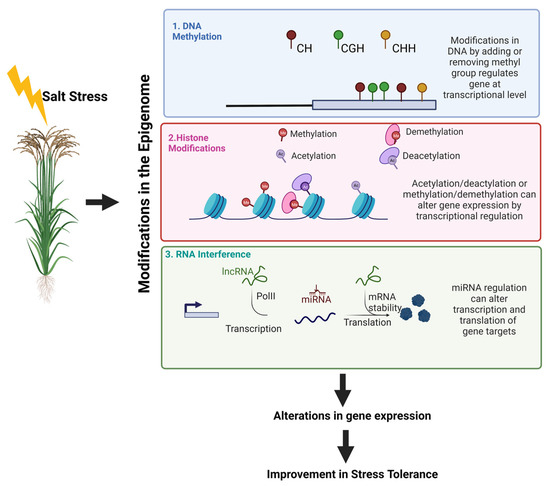
Figure 2.
A summary of how epigenetics and salinity are related in terms to bring in modifications is given here. Modes of plant epigenetic modifications include (a) DNA methylation (b) alterations in histone modifications, and (c) RNA-directed DNA methylation. (a) Changes in DNA methylation is due to changes in the addition or removal of the methylated group, which in turn regulates at the transcriptional level. In plants, three types of methylation patterns are prevalent namely CG, CHH, and CHG. (b) Alterations in histone modifications bring about changes in the core histones of the nucleosomes and which occur through multiple reactions such as acetylation, deacetylation, and methylation, demethylation of a specific set of amino acids. (c) RNA-directed DNA methylation for these miRNAs arises from primary miRNA transcripts. DNA polymerase II transcribes them from miRNA genes. Within miRNA genes, pre-miRNAs present form a hairpin loop-like which later forms miRNA–miRNA* duplex structure. The miRNA* strand is degraded and the miRNA remains intact. This duplex upon transfer gets unwound, and then miRNA shuttles into RISC and this evenually binds to related target transcripts based on complementarities of the sequences affecting gene expression at the translational level. Image developed using Biorender.com (accessed on 7 September 2022).
4. DNA Methylation and Demethylation Pattern Affecting Tolerance to Salinity Stress in Plants
DNA methylation is one of the most well-versed means to bring in epigenomic modifications. DNA methylation occurs through covalent modifications in the DNA sequence. In a methylation-based study, it was noticed that the incorporation of methyl groups takes place at the 5C’ position of the cytosine ring that forms 5 methyl-cytosine. The addition and removal of these methyl groups are undertaken by two active enzymes such as methyltransferases and demethylases respectively. N6-methyladenine (6mA) and 5-methylcytosine (5mC) are the most dominant forms of methylation patterns. In plants, three types of methylation patterns are prevalent namely CG, CHH, and CHG (where H is any base nucleotide except G) of which CG methylation occurs most frequently and CHG methylation is the least recurrent [19]. Upkeeping DNA methylation subsequently of each turn of DNA replication, is facilitated by methyltransferase 1 (MET1) in the context of CG methylation. Chromomethylase 3 holds for CHG methylation [20,21,22]. De-novo methylation of DNA is carried out by METHYLTRANSFERASE1/2 (MET1/2) and CHROMOMETHYLASE 2 (CMT2) [23].
A correlation between actively expressed genes and their related DNA methylation pattern exists under salinity stress in plants. DNA methylation and its impact on the expression of a gene differ, based on the placement of the cytosine nucleotides which are methylated within the genome, and also the sequence pattern of methylation [24]. mCG methylation within the promoter region stops gene expression, but the same methylation pattern within the exonic regions promotes active expression of the gene in some species. [25] in their study showed that there exist a putative small RNA target segment 2600 base pairs upstream of the initiation codon of transporter gene AtHKT1, which is greatly methylated. This stretch impedes the transcription of the gene AtHKT1. But the removal of this fraction of 5mC in this stretch in met1-3 Arabidopsis mutants showed a different profile of AtHKT1 and sensitivity to salinity. This observation testifies that small RNAs are essential for the modulation of expressions of genes to provide salt tolerance [25]. In plants, several reports on altered methylation patterns for bringing in epigenetic changes have been published. Plants of Arabidopsis thaliana show epigenetic regulation of the flavonoid biosynthetic pathways along with the antioxidant regulating networks during salt treatment [26]. Genes that are exclusively involved in the biosynthesis of flavonoids such as flavone-3-hydroxylase, chalcone synthase and isomerase, flavonol synthase, and also genes involved in antioxidant defense including glutathione S-transferase, glutathione peroxidase and reductases show increased fates of demethylation at not only the coding regions but also their respective promoters, especially under salinity in plants of Nicotiana tabacum overexpressing Repressor of silencing1 (AtROS1) [26]. MSAP technique was used to show the undeviating interactivity of chromatin-remodeling complexes with the members involved in ABA signaling [27]. Hypersensitive to ABA1 (HAB1) is a protein phosphatase 2C (PP2C) which acts as negative regulators of the ABA-dependent signaling cascade while SWI3B is a part of SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex which positively regulates ABA signaling. However, HAB1 interacts with and modulates the activity of SWI3B [27]. WGBS study on salt-treated and untreated plants of date palms showed that there was a significant difference regarding the level of methylation specifically at the CHG and CHH sites [28]. It was also found that mCG methylation at promoters reduced the expression of the corresponding gene. Bisulphite sequencing of the roots of soybeans showed more CHH methylation in the resilient salt-stress plants [29]. During salinity stress, demethylation was found to be more widespread in a salt-tolerant genotype of rapeseed while a more widespread methylation trend was found in the salt-sensitive variety [30]. Different dosages of salt cause different types of alteration in the epigenome. Concentrations of salt such as 50 and 100 mM instigated an entirely different set of methylation patterns in plants. In summary, a higher rate of demethylated DNA in stress-tolerant varieties may be ascertained. The genotypes that keep away cytosine methylation might be agriculturally superior as compared to the ones that are prone to faster methylation [31]. Hence, the degree of methylation of a crop genotype is important for crop improvement for amplified abiotic stress tolerance. In Arabidopsis thaliana, methylation level under stress in the roots was studied. It was observed that under 20 and 75 mM NaCl treatment conditions, there was a significant drop in CG methylation in the epidermis which was 33% more in severely stressed ones. This change was found to be related to an increase in the number of cells that formed root hairs called trichoblasts [32]. High-affinity potassium transporters (HKTs) provide stress tolerance by working on ion homeostasis in plants. In Triticum aestivum, change in cytosine methylation pattern and its direct role in this family of genes was studied. The relative expressions of TaHKT2-1 and TaHKT2-3 in the shoot and root tissues of the wheat variety Kharchia 65 were low, rendering these two genes important for improved salt tolerance [33]. Salinity makes varied effects on 5mC patterns in various other species as well. For instance, salt stress reduced the 5mC level in the salinity-tolerant wheat cultivar SR3 than its progenitor-sensitive parent JN177. These two genotypes exhibited a difference in methylation pattern with reference to salt-responsive genes [33]. Salinity stress induces upregulation of 6mA in rice cultivars [34]. Salinity improved the expression of DNA demethylases in Pokkali, which is a well-known salt-tolerant rice genotype. Methylome study between Pokkali and IR29 cultivar also reinforces the previous fact that methylation pattern under salt-stress conditions is different for different genotypes. Salinity-induced demethylation may be attributed to the rapid expression of DNA demethylases under stress. They also showed that mutations in the epigenetic regulators affected specific parameters such as the length of the root and total biomass under salinity stress [35]. In another case study, in rice, salt stress-induced expressed HUO proteins, which are a set of long terminal retrotransposons. Multiple copies of HUO activate genomic variability by altering the degree of methylation of the DNA sstretches and sRNA biogenesis pattern [35]. Even in cotton plants methylation trend was significantly changed under salt stress compared to the control CCRI 29 cotton hybrid, which is a salt-tolerant variety [36]. Genes that showed upregulation were alcohol dehydrogenase, ATP synthase, peroxidase, and glycosyltransferases [36]. Salinity stress in soybean strikingly reduced the 5mC methylation at the promoter of MYB domain protein 84 coding gene (GmMYB84). As observed for conferring stress tolerance GmMYB84 binds to the cis-regulatory elements of the K+ TRANSPORTER 1 (GmAKT1) gene [37]. Salinity causes rapid withdrawal of 5mCs from the promoter region of the OsMYB91 gene encoding MYB domain protein 91 which results in the expression of this gene. As reported in maize by [38], ZmPP2C gene encoding protein phosphatase 2C, which negatively regulates ABA signaling, is repressed but ZmGST encoding glutathione S transferase is upregulated depending on the methylation status of these genes [38]. A gene SpPKE1 (proline (P), lysine (K), and glutamic (E)) showed decreased expression under stress in plants of an abiotic stress-tolerant genotype of Solanum pennelli [39]. The coding region of this gene showed increased methylation. Interestingly, overexpression of this gene using 35S promoter provided salt tolerance in tomatoes and tobacco. The reason behind the tolerance mechanism might be a variation in the methylation pattern in the promoter region [39]. Even though, salt stress induces changes in the expression of methylated genes and transposable elements, the changes in methylation/demethylation patterns in stress responses may vary from plant to plant. Hence, studying them in their entirety is a challenge. Halophytes such as Mesembryanthemum crystallinum, under salt stress conditions, had increased methylation of CpNpG sites present in the satellite DNA. It helped in adaptation to salinity stress and switching over to crassulacean acid metabolism from the C3 pathway of carbon dioxide assimilation [40]. Studies pertaining to OsHKT1;5, encoding a sodium transporter, revealed that its cis-regulatory region has a transposable element (MITE) that undergoes CHH and CHG methylation under salinity stress. Suppressor of variegation 3-9 homolog (OsSUVH7) binds to this methylated transposable element upstream of OsHKT1;5. Thus, OsSUVH7 might be a regulator of OsHKT1-5 in response to salt stress. A list of recent reports on DNA methylation and its role in salinity stress is given in Table 1.

Table 1.
DNA methylations mediated epigenetic changes under salt stress. A summary of recently reported modifications and their corresponding genes are enlisted here.
5. Histone Modifications Affecting Tolerance to Salinity Stress in Plants
The configuration of chromatin affects the pattern of gene expression as well as the recombination of the DNA. Chromatin configures itself by two types of mechanisms, firstly the enzymatic actions that either aid in introducing covalent changes in DNA and histone proteins and secondly, by the usage of ATP to disorder interactions between histone and DNA. Epigenetic events can have a positive or a negative effect on histone modifications and largely depend on internal and external factors. The modifications which bring about changes in the core histones (H2A, H2B, H3, H4) of the nucleosomes and which occur through multiple reactions such as phosphorylation, ubiquitinylation, acetylation and methylation of a specific set of amino acids at the tail of the histone proteins, discern the extent of condensation at the chromatin region [2]. Acetylation and deacetylation of the histone proteins seem to occur at the N terminal lysine residues of H4, H3, H5 of H2A and H2B [43,44]. These two modifications are the most studied processes in the context of histone modifications. Histone acetyltransferase (HATs) causes acetylation and histone deacetylase HDAC has the antagonistic property of acetylation. Both of these in turn signal for transcriptional activation. HDACs have been widely reported in organisms such as yeast, plants, and mammals [45]. In plants, HDACs fall into three categories (1) reduced potassium dependence 3/histone deacetylase 1 (RPD3/HDA1), (2) silent information regulator 2 (SIR2) family, and (3) histone deacetylase 2 (HD2) family [45,46]. So far, there are reports of eighteen HDACs from the genomic repertoire of Arabidopsis, twelve of which belong to the RPD3/HDA1 superfamily, four belong to HD2 family, and two of them fall in the class of SIR2 family [47]. In tomatoes, fourteen HDACs are reported, of which nine belong to RPD3/HDA1 superfamily, three to the HD2 family, and two fall in SIR2 family [48].
Coming to the bigger picture of chromatin modification and its relation to salinity stress tolerance, there are several reports. In Zea mays, on salt treatment, there was an upregulation of genes such as xyloglucan endotransglucosylase (ZmXET1) and expansin β2 (ZmEXPB2), especially in the root system. This upregulation was a result of increased H3K9 acetylation in the region in proximity to the promoter and the reading frames of these genes. In maize, two acetyltransferases ZmHATB and ZmGCN5 control acetylation in its salt-tolerant variety [49]. General control non-repressed protein 5 (GCN5), a histone acetyltransferase, affects the transcription of cellulose biosynthesis genes which helps in increasing cell membrane integrity and thereby stress tolerance [50]. Using RNA sequencing and Co-IP experiments, genes such as expansion-3 (PGX3), polygalacturonase, MYB domain protein 54 (MYB54), and chitinase-like gene (CTL1) were identified which were regulated by GCN5 in wheat. GCN5 caused the acetylation of H3K9 and H3K14 under salinity stress [51]. Thus, the degree of acetylation during stress is related to the activation and functioning of a gene.
In BY2 tobacco cell suspension lines and T87 culture cells of Arabidopsis, quick transitory modification of histone such as phosphorylation of H3 Ser-10 occurred, with further changes into H3 phosphoacetylation and H4 acetylation [52]. However, chromatin modifications are subjected to changes as per the type of stress, suggesting that these modifications are governed selectively by the stress the plant encounters. Furthermore, there is a balance between the positive and negative regulators for any kind of epigenetic regulation to maintain the plant’s physiology [53]. For example, transcriptional adaptor 2b protein (ADA2b) was noted to affect acetyltransferase activity in Arabidopsis, as its mutant plants showed less tolerance to salinity [53]. Mutation in HD2C, HDA6, and HDA19 in Arabidopsis, upturned acetylation level, and their mutants exhibited hypersensitiveness towards salinity [50]. HDA710 or OsHDAC2, belonging to the HDAC RPD3/HDA1 superfamily, causes the acetylation of H3 and H4 histone proteins, specifically at H4K5 and H4K16 under nonstress circumstances. But, there was a notable increase in mRNA of HDA710 under drought and salinity, as well as treatment of abscissic acid and jasmonic acid [54]. Knockout mutant hda710 gave better endurance to salinity tolerance and reduced sensitivity towards abscissic acid, however, the overexpressing plants had contrary phenotypes [54]. An Arabidopsis hdc1 (histone deacetylase complex I) mutant had a notable amount of expression of ABA deficient 1, 3 (ABA1, 3, ABA biosynthesis gene) and also of Response to ABA18 (RAB18 which encodes a dehydrin protein) transcripts under salinity stress. On the other hand, HDC1 overexpression led to transcriptional suppression of ABA1, RAB18, and Responsive desiccation 29A (RD29A) under similar circumstances [55]. Expression of gene PtHDT902 encoding histone deacetylase 2 in Populus trichocarpa is regulated by abiotic stresses. Overexpression of PtHDT902 in Populus and Arabidopsis caused gibberellin (GA) biosynthetic genes to have elevated expression and also brought in GA-linked phenotypic changes [56]. Enhanced primary root growth in Arabidopsis and inhibition of adventitious root formation in poplar were observed by its overexpression. In addition, the poplar transgenic plants overexpressing this gene were more sensitive to salinity than control plants. This represented another HD2 gene as a regulator of adventitious root development and stress tolerance [56].
Sani et al. [57] observed epigenetic change last as long-term somatic memory for salinity response. Whole-genome ChIP-seq analysis recognized H3K27me3 histone methylation under salt treatment [58]. Another case of epigenetic regulation in the form of stress memory was studied in Arabidopsis under salt stress. In this study, a higher expression of proline biosynthesis gene P5CS encoding Δ 1-1-pyrroline-5-carboxylate synthetase was observed under salinity stress [59]. Demethylation at the promoter region of this gene led to overexpression of P5CS which caused the accumulation of proline, an osmoprotectant [59]. Most reports regarding salt stress response in plants have highlighted the acetylation of histones in relation to expressions of genes. It has often been that histone methylation was graphed to figure out methylation and other modifications as well as their combination in relation to stress. A very recent study in barley showed evidence of trimethylation of histone proteins. Salt-tolerant barley variety (Z0119) and salt-sensitive variety (Z0226) were exposed to salinity stress. On ChIP, they found that. H3K4 and H3K27 trimethylation had occurred [60]. A list of recently studied cases regarding histone modifications has been provided in Table 2.

Table 2.
Modifications of histone proteins bring in epigenetic changes under salt stress. A summary of recently reported histone modifications and their target genes are enlisted here.
6. RNA Interference in Plants Affecting Salinity Stress
The third important means to bring in epigenetic change is post-transcriptional gene silencing (PTGS) via RNA interference (RNAi). RNAi molecules are classified on the ground on their mode of biogenesis and are termed microRNAs (miRNAs) or short interfering RNAs (siRNAs). A list of recently studied miRNAs in context to salinity stress is given in Table 3. miRNAs arise from primary miRNA transcripts. DNA polymerase II transcribes them from miRNA genes [61]. Within their stretch, these pri-miRNAs contain a hairpin loop-like precursor RNAs called pre-miRNA. The pre-miRNA hairpin precursor contains 20 - 22 nucleotide long miRNA–miRNA* duplex structure. The miRNA* strand is degraded by Dicer-like 1 (DCL1) protein while miRNA remains intact. Methylation occurs at the 3′ end of the duplex by enzyme HUA ENHANCER1 (HEN1) and exportin protein HATSY transfers pre-miRNAs from the nucleus to the cytoplasm. This duplex upon transfer gets unwound by the helicases, and then miRNA shuttles into the RNA-induced silencing complex (RISC) and guides RISC to bind to related target transcripts based on complementarities of the sequences [62]. Several miRNA species have been found to be operative under environmental stress conditions. In plants, the level of miRNAs transcripts under stress conditions, cannot be consistently correlated with adaptive machinery operative under stress [63]. Salinity-inducible miRNAs have been identified in numerous plants. [64], identified 22 new miRNAs in Raphanus sativus under salt stress. These miRNAs were related to growth modulation and ion homeostasis categories. In maize under salinity stress, mir17, mir205, and mir250 genes were downregulated in leaves. mir330 gene showed downregulation in roots. These miRNAs target genes such as casein kinase II, glutathione peroxidase, pyrroline-5-carboxylate synthase, and translation initiation factors (IF-1) along with other ones which are related to better survival strategies [65]. In Saccharum species, expression of miRNAs relies on plant genre and also on the type of stress prevailing. miR156, miR159, miR166, miR167, miR168, miR169, miR396, miR397 and miR398 were found to be upregulated [66,67].

Table 3.
miRNAs mediated epigenetic changes under salinity stress. A summary of recently reported miRNAs and their targeted genes are enlisted here.
High throughput sequencing of sRNAs led to the identification of two hundred novel miRNAs and seventy-five conserved miRNAs in Pokkali rice under salinity stress. Several novel miRNAs and conserved miRNAs are expressed differentially in root or shoot. They targeted the certain largest family of transcription factors such as Apetala 2 (AP2/EREBP) domain protein, auxin response factor (ARF), NAC protein transcription factor (NAC), MYB transcription factors (MYB), Nuclear transcription factor Y (NF-YA), homeodomain-leucine zipper class III transcription factors (HD-Zip III), Teosinte Branched/Cycloidea/Proliferating Cell Factors (TCP) and SQUAMOSA-promoter binding like proteins (SBP). These genes are known to provide tolerance for different abiotic stresses including salinity [72]. Overexpression of OsmiR528 in Agrostis stolonifera improved salt tolerance by improving characteristics such as membrane integrity, water retention capacity, potassium ion homeostasis, catalase activity, and chlorophyll content [73]. Another, osmiR319 (Osa-miR319a) when expressed improved traits such as wax content, water retention ability, and ion balance [74]. Putative targets of miR319 are TCP and auxin-responsive genes such as transport inhibitor response protein 1 (TIR1), auxin signaling F box protein 2,3 encoding gene (AFB2,3), which when downregulated could make plants withstand salt stress. Higher expression of miR393-resistant TIR1 gene improved salinity tolerance in Arabidopsis. Transgenic plants showed osmoregulation and sodium exclusion. They also showed a higher rate of seed germination, reduced loss of water content, retained chlorophyll, and senescence avoidance under salinity [75]. Overexpression of miR408 elevated tolerance against salinity, oxidative, and also cold stresses in Arabidopsis plants. This miRNA targets genes encoding copper-binding proteins and some laccases [76]. In Suaeda maritime, eleven salt-responsive miRNAs were identified [77]. miR172 family members play important role in stress tolerance in Arabidopsis and Glycine max [71]. In Arabidopsis, overexpression of gma-miR172 caused an increase in root length and seed germination rate and improved tolerance towards salinity. Pan et al. [69] studied the function of miR172a in improving salt tolerance. miR172a targets the SSAC1 gene (salt-suppressed AP2 domain-containing) which encodes a repressor protein. SSAC1 inhibits the thiamine biosynthesis pathway gene (THI1). Overexpression of miR172a increases thiamine biosynthesis to enhance salt tolerance. A wheat miRNA named TaemiR408 was identified for combating salt stress as well as starvation of Pi. This miRNA targeted genes such as NtPYL2, NtSAPK3, SnRK2, NtNPT2, and NtABR2 which are involved in microtubule organization and ABA perception [71].
Discussing the second form of RNAi, siRNAs are just 20 to 25 base-paired long double-stranded RNA molecules made from a long stretch of dsRNAs. These dsRNAs can have an endogenous and exogenous origin and mediate gene silencing [78]. These are made through naturally occurring cis-antisense genes and from DNA repeats [1]. siRNAs produced through miRNA-directed cleavage of ssRNAs or transgene mRNAs act as trans-acting (ta) siRNAs. Naturally generated antisense siRNAs (nat-siRNAs) are made from dsRNAs which are formed by the mRNA transcribed from naturally happening cis-antisense gene pairs. The dsRNAs are further fine-tuned by RDRs and DCL-like proteins. siRas mobile markers, causing epigenetic variations in plants. The role of siRNAs in abiotic stress response in plants was first identified by Sunkar and Zhu [79]. Such examples were repeatedly reported in various cases. The silencing of SlHDA5 made the transgenic tomato plant susceptible to elevated temperature, salinity, and water dehydration [80]. nat-siRNA under saline conditions prevents oxidative damage and provides osmotic adjustment as studied by [66]. A 24-nucleotide long nat-siRNA is formed in Arabidopsis, when transcripts of a stress-induced gene of unknown function (SRO5) and pyrroline-5-carboxylate dehydrogenase (P5CDH) are produced. P5CDH and SRO5 genes act as natural cis-antisense pairs. The nat-siRNA caused downregulation of P5CDH which led to higher accumulation of the osmoprotectant proline and also enhanced stimulation of ROS. ROS was counterbalanced by SRO5 which is an uncharacterized stress-related protein [66]. Thus, there are numerous pieces of evidence that show the pertinent role of PTGS to bring in epigenetic response.
7. Long Noncoding RNA in Plants Affecting Salinity Stress
Gene regulation by non-coding RNAs, has recently gained ground as another mode of epigenetic regulation. LncRNAs are a type of non-coding RNA that does not bear protein-coding ability while being able to regulate gene expressions at epigenetic, transcriptional, and post-transcriptional reach [81]. Plant lncRNAs are transcribed by RNA polymerases PolII, PolIV, and PolV. According to the positioning relative to the protein-coding genes in the genome, lncRNAs can be defined as natural antisense transcripts or lncNATs, intronic lncRNAs or intergenic lncRNAs lincRNAs, and sense lncRNAs [81]. lncRNA transcripts are shorter in size around 200 bp and lack motifs, such as ORFs and Kozak consensus sequences [81]. lncRNAs generally have a 50 m7G cap and a 30 poly(A) tail and are processed as mRNA mimics. However, lncRNAs have shown low conservation in sequences among species and have low expression levels with tissue-specific expression patterns responding to various stresses in plants [82]. The existence of lnc and studies related to its role in stress is upcoming. A list of recently studied miRNAs in context to salinity stress is given in Table 4. Huanca-Mamani et al. in a study portrayed the role of lnRNA’s to be differentially expressed than the protein-coding genes in maize, when they were exposed to a combination of boron and salinity stress [80]. Similar studies have been carried out in Chenopodioum quinoa under salt stress conditions, highlighting the role of lncRNAs [83]. However, one of the earliest known lncRNA was DROUGHT-INDUCED LNCRNA (DRIR) identified in Arabidopsis, which was noted to positively regulate drought and salinity stress responses via ABA-mediated pathway [84]. Plants overexpressing DRIR display enhanced salt and drought tolerance through functioning at or upstream of the stage of gene transcription in the stress or ABA signaling. In cotton, lncRNA973 affects miR399 and its target gene PHO2 expression which are involved in response to salt stress. lncRNA973 can also modulate the expression of reactive-oxygen-species-scavenging (ROS) genes, transcription factors (TF’s), and stress-related processes, especially under saline conditions [85]. On the other hand, lncRNA354 works as a competing endogenous RNA of miR160b to regulate GhARF17/18 genes which affect plant growth and development, affecting auxin signaling under salt stress in cotton [86]. Since very less work has been carried out in terms of stress tolerance. This field of lcRNAs is an open avenue.

Table 4.
lncRNAs mediated epigenetic changes under salinity stress. A summary of recently reported lncRNAs and their targeted genes are enlisted here.
8. Cross Talk between the Major Modes of Epigenetic Changes
The disparity in response between tolerant versus susceptible varieties of plants towards salinity ought to be a collective effect of genetic and epigenetic perturbations. So, there is a cross-talk between these phenomena. A comparison was done between salt-tolerant Nonabokra and salt-sensitive IR64 rice varieties in relation to the salinity-induced chromatin modifications. OsBZ8 gene, a bZIP class abscisic acid-responsive element (ABRE) binding factor, showed heightened expression in Nonabokra even without stress, however, in IR64, expression considerably increased only under stress treatment. DNA sequence and arrangement of the nucleosomes within the region −2000 to +1000 of this OsBZ8 locus showed no integral difference between the studied rice varieties [87]. But a significant difference existed regarding modifications of histones and methylation patterns at the locus of this gene. In the salt-tolerant Nonabokra, the region upstream of this locus was hyperacetylated at H3K9 and H3K27 residues and it was also found that this pattern did not undergo any alteration under salt-stressed conditions. However, in the case of the sensitive variety IR64, acetylation of histones was noted only under salinity stress treatment. Besides, the nucleosome organization upstream of the OsBZ8 gene was highly vigorous in Nonabokra than IR64. Furthermore, reduced DNA methylation was found at the OsBZ8 locus in Nonabokra plants, along with lower H3K27me3 and higher H3K4me3 patterns. IR64 plants had denser DNA methylation and were enriched in H3K27me3 under control conditions. This study gave a heads-up for a noteworthy variation prevailing in the chromatin of the two rice genotypes and emphasized an interplay between kinds of epigenetic changes [88]. In another study, miR403 was shown to tune in the expression of argonaute family genes, AGO1 and AGO2 [89]. AGOs proteins work in small RNA pathways involved in PTGS [90]. The AGO1 gene is also controlled by miR172 [88]. miR403 and miR172 regulate stress-specific epigenetic response in Helianthus. The expression of AGO1 and AGO2 genes is also regulated by the action of DNA methylases, such as DML1 and DML3. Thus, both miRNAs and DNA methylation play a role together to put out epigenetic changes [89]. These intricate controls aid Helianthus plants to withstand abiotic stresses. miR61 and miR173 were identified as salinity-induced miRNAs in Arabidopsis [91]. AGO1 was found to interact with the loci of these two miRNAs and was accountable for the transcriptional complex disassembly and delivery of short transcripts [92]. In a study on three rice cultivars having a different degrees of salt tolerance, the profusion of sRNAs was found to be greater in the differentially methylated region (DMR) in all three varieties [93]. A greater rate of sRNA enrichment in the hypermethylated genes while a considerable loss of sRNAs in the hypo-methylated genes was found to be correlated to the salt enduring capacity of the varieties. These outcomes put forward a positive correlation between sRNAs abundance and the level of DNA methylation in rice varieties [93]. The methylation of DNAs at CG, CHG, and CHH sites is retained by diverse mechanisms in plants [94]. Zhong et al. [94] observed the existence of a link between various modes of epigenetic changes, chromatin accessibility, and genomic architecture in plants. Their study critically commented on an interplay between CG and non-CG methylation which influences the heterochromatin organization. From a follow-up study of the AtROS1 gene, another putative acetyltransferase AtROS4 was identified. The amino acid sequence within the AtROS4 protein showed similarity with that of the acetyltransferase domain, i.e PHD finger domain which binds to unmethylated histone H3K4. Mutation of AtROS4 resulted in reduction in acetylation of H3K18 and H3K23. AtROS4 contributes to the anti-silencing of the RNA-directed DNA methylation pathway [68]. Though the research in such an area is still growing, one cannot rule out the option of the interplay between various modes of epigenomic changes to bring about a change in the genome of a plant.
9. Application of Epigenetics in Raising Salinity Stress-Tolerant Plant
The epigenetic study provides notable ways and means to design strategies for the improvement of specific traits which have paved the way for epigenetic breeding. It is rapidly proliferating among various other modern crop improvement approaches. Employment of epigenetic modifications is easier in plants. Researchers have been successful in generating transgenic crops that can withstand stress conditions through these modes of alterations. Using the epigenetic approach, the gene AtROS1 was overexpressed and it provided stress tolerance in tobacco plants [26]. There are reports that state that plants are regenerated using tissue culture protocols and have stable integration of epigenome modifications as seen in rice [23]. Keeping up with the epigenetic regulations, miRNAs and siRNAs has outperformed as key performers in the regulation of plant responses to salt stress. Although an ambitious number of salinity-related genes have been recognized using epigenomic studies, their precise role remains to be verified, along with their finely tuned pathways and networks. Brassica napus which is a known model crop for its complexity in genomic architecture has given a promising insight into epigenetic breeding. One such example was with improved energy use efficiency (EUE) trait by recurrent selection for plants having the lowest and highest cellular respiration rate raised in an isogenic doubled haploid line [92,95]. The raised plants had a higher EUE index, positively correlated with a higher yield of around 8% and 20% higher seed yield under normal and drought conditions, respectively, than control plants. Methylated AFLP analysis revealed differentially methylated fragments in the sequences of genes which were maintained across 8 filial generations. This epigenetic EUE component was stable even after 3 years of elaborate field trials, and was also seen as an effect of heterosis with a yield increase of 6% than the control hybrids plants [92]. In another report where canola plants were searched with a reduced respiration rate under drought stress conditions by growing explants from canola hypocotyls in drought-conditioned medium [96]. When this shooting explants were kept in potting conditions and raised in the next generation. These lines showed enhanced performance under drought and low nitrogen conditions as they previously went through preconditioning to drought conditions. On further analysis, these epigenetic variants displayed changes in H3 Lys4 trimethylation in the coding sequences of stress-responsive genes. The epigenetically raised variant had this phenotype over a series of 7 generations. Thus, this kind of epigenetic study has gained superior consideration in a post-genomic era as a tool to fish out future prospective suitable candidates for enhancing stress tolerance in plants. Raju et al. [97] raised mutants of the MutS Homolog 1 (MHS1) gene. This gene is a plant-specific homolog of the bacterial DNA repair gene MutS, showing enhanced tolerance for salt and drought stress. The studied degree of nonCG methylation was more in msh1 mutants than in the wild type control plants. They also analyzed, that CHG changes are centered and changes with CHH more scattered [98]. The silencing of the MSH gene tames soybean plants to grow under harsh environmental conditions, in a way that makes them tolerant by triggering their other existing survival mechanisms, influencing more robust growth of the plant and ultimately providing greater yield [99]. This report suggests that some genes can also be used for long-term activation or repression of a chosen gene or a pathway for trait improvement purposes in crops. In the recent decade, there has been an increase in studies focusing on the epigenetic modifications of plant growth processes and raising likewise epialleles through breeding epirecombinant inbred lines(epiRILs). These epiRILs are thus populations of plants that undergo inbreeding as they are generated from parental strains with subtle changes in their DNA methylation pattern. In these lines, phenotypic changes occur in plants towards abiotic stress tolerance as well. EpiRILs are used for studying and quantifying epigenetic variations and their association with phenotypes. Similar to RIL, in epiRIL, a linkage map is needed to associate epigenetic loci with phenotypic variation in an [68] epiRIL. In the case of epiRILs, markers that are used for the linkage maps are the differentially methylated regions (DMRs). Analysis of the genome-wide DNA methylation of the parent and those of the epiRILs are done to check the status of these marker DMRs which are stably inherited. These comparisions are used to determine those DMRs which are stably inherited from either parent.
Next in such an epigenetic study are Epi-fingerprinting. Epi-fingerprinting not only allows to study of the relationships between environmental factors and epigenetic variations, but it helps in tracking plant health for yield. Methylation markers are studied for understanding epi-fingerprinting. These markers can be used as pre-stress interventions to control the overall productivity of crops during increased stress periods [100]. Thus, epi-fingerprinting has a tremendous upsurge in achieving sustainability in food production and quality evaluation under stress conditions.
Though this is a blind technique but there are chemical reagents such as 5-azacytidine, 5-aza 2′-deoxycytidine, and zebularine which can usher epigenetic modifications such as modifications in the histone proteins and changes in DNA methylation patterns. These chemicals are generally methyltransferase inhibitors which prevent the transfer of methyl group from S-adenosylmethionine to cytosine [96,101]. In Brassica rapa, using the chemical 5-azacytidine, hypomethylated epialleles were generated by Amoah et al., [96]. HDAC inhibitors such as trichostatin-A, nicotinamide diallyl disulfide sodium butyrate, and Helminthosporium carbonum toxins are also potent drugs for bringing in epigenetic variations in non-model plants [102]. A chemical mutagenesis is thus a powerful tool that can be used as a source for epibreeding populations for improving abiotic stress tolerance.
Even newer strategies are taking place for inducing epigenetic changes which include both targeted and nontargeted epigenetic modifications. Other methods of epigenetic editing include DNA-binding domains such as Zinc fingers proteins (ZFs), transcription activator-like effectors (TALEs), and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) proteins to known chromatin modifiers. These modifiers would be a potential tool to bring in epigenetic modifications to the selected loci then automate stress tolerance in the next progeny [98]. A newly discovered technique of engineered DNA-binding domains could enable precise epigenome engineering at specific loci. If epigenetic marks associated with specific stressors could be identified, they could allow the generation of stress-tolerant plants [99].
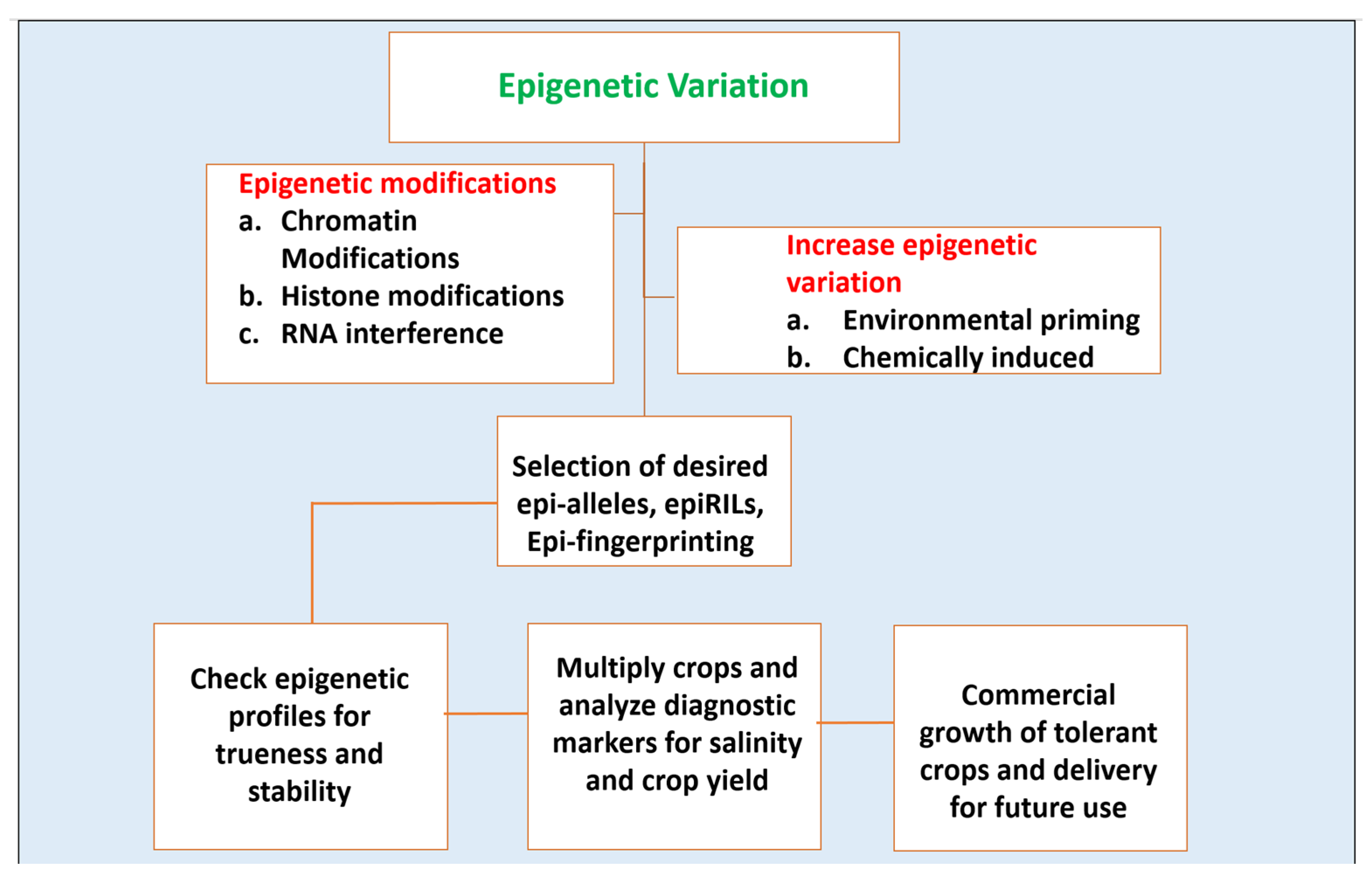
But most of such aforementioned studies and their related strategies were based on Arabidopsis and genes related to plant growth and development and much is not interpreted in the case of crops. The classical trend of crop breeding is a powerful method to obtain crops with valued agronomical potentials, but it has its own cons i.e limitation of genetic variation. Epigenome in this course is a way out as it gives out a source of variation which can be brought about by chemical, tissue culture, grafting, or any stress or phytohormone treatment [103]. Classical crop breeding is a powerful method to obtain crops with valued agronomical traits, but its potential is gradually being compromised by the limitation of genetic variation. Resorting to the epigenome as a source of variation could serve as a promising alternative. Dealing with abiotic stress with the help of epigenetic breeding is still at its infancy. The usage of these epigenetic tools for trait improvement of crops will also be beneficial for the GMO-free raising of crop plants. A future strategy for ways of generating salinity-tolerant plants using epigenetic breeding is given in Figure 3. Epigenetic breeding will provide some advantages over classical breeding in terms of the selection of elite varieties, providing a broad spectrum of resistance to abiotic stress and also a balance between important agronomic traits [102]. This kind of breeding will be time and cost-effective and most importantly will have more acceptance than GMO’s to be grown worldwide [102].
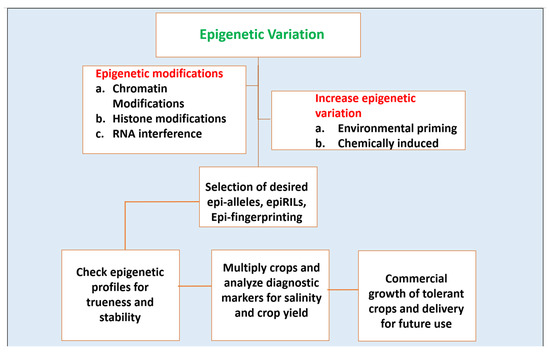
Figure 3.
A schematic representation of the idea of epigenetic breeding for raising salinity-tolerant crop plants.
10. Conclusions
Epigenetics studies can be implemented in wide areas. These studies have appeared as a sound platform to develop salinity-tolerant crops. Using these studies, plants can be generated which can sustain in a new habitat with magnified stress adaptive properties and this can be generated in a time-effective manner. The growing rate of soil salinization of arable lands and its consequences has raised the need of the hour to recognize novel ways and means to increase salt tolerance in major food as well as cash crops. Numerous studies have underlined epigenetics for figuring out a way to mitigate salt stress and its notorious impact on plants. These epigenetic pedals are often allied by tightened regulation of the stress-responsive genes in plants when exposed to sodicity and salinity. The challenges which are yet to be overcome are associated with the prediction models for the epigenetic performance of crops which includes facts such as correct identification of epibreeding markers for studying the epigenetic patterns, characterization of the mode of transmission which can be possibly either meiotic or mitotic, and quantification of elaborate linkage maps between epigenetic and phenotypic variations. [104]. Figure 3 gives a schematic representation of the idea of epigenetic breeding for raising salinity-tolerant crop plants.
Author Contributions
S.R. and P.S. conceptualized the topic. S.R. prepared the draft and wrote the review. P.S. and S.R. edited the draft. All authors have read and agreed to the published version of the manuscript.
Funding
The research involved no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
S.R. would like to thank, CMM, UC San Diego. P.S. would like to acknowledge the basic infrastructure and research facility provided by the Department of Botany, University of Rajasthan, Jaipur. P.S. would also like to thank the Departmental Research Support (DRS Phase-II) Department of Botany, University of Rajasthan.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
3C, Chromosome capture confirmation; 4C, Circular Chromosome Conformation Capture; 5C, Chromosome Conformation Capture Carbon Copy; DRM, DOMAINS REARRANGED METHYLASE; HRM, High-Resolution Melt Analysis/High-resolution melt; HATs, Histone acetyltransferase; MeDIP-Seq, Methylated DNA immunoprecipitation sequencing; MSAP, Methylation Sensitive Amplification Polymorphism; 5Mc, 5-methylcytosine; WGBS, Whole Genome Bisulfite Sequencing; 6mA-IP, N6-methyladenine Immunoprecipitation
References
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Huang, K.; Fan, G. DNA methylation in cell differentiation and reprogramming: An emerging systematic view. Regen. Med 2010, 5, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Stirzaker, C.; Taberlay, P.C.; Statham, A.L.; Clark, S.J. Mining cancer methylomes: Prospects and challenges. Trends Genet. 2014, 30, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Wojdacz, T.K.; Dobrovic, A. Methylation-sensitive high resolution melting (MS-HRM): A new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007, 35, e41. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, P.; Chaudhuri, S. Analysis of DNA Methylation Profile in Plants by Chop-PCR. In Plant Innate Immunity: Methods and Protocols; Gassmann, W., Ed.; Springer: New York, NY, USA, 2019; pp. 79–90. [Google Scholar]
- Chwialkowska, K.; Korotko, U.; Kosinska, J.; Szarejko, I.; Kwasniewski, M. Methylation Sensitive Amplification Polymorphism Sequencing (MSAP-Seq)—A Method for High-Throughput Analysis of Differentially Methylated CCGG Sites in Plants with Large Genomes. Front. Plant Sci. 2017, 8, 2056. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Davies, J.J.; Wittig, D.; Oakeley, E.J.; Haase, M.; Lam, W.L.; Schubeler, D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005, 37, 853–862. [Google Scholar] [CrossRef]
- Guevara, M.Á.; de María, N.; Sáez-Laguna, E.; Vélez, M.D.; Cervera, M.T.; Cabezas, J.A. Analysis of DNA Cytosine Methylation Patterns Using Methylation-Sensitive Amplification Polymorphism (MSAP). In Plant Epigenetics: Methods and Protocols; Kovalchuk, I., Ed.; Springer: Boston, MA, USA, 2017; pp. 99–112. [Google Scholar]
- Brind’Amour, J.; Liu, S.; Hudson, M.; Chen, C.; Karimi, M.M.; Lorincz, M.C. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Commun. 2015, 6, 6033. [Google Scholar] [CrossRef] [PubMed]
- Zentner, G.E.; Kasinathan, S.; Xin, B.; Rohs, R.; Henikoff, S. ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nat. Commun. 2015, 6, 8733. [Google Scholar] [CrossRef] [PubMed]
- Gavrilov, A.; Eivazova, E.; Priozhkova, I.; Lipinski, M.; Razin, S.; Vassetzky, Y. Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods Mol. Biol. 2009, 567, 171–188. [Google Scholar] [CrossRef]
- Ohlsson, R.; Gondor, A. The 4C technique: The ’Rosetta stone’ for genome biology in 3D? Curr. Opin. Cell Biol. 2007, 19, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Dostie, J.; Richmond, T.A.; Arnaout, R.A.; Selzer, R.R.; Lee, W.L.; Honan, T.A.; Rubio, E.D.; Krumm, A.; Lamb, J.; Nusbaum, C.; et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006, 16, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Budak, H.; Khan, Z.; Kantar, M. History and current status of wheat miRNAs using next-generation sequencing and their roles in development and stress. Brief. Funct. Genom. 2014, 14, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Ci, D.; Song, Y.; Tian, M.; Zhang, D. Methylation of miRNA genes in the response to temperature stress in Populus simonii. Front. Plant Sci. 2015, 6, 921. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect Biol. 2014, 6, a019471. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; He, Z. Small DNA Methylation, Big Player in Plant Abiotic Stress Responses and Memory. Front. Plant Sci. 2020, 11, 595603. [Google Scholar] [CrossRef] [PubMed]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.Q.; Pearson, J.K.; Hsieh, T.-F.; An, Y.-Q.C.; et al. Dynamic DNA Methylation in Plant Growth and Development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Jacobsen, S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci USA 2002, 99 Suppl. 4, 16491–16498. [Google Scholar] [CrossRef]
- Kankel, M.W.; Ramsey, D.E.; Stokes, T.L.; Flowers, S.K.; Haag, J.R.; Jeddeloh, J.A.; Riddle, N.C.; Verbsky, M.L.; Richards, E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 2003, 163, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Baek, D.; Jiang, J.; Chung, J.S.; Wang, B.; Chen, J.; Xin, Z.; Shi, H. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 2011, 52, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Bharti, P.; Mahajan, M.; Vishwakarma, A.K.; Bhardwaj, J.; Yadav, S.K. AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J Exp. Bot. 2015, 66, 5959–5969. [Google Scholar] [CrossRef]
- Saez, A.; Rodrigues, A.; Santiago, J.; Rubio, S.; Rodriguez, P.L. HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 2008, 20, 2972–2988. [Google Scholar] [CrossRef]
- Al-Harrasi, I.; Al-Yahyai, R.; Yaish, M.W. Differential DNA methylation and transcription profiles in date palm roots exposed to salinity. PLoS ONE 2018, 13, e0191492. [Google Scholar] [CrossRef]
- Chen, J.; Haanpää, M.K.; Gruber, J.J.; Jäger, N.; Ford, J.M.; Snyder, M.P. High-Resolution Bisulfite-Sequencing of Peripheral Blood DNA Methylation in Early-Onset and Familial Risk Breast Cancer Patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 5301–5314. [Google Scholar] [CrossRef]
- Marconi, G.; Pace, R.; Traini, A.; Raggi, L.; Lutts, S.; Chiusano, M.; Guiducci, M.; Falcinelli, M.; Benincasa, P.; Albertini, E. Use of MSAP markers to analyse the effects of salt stress on DNA methylation in rapeseed (Brassica napus var. oleifera). PLoS ONE 2013, 8, e75597. [Google Scholar] [CrossRef]
- Gao, G.; Li, J.; Li, H.; Li, F.; Xu, K.; Yan, G.; Chen, B.; Qiao, J.; Wu, X. Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed. Sci. 2014, 64, 125–133. [Google Scholar] [CrossRef]
- Beyrne, C.C.; Iusem, N.D.; Gonzalez, R.M. Effect of Salt Stress on Cytosine Methylation within GL2, An Arabidopsis thaliana Gene Involved in Root Epidermal Cell Differentiation. Absence of Inheritance in the Unstressed Progeny. Int. J. Mol. Sci. 2019, 20, 4446. [Google Scholar] [CrossRef]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Salt-Induced Tissue-Specific Cytosine Methylation Downregulates Expression of HKT Genes in Contrasting Wheat (Triticum aestivum L.) Genotypes. DNA Cell Biol. 2017, 36, 283–294. [Google Scholar] [CrossRef]
- Ferreira, L.J.; Azevedo, V.; Maroco, J.; Oliveira, M.M.; Santos, A.P. Salt Tolerant and Sensitive Rice Varieties Display Differential Methylome Flexibility under Salt Stress. PLoS ONE 2015, 10, e0124060. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, Y.; Gui, Y.; An, D.; Liu, J.; Xu, X.; Li, Q.; Wang, J.; Wang, W.; Shi, C.; et al. Elimination of a Retrotransposon for Quenching Genome Instability in Modern Rice. Mol. Plant 2019, 12, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, M.; Fu, R.; Qian, X.; Rong, P.; Zhang, Y.; Jiang, P.; Wang, J.; Lu, X.; Wang, D.; et al. Epigenetic mechanisms of salt tolerance and heterosis in Upland cotton (Gossypium hirsutum L.) revealed by methylation-sensitive amplified polymorphism analysis. Euphytica 2016, 208, 477–491. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, N.; Yang, J.; Guo, H.; Liu, Z.; Zheng, X.; Li, S.; Xiang, F. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. Int. J. Exp. Plant Biol. 2020, 291, 110326. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.; Lu, Y.; Jiang, W.; Wu, T.; Zhang, R.; Zhao, Y.; Zhou, D.X. DDM1 Represses Noncoding RNA Expression and RNA-Directed DNA Methylation in Heterochromatin. Plant Physiol. 2018, 177, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, C.; Wei, J.; Pan, Y.; Su, C.; Zhang, X. SpPKE1, a Multiple Stress-Responsive Gene Confers Salt Tolerance in Tomato and Tobacco. Int. J. Mol. Sci. 2019, 20, 2478. [Google Scholar] [CrossRef] [PubMed]
- Dyachenko, O.V.; Zakharchenko, N.S.; Shevchuk, T.V.; Bohnert, H.J.; Cushman, J.C.; Buryanov, Y.I. Effect of hypermethylation of CCWGG sequences in DNA of Mesembryanthemum crystallinum plants on their adaptation to salt stress. Biochem. (Mosc. ) 2006, 71, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.-X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Nan, N.; Li, N.; Liu, Y.; Wang, T.-J.; Hwang, I.; Liu, B.; Xu, Z.-Y. A DNA Methylation Reader–Chaperone Regulator–Transcription Factor Complex Activates OsHKT1;5 Expression during Salinity Stress. Plant Cell 2020, 32, 3535–3558. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Loidl, P. A plant dialect of the histone language. Trends Plant Sci. 2004, 9, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Seto, E. HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007, 26, 5310–5318. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef]
- Alinsug, M.V.; Yu, C.W.; Wu, K. Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol. 2009, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lu, J.; Zhang, J.; Wu, P.-Y.; Yang, S.; Wu, K. Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum). Front. Plant Sci. 2015, 5, 760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, S.; Zhao, L.; Tan, J.; Zhang, Q.; Gao, F.; Wang, P.; Hou, H.; Li, L. Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 2014, 14, 105. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wang, Y.-Y.; Liu, X.; Yang, S.; Lu, Q.; Cui, Y.; Wu, K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012, 63, 3297–3306. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Liu, X.; Lin, J.; Liu, X.; Wang, Z.; Xin, M.; Yao, Y.; Peng, H.; Zhou, D.-X.; Ni, Z.; et al. Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J. 2019, 97, 587–602. [Google Scholar] [CrossRef] [PubMed]
- Sokol, A.; Kwiatkowska, A.; Jerzmanowski, A.; Prymakowska-Bosak, M. Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 2007, 227, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Kaldis, A.; Tsementzi, D.; Tanriverdi, O.; Vlachonasios, K.E. Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta 2011, 233, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Ullah, F.; Xu, Q.; Zhao, Y.; Zhou, D.X. Histone deacetylase HDA710 controls salt tolerance by regulating ABA signaling in rice. J. Integr. Plant Biol. 2020, 63, 451–467. [Google Scholar] [CrossRef]
- Perrella, G.; Lopez-Vernaza, M.A.; Carr, C.; Sani, E.; Gosselé, V.; Verduyn, C.; Kellermeier, F.; Hannah, M.A.; Amtmann, A. Histone Deacetylase Complex1 Expression Level Titrates Plant Growth and Abscisic Acid Sensitivity in Arabidopsis. J. Plant Cell 2013, 25, 3491–3505. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liang, X.; Lv, S.; Guan, T.; Jiang, T.; Cheng, Y. Histone deacetylase gene PtHDT902 modifies adventitious root formation and negatively regulates salt stress tolerance in poplar. Plant Sci. 2020, 290, 110301. [Google Scholar] [CrossRef]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, R59. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Song, G.; Guo, W.; Wang, W.; Zhao, H.; Gao, T.; Lv, Q.; Yang, X.; Xu, F.; Dong, Y.; et al. Dynamic Changes in Genome-Wide Histone3 Lysine27 Trimethylation and Gene Expression of Soybean Roots in Response to Salt Stress. Front. Plant Sci. 2019, 10, 1031. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhury, A.; Banerjee, A.; Lahiri, V. Metabolic and molecular-genetic regulation of proline signaling and itscross-talk with major effectors mediates abiotic stress tolerance in plants. Turk. J. Bot. 2015, 39, 887–910. [Google Scholar] [CrossRef]
- Yuzhen, B.; Sang, Z.; Mu, W.; Yu, M.; Wang, Y.; Yuan, H.; Xu, Q. Whole-genome analysis of the trimethylation of histone H3 lysine 4 and lysine 27 in two contrasting Tibetan hulless barley genotypes under salinity stress. Acta. Physiol. Plant. 2021, 43, 89. [Google Scholar] [CrossRef]
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 2007, 12, 301–309. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef] [PubMed]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. MicroRNAs As Potential Targets for Abiotic Stress Tolerance in Plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef]
- Sun, X.; Xu, L.; Wang, Y.; Yu, R.; Zhu, X.; Luo, X.; Gong, Y.; Wang, R.; Limera, C.; Zhang, K.; et al. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genom. 2015, 16, 197. [Google Scholar] [CrossRef]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of Salt Tolerance-related microRNAs and Their Targets in Maize (Zea mays L.) Using High-throughput Sequencing and Degradome Analysis. Front. Plant Sci. 2017, 8, 864. [Google Scholar] [CrossRef]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef]
- Gentile, A.; Dias, L.I.; Mattos, R.S.; Ferreira, T.H.; Menossi, M. MicroRNAs and drought responses in sugarcane. Front. Plant Sci. 2015, 6, 58. [Google Scholar] [CrossRef]
- Li, X.; Qian, W.; Zhao, Y.; Wang, C.; Shen, J.; Zhu, J.K.; Gong, Z. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 11425–11430. [Google Scholar] [CrossRef]
- Pan, W.J.; Tao, J.J.; Cheng, T.; Bian, X.H.; Wei, W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Soybean miR172a Improves Salt Tolerance and Can Function as a Long-Distance Signal. Mol. Plant 2016, 9, 1337–1340. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Yan, J.; Wang, K.; Luo, H.; Zhang, W. MiR319 mediated salt tolerance by ethylene. Plant Biotechnol. J. 2019, 17, 2370–2383. [Google Scholar] [CrossRef]
- Bai, Q.; Wang, X.; Chen, X.; Shi, G.; Liu, Z.; Guo, C.; Xiao, K. Wheat miRNA TaemiR408 Acts as an Essential Mediator in Plant Tolerance to Pi Deprivation and Salt Stress via Modulating Stress-Associated Physiological Processes. Front. Plant Sci. 2018, 9, 499. [Google Scholar] [CrossRef]
- Parmar, S.; Gharat, S.A.; Tagirasa, R.; Chandra, T.; Behera, L.; Dash, S.K.; Shaw, B.P. Identification and expression analysis of miRNAs and elucidation of their role in salt tolerance in rice varieties susceptible and tolerant to salinity. PLoS ONE 2020, 15, e0230958. [Google Scholar] [CrossRef]
- Yuan, S.; Li, Z.; Li, D.; Yuan, N.; Hu, Q.; Luo, H. Constitutive Expression of Rice MicroRNA528 Alters Plant Development and Enhances Tolerance to Salinity Stress and Nitrogen Starvation in Creeping Bentgrass. Plant Physiol. 2015, 169, 576–593. [Google Scholar] [CrossRef]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013, 161, 1375–1391. [Google Scholar] [CrossRef]
- Chen, Z.; Hu, L.; Han, N.; Hu, J.; Yang, Y.; Xiang, T.; Zhang, X.; Wang, L. Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 73–83. [Google Scholar] [CrossRef]
- Ma, C.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef]
- Gharat, S.A.; Shaw, B.P. Novel and conserved miRNAs in the halophyte Suaeda maritima identified by deep sequencing and computational predictions using the ESTs of two mangrove plants. BMC Plant Biol. 2015, 15, 301. [Google Scholar] [CrossRef]
- Siomi, H.; Siomi, M.C. On the road to reading the RNA-interference code. Nature 2009, 457, 396–404. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Huanca-Mamani, W.; Arias-Carrasco, R.; Cárdenas-Ninasivincha, S.; Rojas-Herrera, M.; Sepúlveda-Hermosilla, G.; Caris-Maldonado, J.C.; Bastías, E.; Maracaja-Coutinho, V. Long Non-Coding RNAs Responsive to Salt and Boron Stress in the Hyper-Arid Lluteño Maize from Atacama Desert. Genes 2018, 9, 170. [Google Scholar] [CrossRef]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef]
- Deng, P.; Liu, S.; Nie, X.; Weining, S.; Wu, L. Conservation analysis of long non-coding RNAs in plants. Sci. China. Life Sci. 2018, 61, 190–198. [Google Scholar] [CrossRef]
- Luo, C.; He, B.; Shi, P.; Xi, J.; Gui, H.; Pang, B.; Cheng, J.; Hu, F.; Chen, X.; Lv, Y. Transcriptome dynamics uncovers long non-coding RNAs response to salinity stress in Chenopodium quinoa. Front. Plant Sci. 2022, 13, 3445. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A Nucleus-Localized Long Non-Coding RNA Enhances Drought and Salt Stress Tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, J.; Deng, F.; Wang, W.; Cheng, Y.; Song, L.; Hu, M.; Shen, J.; Xu, Q.; Shen, F. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019, 19, 459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shen, J.; Xu, Q.; Dong, J.; Song, L.; Wang, W.; Shen, F. Long noncoding RNA lncRNA354 functions as a competing endogenous RNA of miR160b to regulate ARF genes in response to salt stress in upland cotton. Plant Cell Environ. 2021, 44, 3302–3321. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Dasgupta, P.; Roy, D.; Chaudhuri, S. Comparative analysis of Histone modifications and DNA methylation at OsBZ8 locus under salinity stress in IR64 and Nonabokra rice varieties. Plant Mol. Biol. 2017, 95, 63–88. [Google Scholar] [CrossRef] [PubMed]
- Ronemus, M.; Vaughn, M.W.; Martienssen, R.A. MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 2006, 18, 1559–1574. [Google Scholar] [CrossRef]
- Ebrahimi Khaksefidi, R.; Mirlohi, S.; Khalaji, F.; Fakhari, Z.; Shiran, B.; Fallahi, H.; Rafiei, F.; Budak, H.; Ebrahimie, E. Differential expression of seven conserved microRNAs in response to abiotic stress and their regulatory network in Helianthus annuus. Front. Plant Sci. 2015, 6, 741. [Google Scholar] [CrossRef]
- Schraivogel, D.; Meister, G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 2014, 39, 420–431. [Google Scholar] [CrossRef]
- Dolata, J.; Bajczyk, M.; Bielewicz, D.; Niedojadlo, K.; Niedojadlo, J.; Pietrykowska, H.; Walczak, W.; Szweykowska-Kulinska, Z.; Jarmolowski, A. Salt Stress Reveals a New Role for ARGONAUTE1 in miRNA Biogenesis at the Transcriptional and Posttranscriptional Levels. Plant Physiol. 2016, 172, 297–312. [Google Scholar] [CrossRef]
- Hauben, M.; Haesendonckx, B.; Standaert, E.; Van Der Kelen, K.; Azmi, A.; Akpo, H.; Van Breusegem, F.; Guisez, Y.; Bots, M.; Lambert, B.; et al. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc. Natl. Acad. Sci. USA 2009, 106, 20109–20114. [Google Scholar] [CrossRef]
- Garg, R.; Narayana Chevala, V.V.S.; Shankar, R.; Jain, M. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 2015, 5, 14922. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Feng, S.; Duttke, S.H.; Potok, M.E.; Zhang, Y.; Gallego-Bartolomé, J.; Liu, W.; Jacobsen, S.E. DNA methylation-linked chromatin accessibility affects genomic architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2023347118. [Google Scholar] [CrossRef]
- Verkest, A.; Byzova, M.; Martens, C.; Willems, P.; Verwulgen, T.; Slabbinck, B.; Rombaut, D.; Van de Velde, J.; Vandepoele, K.; Standaert, E.; et al. Selection for Improved Energy Use Efficiency and Drought Tolerance in Canola Results in Distinct Transcriptome and Epigenome Changes. Plant Physiol. 2015, 168, 1338–1350. [Google Scholar] [CrossRef] [PubMed]
- Amoah, S.; Kurup, S.; Rodriguez Lopez, C.M.; Welham, S.J.; Powers, S.J.; Hopkins, C.J.; Wilkinson, M.J.; King, G.J. A hypomethylated population of Brassica rapa for forward and reverse epi-genetics. BMC Plant Biol. 2012, 12, 193. [Google Scholar] [CrossRef]
- Kenchanmane Raju, S.K.; Shao, M.R.; Wamboldt, Y.; Mackenzie, S. Epigenomic plasticity of Arabidopsis msh1 mutants under prolonged cold stress. Plant Direct 2018, 2, e00079. [Google Scholar] [CrossRef] [PubMed]
- Waryah, C.B.; Moses, C.; Arooj, M.; Blancafort, P. Zinc Fingers, TALEs, and CRISPR Systems: A Comparison of Tools for Epigenome Editing. Methods Mol. Biol. 2018, 1767, 19–63. [Google Scholar] [CrossRef]
- Bilichak, A.; Kovalchuk, I. Transgenerational response to stress in plants and its application for breeding. J. Exp. Bot. 2016, 67, 2081–2092. [Google Scholar] [CrossRef]
- Rodríguez López, C.M.; Wilkinson, M.J. Epi-fingerprinting and epi-interventions for improved crop production and food quality. Front. Plant Sci. 2015, 6, 397. [Google Scholar] [CrossRef]
- Akone, S.H.; Ntie-Kang, F.; Stuhldreier, F.; Ewonkem, M.B.; Noah, A.M.; Mouelle, S.E.M.; Müller, R. Natural Products Impacting DNA Methyltransferases and Histone Deacetylases. Front. Pharmacol. 2020, 11, 992. [Google Scholar] [CrossRef]
- Tirnaz, S.; Batley, J. Epigenetics: Potentials and Challenges in Crop Breeding. Mol. Plant 2019, 12, 1309–1311. [Google Scholar] [CrossRef]
- Dalakouras, A.; Vlachostergios, D. Epigenetic approaches to crop breeding: Current status and perspectives. J. Exp. Bot. 2021, 72, 5356–5371. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chaudhary, P.; Taunk, J.; Kumar Singh, C.; Sharma, S.; Singh, V.J.; Singh, D.; Chinnusamy, V.; Yadav, R.; Pal, M. Plant epigenomics for extenuation of abiotic stresses: Challenges and future perspectives. J. Exp. Bot. 2021, 72, 6836–6855. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).