Abstract
Background: To describe trends and clinical experiences in applying commercial pharmacogenetic testing among pediatric patients with neuropsychiatric disorders. Methods: Demographic and clinical data of patients receiving GeneSight® testing from January 2015 to November 2016 at an urban pediatric hospital were retrospectively extracted from medical charts. Outcome data included pharmacogenetic test results and medication prescriptions before and after the test. Results: A total of 450 patients (12.1 ± 4.3 years) diagnosed with anxiety disorder, attention deficit hyperactivity disorder, developmental disorders including autism, and/or a mood disorder received testing, and 435 of them were prescribed medications. Comparing data before and after testing, the total number of psychotropic prescriptions were reduced by 27.2% and the number of prescribed medications with severe gene-drug interactions decreased from 165 to 95 (11.4% to 8.9% of total medications prescribed). Approximately 40% of actionable genetic annotation were related to CYP2CD6 and CYP2C19. Patients of Asian descent had significantly higher likelihood than other races of being classified as poor to intermediate metabolizers of antidepressants, mood stabilizers, and antipsychotics (p = 0.008, 0.007, and 0.001, respectively). Diagnoses, including autism spectrum disorder, were not associated with increased risks of severe gene-drug interactions. Conclusions: Pharmacogenetic testing in child and adolescent psychiatry is currently based on few clinically actionable genes validated by CPIC and/or FDA. Although this approach can be moderately utilized to guide psychotropic medication prescribing for pediatric patients with psychiatric disorders, clinicians should cautiously interpret test results while still relying on clinical experience and judgment to direct the final selection of medication.
1. Introduction
Approximately 16.5% (7.7 million) of youth aged 6- to 17-years old in the United States are diagnosed with at least one mental health disorder [1]. Among those aged 2 to 17 years, 9.4% have Attention Deficit Hyperactivity Disorder (ADHD) [2], 7.1% have anxiety disorder, and 3.2% have major depressive disorder [3]. These disorders are associated with significant morbidity [4]. As their prevalence increases, clinicians are challenged to find effective early treatments to avert disease progression. While an armamentarium of psychotropic medications is available, heterogeneity exists with treatment response and medication tolerance as attributed to factors such as gender, accuracy of diagnosis, and comorbidities. Additionally, genetics accounts for about 40% of the variability in antidepressant response within major depression [5]. As such, pharmacogenetics, which studies the impact of genetic variations on drug responses, is an area of increasing interest among clinicians seeking to incorporate genetics to personalize psychotropic treatments and to reduce the trial-and-error approach to prescribing.
Conflicting evidence exists regarding the clinical utility of pharmacogenetic testing in psychiatry despite a strong interest in adopting this approach to guide medication prescribing for adults with mental health disorders [6,7,8,9,10]. The evidence for pharmacogenetic-guided treatment of pediatric mental health disorders is more limited [11,12,13]. Further, while the Clinical Pharmacogenetics Implementation Consortium (CPIC) has pharmacogenetic guidelines for CYP2D6 and atomoxetine, which are specifically based on pediatric studies, they added a cautionary statement for applying guidelines for other medications (e.g., SSRIs) in pediatric patients since most of the data were derived from adult study populations [14]. The ontologic impact of the expression of CYP2D6 and CYP2C19 drug metabolizing enzyme activity also adds another level of complexity that clinicians need to consider when applying pharmacogenetic results [15]. Despite this, the use of pharmacogenetic testing to guide treatment decision in pediatric psychiatric disorders is gaining traction [16,17,18], as more pediatric hospitals in the United States are adopting the test with some places testing specific genes while others use panel-based test [12,19,20,21,22].
While prior studies have evaluated panel-based pharmacogenetic testing in adults with mental health disorders, to our knowledge, there is little published literature on its use in children and adolescents with mental health disorders [18,23,24,25,26,27,28]. This study seeks to address this knowledge gap by evaluating the application of such testing among pediatric patients with neuropsychiatric disorders treated at a tertiary care hospital through a retrospective chart review. Demographic and diagnostic factors were examined to see if certain patient types are at higher risk to have severe gene-drug interactions, potentially suggesting which groups may benefit most from pharmacogenetic testing. Findings were described as trends and experiences in applying commercial pharmacogenetic testing among pediatric patients with neuropsychiatric disorders.
2. Materials and Methods
2.1. Study Design and Study Population
This is a retrospective analysis of pediatric patients receiving care for a psychiatric disorder in a large urban academic children’s hospital. Patients were seen in outpatient specialty clinics in neurology, developmental/behavioral pediatrics, adolescent medicine, and psychiatry, as well as in inpatient psychiatry units; all of which are operating within the same institution. During the study period, clinicians had started to use Genesight® testing for patients who were receiving polypharmacy, experiencing medication side effects, or were not responding to current medication regimen. Inclusion criteria were patients aged 1 to 22 years (maximum age of patients seen in specialty clinics), with a diagnosis of psychiatric disorder, who had undergone psychiatric pharmacogenetic testing using GeneSight® Psychotropic (Myriad Neuroscience) between January 2015 to November 2016. Pharmacogenomic test reports of patients meeting the inclusion criteria were obtained from Myriad Neuroscience who housed the data. This dataset was then merged with clinical dataset (e.g., basic demographic information, psychotropic medication prescribed any time before and after the pharmacogenomic test date until the end of the study period) that was extracted from the study site’s electronic medical database. All patient identifiers were removed after both pharmacogenomic and clinical datasets were merged, with the data managed using the Research Electronic Data Capture (REDCap) [29]. All data analyses were completed using the deidentified dataset.
2.2. Pharmacogenetic Testing
GeneSight® Psychotropic test from Myriad Neuroscience (formerly Assurex Health, Inc., Mason, OH) tested 59 alleles and variants across 8 genes (CYP1A2, CYP2C9, CYP2C19, CYP3A4, CYP2B6, CYP2D6, HTR2A, SLC6A4) as reported elsewhere [30]. A proprietary algorithm that weighted the combined influence of a patient’s genotype results was applied to provide pharmacogenetic recommendations for 38 psychotropic medications based on three levels of gene-drug interaction: (i) ‘use as directed’ (no gene-drug interaction detected; annotated by green label in the test report); (ii) ‘use with caution’ (moderate gene-drug interaction and drug may be effective with dose modification; yellow label); and (iii) ‘use with increased caution and with more frequent monitoring’ (severe gene-drug interaction, which may significantly impact drug safety and/or efficacy; a red label) [30].
2.3. Prescription Trends Analysis
Psychotropic medications prescribed in this study were categorized into antidepressants, antipsychotics, medications to treat ADHD, and mood stabilizers. These medications were analyzed for pharmacogenetic annotations based on CPIC guidelines [14] and the Food and Drug Administration (FDA) list of drugs with pharmacogenomic biomarkers in the drug label [31]. Psychotropic medications prescribed before and after the pharmacogenetic test within the study period were compared against the Genesight® report to determine the type of gene-drug interaction. The study then focused on analyzing antidepressants to illustrate how pharmacogenetic results were applied clinically. Specifically, an analysis was performed on the selective serotonin reuptake inhibitors (SSRI; citalopram, escitalopram, fluvoxamine, paroxetine, fluoxetine and sertraline), serotonin norepinephrine reuptake inhibitors (SNRI; desvenlafaxine, duloxetine, levomilnacipran, venlafaxine), and dopamine norepinephrine reuptake inhibitor (DNRI; bupropion). Tricyclic antidepressants were excluded as they were less frequently prescribed clinically given their side effect profile and low utilization for depression in pediatric patients.
2.4. Statistical Analysis
De-identified dataset was analyzed using descriptive statistics (mean, standard deviation, frequency as count and percentage). The number and type of psychotropic medications prescribed for each subject before and after the pharmacogenetic test (within the study period) were compared. Correlational statistics (chi-square) was applied to determine whether pertinent predictors (age, gender, race, and psychiatric diagnoses) were related to patients receiving prescription for medications in the severe gene-drug interaction category. All analyses were performed using JMP® Pro (version 14.1.0), and significance level was set at p < 0.05.
3. Results
3.1. Demographic and Medical Profile of the Study Cohort
A total of 28 clinicians tested 450 patients during the study period. These clinicians consisted of 12 child and adolescent psychiatrists, 7 child and adolescent psychiatry fellows, 3 pediatricians specialized in adolescent medicine, 3 developmental pediatricians, and 3 pediatric neurologists. The study cohort (N = 450) was 64% male, 76% Caucasian, with a mean age (SD) of 12 (4.3) years. The most common mental health diagnosis was an anxiety disorder (44%), followed by ADHD (38%), and autism spectrum disorder (ASD) (35%). Major depression was present in 15% of the cohort. Other depressive disorders such as disruptive mood dysregulation disorder, persistent depressive disorder (dysthymia), substance/medication-induced depressive disorder, depressive disorder due to another medical condition, other specified depressive disorder, and/or unspecified depressive disorder were present in 25% of the cohort. The majority of the patients (42%) had two concurrent psychiatric diagnoses, and 18% had at least three concurrent psychiatric diagnoses. All diagnoses were based on DSM–5 criteria [32]. Table 1 lists the basic demographic and medical profile of the study cohort.

Table 1.
Basic demographic and medical profiles of study cohort (N = 450).
3.2. Prescription Trends of Psychotropic Medications
Among the 450 patients who underwent psychiatric pharmacogenetic testing, only 435 (97%) were prescribed psychotropic medications. A total of 47 different psychotropic medications were prescribed to these patients during the study period (Table 2). Most of the medications were antidepressants (40%), followed by antipsychotics (28%), ADHD medications (17%), and mood stabilizers (15%). The top 10 psychotropic medications prescribed included guanfacine (50%), sertraline (43%), risperidone (35%), methylphenidate (33%), aripiprazole (33%), fluoxetine (29%), mixed amphetamine salts (25%), lamotrigine (19%), atomoxetine (16%), and lisdexamfetamine (15%) (Table 2). Data collected from the time before the pharmacogenetic test was conducted showed that there were 1469 prescriptions written for these patients (an average of 3.3 prescriptions per patient). Data from time period after the test indicated that there were 1070 prescriptions (an average of 2.5 prescriptions per patient). Overall, this reflected a decrease of 27.2% in the number of prescriptions. Additionally, the number of prescribed medications with severe gene-drug interactions (as rated by GeneSight® algorithm) decreased from 165 prior to testing to 95 following the testing (i.e., from 11.4% to 8.9% of total medications prescribed).

Table 2.
Frequency distribution of psychotropic medications (N = 47; as percentage; in descending order of prescribing frequency) being prescribed to the study cohort (N = 435) at any time during the study period, and their respective pharmacogenetics information.
3.3. Predictors of Severe Gene-Drug Interactions
Based on their pharmacogenetic test results, Asians had significantly more severe gene-drug interactions for antidepressants and mood stabilizers than patients of other races (p = 0.008 and p = 0.007, respectively). A similar trend was observed when comparing Asians with Caucasians for antipsychotics (p = 0.001). However, age, gender, and psychiatric diagnoses were not significant predictors of which patients would be at risk for severe gene-drug interactions (p > 0.05 for all comparisons). Notably, a diagnosis of ASD did not predict a higher likelihood of severe gene-drug interactions.
3.4. Psychotropic Medications with Pharmacogenetic Annotation
Approximately half of the medications listed in the GeneSight® algorithm (53%) did not have any pharmacogenetic annotation from CPIC or the FDA; while 11 (24%) had information from CPIC and FDA, 10 (21%) had information from FDA alone, and 1 (2%) had information from CPIC alone (N = 47; Figure 1).
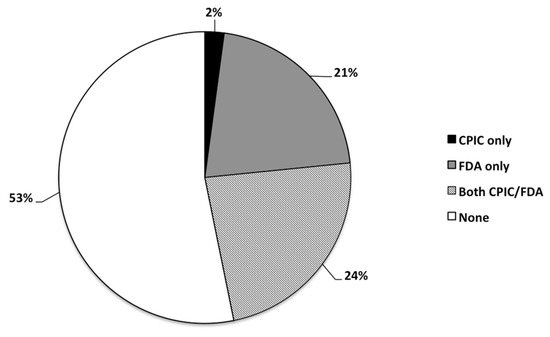
Figure 1.
Frequency distribution (as percentage) of psychotropic medications (N = 47) being prescribed to the study cohort (N = 435) having CPIC and/or FDA guidelines, or none at all.
The majority of the pharmacogenetic annotations were related to CYP2CD6 and CYP2C19 (40%), followed by HLA-A and HLA-B genes for drug hypersensitivity reaction (4%) and POLG gene, which predicted the risk for valproate-induced hepatotoxicity (2%) (Table 2). Since CYP2D6 and CYP2C19 were the most frequently noted metabolizer phenotypes, the rates of these phenotypes of the study cohort (as derived from the Genesight® reports) were compared against trends of general population published in literature [33,34]. Only trends for Caucasians were analyzed due to sample size. This comparative analysis was performed to evaluate whether CYP2D6 and CYP2C19 metabolizer frequency distribution of our study cohort was comparable to the general population, considering that the patients in our cohort were potentially more likely to have comorbidities and treatment failures since they were seen at a tertiary care center. When compared to data from published literature [33,34], Caucasians in this study cohort had similar CYP2D6 and CYP2C19 metabolizer frequency distribution as the general population. For CYP2D6 metabolizer phenotype, the majority of the study cohort were normal metabolizers (80.3%; compared to 80.9% in general population), followed by intermediate (9.7% vs. 7.8%), poor (7.7% vs. 6.6%), ultrarapid (2.4% vs. 3.5%), and normal (0% vs. 1.2%). For CYP2C19 metabolizer phenotype, the study cohort was mostly of normal metabolizer phenotype (40.1%; compared to 42.0% in general population), followed by rapid (27.0% vs. 27.0%), intermediate (26.7% vs. 19.0%), ultrarapid (4.8% vs. 4.2%), and poor (1.5% vs. 2.8%).
3.5. Pharmacogenetic Results for Antidepressants (SSRIs, SNRIs, DNRIs)
Among the SSRIs, paroxetine (54%) had the highest proportion of patients with severe gene-drug interactions (red label) followed by fluvoxamine (42%), then fluoxetine (29%). Among the SNRIs, duloxetine (42%) had the largest proportion of patients with severe gene-drug interactions followed by venlafaxine (20%). In contrast, desvenlafaxine (an SNRI) had no gene-drug interaction (green label). For bupropion (a DNRI), 19% of the study cohort had severe gene-drug interaction. These results are summarized in Figure 2.
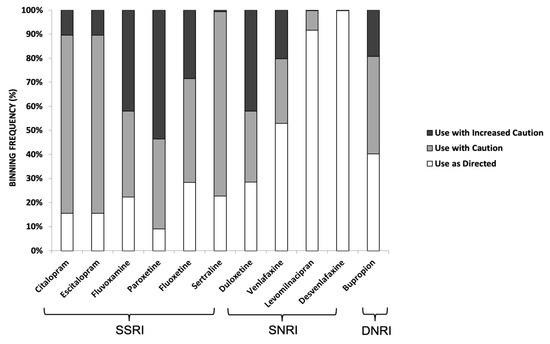
Figure 2.
Binning frequency distribution of SSRI, SNRI, and DNRI, which were prescribed to the study cohort based on their GeneSight® pharmacogenetics reports. Color codes for respective bins were based on the classification outlined by Jablonski et al. [30]. These are: (i) ‘use as directed’ (no gene-drug interaction detected; annotated by green label in the test report); (ii) ‘use with caution’ (moderate gene-drug interaction and drug may be effective with dose modification; annotated by a yellow label); and (iii) ‘use with increased caution and with more frequent monitoring’ (severe gene-drug interaction that may significantly impact drug safety and/or efficacy; as annotated by a red label).
4. Discussion
Increasing prevalence of mental health disorders among youth has created a relative shortage of pediatric specialists (e.g., child and adolescent psychiatrists, pediatric neurologists, developmental and behavioral pediatricians, among others) to provide evaluation and treatment. Commercial panel-based pharmacogenetic testing has been introduced to the field of psychiatry as a way to guide prescribing practices. Currently, the use of pharmacogenomic testing prior to prescribing psychotropic medications in pediatric patients is not mandatory, nor it is a routine practice. Such testing is typically performed once a pediatric patient has had unanticipated medication side effects, has a history of failed medications, or is initiating a medication for which there is a clearly defined guideline response (e.g., carbamazepine and HLA-B*1502). While its role in child and adolescent psychiatry continues to be defined, many clinicians have adopted such testing clinically, or find their patients’ caregivers bringing in these results for them to interpret. Since there are limited data in this field based on pediatric populations, this retrospective study primarily sought to describe trends observed from a large cohort of pediatric patients with psychiatric disorders seen at a tertiary care center who had undergone such testing. A secondary outcome to this study was to highlight pertinent points garnered from our observation that clinicians could consider when using commercial pharmacogenetic testing to guide prescribing practices in light of current evidence.
There are currently more than 200 medications with pharmacogenetic biomarker information in FDA-approved labels, with 39 of them being prescribed for psychiatric conditions [31,35]. Among psychotropic medications, the main biomarkers involved are the hepatic cytochrome P450 2C19 and 2D6 metabolizing enzymes [36]. CPIC has also published evidence-based peer-reviewed pharmacogenetic guidelines for tricyclic antidepressants and SSRIs based on CYP2D6 and CYP2C19 genotypes, and atomoxetine based on CYP2D6 genotypes [14,37,38]. The interest to personalize medication prescribing in psychiatry has fueled the development of commercial pharmacogenetic tests with several marketed as being psychiatry focused [39]. Many of these tests, however, included other genes of varying pharmacogenetic level of evidence in addition to CYP2D6 and CYP2C19. Patients’ test results may be reported in the form of a pharmacogenetic decision support tool, such as classifying medications in color-coded bins based on the level of gene-drug interaction. However, these categorizations are based on proprietary algorithms that are not standardized across the different companies [39,40,41]. Clinicians need to be aware of these points when selecting a pharmacogenetic test for their patients or when being presented with commercial test reports for clinical interpretation.
Anxiety and mood/depressive disorders are common diagnoses in our study cohort. It has been estimated that more than half of all adult patients with major depressive disorders fail to achieve remission with the initial antidepressant regimen, with the pediatric literature reporting a wider variability (23 to 63%) [42]. This factor has driven the interest for incorporating pharmacogenetics to guide medication selection, especially to aid in identifying medications with a low side effect profile or adjusting dosage to reflect a patient’s metabolic phenotype. While CPIC guidelines provide recommendations on drug selection and dosing based on CYP2C19 and CYP2D6 genetic polymorphisms, which alter the metabolism of certain antidepressants, other pharmacodynamic factors remain to be elucidated since the biology of mental health disorders remains to be fully understood [43,44]. Clinicians who utilize pharmacogenetic tests should also be cognizant of CPIC and FDA guidelines, especially since CPIC recommendations for the pediatric population were formulated based on current pharmacogenetic evidence and continues to be updated [14,31]. Further, a systematic review of drugs with pharmacogenetic information in FDA drug labels found 65 drugs with pharmacogenetic information that have been evaluated in children, but a majority (86%) of the pharmacogenetic information were extrapolated from adult studies [45]. As such, clinicians need to moderate the expectations of patients and their caregivers toward results of pharmacogenetic testing by explaining the role and limitations of these tests, especially when applying adult-derived pharmacogenetic information towards the care of their pediatric patients.
Another practical consideration is related to the clinical interpretation of pharmacogenetic test results for genetic variants that have mixed evidence. The FDA issued a warning statement expressing concerns over pharmacogenetic tests that claim to “predict a patient’s response to specific medications that have not been reviewed by the FDA and may not be supported by clinical evidence” [46]. As a case example, a certain genetic test claimed that their test could identify the extent of efficacy of select antidepressants when compared to others. This is troubling because the relationship between DNA variations and physiological processes influencing antidepressant efficacy has not been established [46]. The FDA also expressed worries that certain software programs that interpreted genetic information made similar claims when current evidence does not support those claims [47]. Reflecting on these cases, the FDA was concerned that clinicians who largely relied on the results of pharmacogenetic tests might make inappropriate medication changes that could inadvertently result in adverse health consequences. Consequently, the FDA has reached out to several pharmacogenetic testing companies who have since removed specific medication names from their promotional material and patient test reports. The FDA also reiterated their commitment to supporting innovation in this area given the evidence available for some gene-drug pairs. While the FDA continues their monitoring efforts, clinicians can consult resources such as FDA-approved drug labels and CPIC guidelines on how to use genetic information to guide their prescribing [14,31].
With regards to the role of pharmacogenetic testing in psychiatry medicine, the International Society of Psychiatry Genetics (ISPG) issued a genetic testing statement that “Pharmacogenetic testing should be viewed as a decision-support tool to assist in thoughtful implementation of good clinical care, enhancing rather than offering an alternative to standard protocols” [48]. While genetics is one piece of information that can be used to guide medication prescribing, other clinical factors (e.g., renal function, and medical and psychiatric comorbidities) must also be considered. The analysis of the antidepressants (SSRIs, SNRIs, and DNRIs) prescribed in our genotyped pediatric cohort showed that a majority of their pharmacogenetic recommendations were in the moderate-to-severe gene-drug interaction category (yellow or red designation in the test reports) (Figure 2), which may alarm some clinicians (and their patients and caregivers). However, clinically in many cases, such classification does not necessarily lead to contraindications that require medication change. Instead, there are other strategies that can be implemented including dosing adjustment and closer monitoring of side effects. Clinicians at the study site may have used these strategies since we observed a reduction in polypharmacy in our study cohort post-testing (M.Z.W.; through personal communications and case discussions). Thus, clinical insights are still necessary to guide prescribing practices instead of completely relying on pharmacogenetic test results.
Many clinicians believe commercial pharmacogenetic testing has moderate clinical utility, despite its limitations and conflicting evidence in the literature [49]. However, these findings may be skewed in favor of pharmacogenetic testing since several of these studies were performed at sites where clinicians were already familiar with pharmacogenetics [50,51,52,53]. A recent survey of clinicians also indicated that 85% of respondents were concerned about the lack of clear guidance for clinical application [54]. This trend is troubling considering that many clinicians who use this type of test to guide clinical decision-making may not have an extensive pharmacogenetic knowledge to be sufficiently proficient in interpreting the test reports, among other perceived barriers [20,49,55]. Considering these factors, a multidisciplinary approach should be considered when interpreting pharmacogenetic test results. Psychiatric pharmacists trained in pharmacogenetics can provide additional clinical support in this way. However, a survey of psychiatric pharmacists indicated that only 36% considered themselves to be knowledgeable in pharmacogenetics [52]. Medical liaisons from pharmacogenetic testing vendors could provide another support resource, although their expertise may not translate to medical realm. Overall, this highlights the need of providing more pharmacogenetic training and other resources for healthcare providers, which is consistent with the findings of Liko et al. [49].
Logistical variables related to ordering the pharmacogenetic test (e.g., where to obtain the test, on whom and when to perform the test, costs to patients, and other pertinent factors) should be discussed at the clinic visit. A recent survey of psychiatrists noted that 94% of respondents were concerned about the cost associated with pharmacogenetic testing [54], which may not be covered by insurance providers. The cost of pharmacogenetic testing may be a barrier to its uptake, as most patients currently pay out-of-pocket for the test. In the United States, many Medicare contractors do not consider the test reasonable or necessary, and private insurers tend to follow Medicare coverage decisions [56]. However, this is gradually changing [57]. In 2019, United Healthcare agreed to reimburse pharmacogenetic panel-based testing, as it is proven and medically necessary to guide antidepressant and antipsychotic prescribing under certain circumstances [58]. Additionally, Medicare expanded their coverage through new Molecular Diagnostic Services (MolDx) local coverage determinations starting in 2020 onwards. A cost-effectiveness study comparing pharmacogenetic guided strategy versus treatment-as-usual in guiding antidepressant treatment in Canadian patients showed that the pharmacogenetic guided strategy was more efficacious and less costly compared to the latter [59]. It should be noted that pediatric data are lacking since most cost-benefit studies on pharmacogenetic testing in psychiatry have been conducted in adult populations. The majority of the pharmacoeconomic evaluation studies are also based on simulations rather than actual clinical data [60]. Therefore, future studies can be conducted to evaluate the cost-benefit of using pharmacogenetic testing to guide drug treatment in pediatric patients with neuropsychiatric disorders, and address payer concerns regarding its clinical utility and benefits.
Meanwhile, clinicians might consider ordering pharmacogenetic test for select patients, such as those with history of psychotropic medication intolerance, and/or repeated treatment failure. Our results indicated that patients of Asian descent were more likely to have significant gene-drug interactions, hence those patients may be candidates for such testing as well. Overall, it is imperative that clinicians communicate these logistical variables, as well as the potential utility and limitations of pharmacogenetic testing when discussing testing plan and subsequent results with their patients and caregivers.
There are several limitations to this study. First, the study did not set out to determine the clinical impact of prescribing decisions as a result of pharmacogenetic testing, but rather provided a brief description of prescribing trends of various psychotropic medications based on the pharmacogenetic results of the study cohort. The retrospective study design that was utilized in this study is certainly not the gold standard for objectively measuring symptomatology and long-term impact of prescribed medications. Our interpretation about the impact of pharmacogenetic testing at the study site was also influenced by the variability in the detailed information recorded in patients’ charts. While the study cohort’s medical record did not systematically document reasons for medication changes, most of the clinicians ordered the pharmacogenetic test when the initial medication regimens were either not effective and/or were causing significant side effects to the patient (M.Z.W.; personal communications and clinical case discussions). Further, the study was conducted in a single tertiary care center and thus the results may not be generalizable to other pediatric medical centers due to differing practice preferences and hospital formularies. This study also included a diverse study population (i.e., patients from both inpatient and outpatient settings), which may have introduced confounders. Despite these limitations, the findings remain valuable to the literature as it reflects a snapshot of ‘real world’ practice where pediatric patients were selected for genotyping based on clinical consideration.
5. Conclusions
In summary, commercial pharmacogenetic tests can be utilized to guide psychotropic medication prescribing for pediatric patients with psychiatric disorders while recognizing their limitations. In the context of pediatric pharmacogenetic testing, clinicians need to be aware of the ontologic effects of CYP450 drug metabolizing enzyme development in children. Clinicians should also use pharmacogenetic resources such as the CPIC and/or FDA guidelines to corroborate gene-drug(s) pairs that have strong evidence to support their use in the pediatric cohort. We anticipate that the debate surrounding the clinical utility of pharmacogenetic test for pediatric patients with psychiatric disorders continues as long as large randomized pharmacogenetic studies based on pediatric cohort remain unexplored, and that clinicians are not adequately trained in pharmacogenetics to effectively interpret test results. Ultimately, the results of commercial panel-based pharmacogenetic testing should not replace clinical experience and judgment for making the final medication choice.
Author Contributions
Conceptualization, M.A. and M.Z.W.; methodology, M.A. and M.Z.W.; validation, M.A.; formal analysis, M.A. and M.Z.W.; investigation, M.A., Y.M.L., D.L.S., S.L. and M.Z.W.; resources, M.A., Y.M.L., D.L.S., and M.Z.W.; data curation, M.A.; writing—original draft preparation, M.A.; writing—review and editing, M.A., Y.M.L., D.L.S., S.L., and M.Z.W.; visualization, M.A.; supervision, M.Z.W.; project administration, M.A.; funding acquisition, M.Z.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Colorado Multiple Institutional Review Board (COMIRB #16-0768, approval date 12 April 2017).
Informed Consent Statement
Patient consent was waived since this was a retrospective study involving data extraction from medical charts.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Whitney, D.G.; Peterson, M.D. US National and State-Level Prevalence of Mental Health Disorders and Disparities of Mental Health Care Use in Children. JAMA Pediatrics 2019, 173, 389–391. [Google Scholar] [CrossRef]
- Danielson, M.L.; Bitsko, R.H.; Ghandour, R.M.; Holbrook, J.R.; Kogan, M.D.; Blumberg, S.J. Prevalence of Parent-Reported ADHD Diagnosis and Associated Treatment Among U.S. Children and Adolescents, 2016. J. Clin. Child Adolesc. Psychol. 2018, 47, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Ghandour, R.M.; Sherman, L.J.; Vladutiu, C.J.; Ali, M.M.; Lynch, S.E.; Bitsko, R.H.; Blumberg, S.J. Prevalence and Treatment of Depression, Anxiety, and Conduct Problems in US Children. J. Pediatr. 2019, 206, 256–267.e253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CDC. Children’s Mental Health. Available online: https://www.cdc.gov/childrensmentalhealth/data.html (accessed on 21 January 2020).
- Tansey, K.E.; Guipponi, M.; Hu, X.; Domenici, E.; Lewis, G.; Malafosse, A.; Wendland, J.R.; Lewis, C.M.; McGuffin, P.; Uher, R. Contribution of common genetic variants to antidepressant response. Biol. Psychiatry 2013, 73, 679–682. [Google Scholar] [CrossRef]
- Bradley, P.; Shiekh, M.; Mehra, V.; Vrbicky, K.; Layle, S.; Olson, M.C.; Maciel, A.; Cullors, A.; Garces, J.A.; Lukowiak, A.A. Improved efficacy with targeted pharmacogenetic-guided treatment of patients with depression and anxiety: A randomized clinical trial demonstrating clinical utility. J. Psychiatr. Res. 2018, 96, 100–107. [Google Scholar] [CrossRef]
- Hall-Flavin, D.K.; Winner, J.G.; Allen, J.D.; Carhart, J.M.; Proctor, B.; Snyder, K.A.; Drews, M.S.; Eisterhold, L.L.; Geske, J.; Mrazek, D.A. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharm. Genom. 2013, 23, 535–548. [Google Scholar] [CrossRef] [Green Version]
- Rosenblat, J.D.; Lee, Y.; McIntyre, R.S. Does Pharmacogenomic Testing Improve Clinical Outcomes for Major Depressive Disorder? A Systematic Review of Clinical Trials and Cost-Effectiveness Studies. J. Clin. Psychiatry 2017, 78, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Zhao, W.; Qiu, Y.; Wu, H.; Chen, J.; Fang, Y.; Guo, W.; Li, L. Preliminary Clinical Investigation of Combinatorial Pharmacogenomic Testing for the Optimized Treatment of Depression: A Randomized Single-Blind Study. Front. Neurosci. 2019, 13, 960. [Google Scholar] [CrossRef]
- Winner, J.G.; Carhart, J.M.; Altar, C.A.; Allen, J.D.; Dechairo, B.M. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov. Med. 2013, 16, 219–227. [Google Scholar]
- Ramsey, L.B.; Bishop, J.R.; Strawn, J.R. Pharmacogenetics of treating pediatric anxiety and depression. Pharmacogenomics 2019, 20, 867–870. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, L.B.; Prows, C.A.; Zhang, K.; Saldana, S.N.; Sorter, M.T.; Pestian, J.P.; Wenstrup, R.J.; Vinks, A.A.; Glauser, T.A. Implementation of Pharmacogenetics at Cincinnati Children’s Hospital Medical Center: Lessons Learned Over 14 Years of Personalizing Medicine. Clin. Pharm. Ther. 2019, 105, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Maruf, A.A.; Greenslade, A.; Arnold, P.D.; Bousman, C. Antidepressant pharmacogenetics in children and young adults: A systematic review. J. Affect. Disord 2019, 254, 98–108. [Google Scholar] [CrossRef] [PubMed]
- CPIC. Clinical Pharmacogenetics Implementation Consortium (CPIC). Available online: https://cpicpgx.org/guidelines/ (accessed on 28 December 2020).
- Hines, R.N. Developmental expression of drug metabolizing enzymes: Impact on disposition in neonates and young children. Int. J. Pharm. 2013, 452, 3–7. [Google Scholar] [CrossRef]
- Aldrich, S.L.; Poweleit, E.A.; Prows, C.A.; Martin, L.J.; Strawn, J.R.; Ramsey, L.B. Influence of CYP2C19 Metabolizer Status on Escitalopram/Citalopram Tolerability and Response in Youth With Anxiety and Depressive Disorders. Front. Pharm. 2019, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.R.; Najjar, F.; Rubin, L.H.; Guter, S.J.; Owley, T.; Mosconi, M.W.; Jacob, S.; Cook, E.H. Escitalopram pharmacogenetics: CYP2C19 relationships with dosing and clinical outcomes in autism spectrum disorder. Pharm. Genom. 2015, 25, 548–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blasco-Fontecilla, H. Clinical utility of pharmacogenetic testing in children and adolescents with severe mental disorders. J. Neural Transm. 2019, 126, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haidar, C.E.; Relling, M.V.; Hoffman, J.M. Preemptively Precise: Returning and Updating Pharmacogenetic Test Results to Realize the Benefits of Preemptive Testing. Clin. Pharm. Ther. 2019, 106, 942–944. [Google Scholar] [CrossRef]
- Brown, J.T.; Ramsey, L.B.; Van Driest, S.L.; Aka, I.; Colace, S.I. Characterizing Pharmacogenetic Testing Among Children’s Hospitals. Clin. Transl. Sci. 2021, 14, 692–701. [Google Scholar] [CrossRef]
- Claudio-Campos, K.; Padrón, A.; Jerkins, G.; Nainaparampil, J.; Nelson, R.; Martin, A.; Wiisanen, K.; Smith, D.M.; Strekalova, Y.; Marsiske, M.; et al. Acceptability, Feasibility, and Utility of Integrating Pharmacogenetic Testing into a Child Psychiatry Clinic. Clin. Transl. Sci. 2021, 14, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Sandritter, T.L.; Dinh, J.C.; Wagner, J.A.; Lowry, J.A. Description of an Innovative Pediatric Individualized Therapeutics Clinic: Working toward Precision Drug Therapy. Children 2019, 6, 35. [Google Scholar] [CrossRef] [Green Version]
- Stutzman, D. Pharmacogenomic testing in child and adolescent psychiatry. Colo. J. Psychiatry Psychol. 2019, 3, 11–30. [Google Scholar]
- Wehry, A.M.; Ramsey, L.; Dulemba, S.E.; Mossman, S.A.; Strawn, J.R. Pharmacogenomic Testing in Child and Adolescent Psychiatry: An Evidence-Based Review. Curr. Probl. Pediatr. Adolesc. Health Care 2018, 48, 40–49. [Google Scholar] [CrossRef]
- Greden, J.F.; Parikh, S.V.; Rothschild, A.J.; Thase, M.E.; Dunlop, B.W.; DeBattista, C.; Conway, C.R.; Forester, B.P.; Mondimore, F.M.; Shelton, R.C.; et al. Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: A large, patient- and rater-blinded, randomized, controlled study. J. Psychiatr. Res. 2019, 111, 59–67. [Google Scholar] [CrossRef]
- Rosenblat, J.D.; Lee, Y.; McIntyre, R.S. The effect of pharmacogenomic testing on response and remission rates in the acute treatment of major depressive disorder: A meta-analysis. J. Affect. Disord 2018, 241, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Shelton, R.C.; Parikh, S.V.; Law, R.A.; Rothschild, A.J.; Thase, M.E.; Dunlop, B.W.; DeBattista, C.; Conway, C.R.; Forester, B.P.; Macaluso, M.; et al. Combinatorial Pharmacogenomic Algorithm is Predictive of Citalopram and Escitalopram Metabolism in Patients with Major Depressive Disorder. Psychiatry Res. 2020, 290, 113017. [Google Scholar] [CrossRef] [PubMed]
- Tanner, J.A.; Davies, P.E.; Voudouris, N.C.; Shahmirian, A.; Herbert, D.; Braganza, N.; Gugila, A.; Dechairo, B.M.; Kennedy, J.L. Combinatorial pharmacogenomics and improved patient outcomes in depression: Treatment by primary care physicians or psychiatrists. J. Psychiatr. Res. 2018, 104, 157–162. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inf. 2009, 42, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Jablonski, M.R.; King, N.; Wang, Y.; Winner, J.G.; Watterson, L.R.; Gunselman, S.; Dechairo, B.M. Analytical validation of a psychiatric pharmacogenomic test. Pers. Med. 2018, 15, 189–197. [Google Scholar] [CrossRef] [Green Version]
- FDA. Table of Pharmacogenomic Biomarkers in Drug Labeling. Available online: https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling (accessed on 9 September 2020).
- APA. American Psychiatric Association: Diagnostic and Statistical Manual of Mental Disorders: Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; APA: Arlington, VA, USA, 2013. [Google Scholar]
- Del Tredici, A.L.; Malhotra, A.; Dedek, M.; Espin, F.; Roach, D.; Zhu, G.D.; Voland, J.; Moreno, T.A. Frequency of CYP2D6 Alleles Including Structural Variants in the United States. Front. Pharm. 2018, 9, 305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strom, C.M.; Goos, D.; Crossley, B.; Zhang, K.; Buller-Burkle, A.; Jarvis, M.; Quan, F.; Peng, M.; Sun, W. Testing for variants in CYP2C19: Population frequencies and testing experience in a clinical laboratory. Genet. Med. 2012, 14, 95–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, L.; Eum, S.; Haga, S.B.; Strawn, J.R.; Zierhut, H. Clinical Utilization of Pharmacogenetics in Psychiatry—Perspectives of Pharmacists, Genetic Counselors, Implementation Science, Clinicians, and Industry. Pharmacopsychiatry 2020, 53, 162–173. [Google Scholar] [CrossRef]
- Bousman, C.; Maruf, A.A.; Muller, D.J. Towards the integration of pharmacogenetics in psychiatry: A minimum, evidence-based genetic testing panel. Curr. Opin. Psychiatry 2019, 32, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.T.; Bishop, J.R.; Sangkuhl, K.; Nurmi, E.L.; Mueller, D.J.; Dinh, J.C.; Gaedigk, A.; Klein, T.E.; Caudle, K.E.; McCracken, J.T.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Cytochrome P450 (CYP)2D6 Genotype and Atomoxetine Therapy. Clin. Pharm. Ther. 2019, 106, 94–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicks, J.K.; Sangkuhl, K.; Swen, J.J.; Ellingrod, V.L.; Muller, D.J.; Shimoda, K.; Bishop, J.R.; Kharasch, E.D.; Skaar, T.C.; Gaedigk, A.; et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharm. Ther. 2017, 102, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Bousman, C.A.; Hopwood, M. Commercial pharmacogenetic-based decision-support tools in psychiatry. Lancet Psychiatry 2016, 3, 585–590. [Google Scholar] [CrossRef]
- Bousman, C.A.; Dunlop, B.W. Genotype, phenotype, and medication recommendation agreement among commercial pharmacogenetic-based decision support tools. Pharm. J. 2018, 18, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Hall-Flavin, D.K.; Winner, J.G.; Allen, J.D.; Jordan, J.J.; Nesheim, R.S.; Snyder, K.A.; Drews, M.S.; Eisterhold, L.L.; Biernacka, J.M.; Mrazek, D.A. Using a pharmacogenomic algorithm to guide the treatment of depression. Transl. Psychiatry 2012, 2, e172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennard, B.D.; Emslie, G.J.; Mayes, T.L.; Nightingale-Teresi, J.; Nakonezny, P.A.; Hughes, J.L.; Jones, J.M.; Tao, R.; Stewart, S.M.; Jarrett, R.B. Cognitive-behavioral therapy to prevent relapse in pediatric responders to pharmacotherapy for major depressive disorder. J. Am. Acad. Child Adolesc. Psychiatry 2008, 47, 1395–1404. [Google Scholar] [CrossRef] [Green Version]
- Corponi, F.; Fabbri, C.; Serretti, A. Pharmacogenetics and Depression: A Critical Perspective. Psychiatry Investig. 2019, 16, 645–653. [Google Scholar] [CrossRef] [Green Version]
- Dubovsky, S.L. The Limitations of Genetic Testing in Psychiatry. Psychother. Psychosom. 2016, 85, 129–135. [Google Scholar] [CrossRef]
- Green, D.J.; Mummaneni, P.; Kim, I.W.; Oh, J.M.; Pacanowski, M.; Burckart, G.J. Pharmacogenomic information in FDA-approved drug labels: Application to pediatric patients. Clin. Pharm. Ther. 2016, 99, 622–632. [Google Scholar] [CrossRef]
- FDA. The FDA Warns against the Use of Many Genetic Tests with Unapproved Claims to Predict Patient Response to Specific Medications: FDA Safety Communication. Available online: https://www.fda.gov/medical-devices/safety-communications/fda-warns-against-use-many-genetic-tests-unapproved-claims-predict-patient-response-specific#actions (accessed on 9 September 2020).
- FDA. FDA Issues Warning Letter to Genomics Lab for Illegally Marketing Genetic Test that Claims to Predict Patients’ Responses to Specific Medications. Available online: https://www.fda.gov/news-events/press-announcements/fda-issues-warning-letter-genomics-lab-illegally-marketing-genetic-test-claims-predict-patients (accessed on 12 December 2020).
- ISPG. Genetic Testing Statement. Available online: https://ispg.net/genetic-testing-statement/ (accessed on 12 December 2020).
- Liko, I.; Lee, Y.M.; Stutzman, D.L.; Blackmer, A.B.; Deininger, K.M.; Reynolds, A.M.; Aquilante, C.L. Providers’ perspectives on the clinical utility of pharmacogenomic testing in pediatric patients. Pharmacogenomics 2021, 22, 263–274. [Google Scholar] [CrossRef]
- Dunbar, L.; Butler, R.; Wheeler, A.; Pulford, J.; Miles, W.; Sheridan, J. Clinician experiences of employing the AmpliChip(R) CYP450 test in routine psychiatric practice. J. Psychopharmacol. 2012, 26, 390–397. [Google Scholar] [CrossRef]
- Hoop, J.G.; Lapid, M.I.; Paulson, R.M.; Roberts, L.W. Clinical and ethical considerations in pharmacogenetic testing: Views of physicians in 3 “early adopting” departments of psychiatry. J. Clin. Psychiatry 2010, 71, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Shishko, I.; Almeida, K.; Silvia, R.J.; Tataronis, G.R. Psychiatric pharmacists’ perception on the use of pharmacogenomic testing in the mental health population. Pharmacogenomics 2015, 16, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.; Steven, P.H.; Catriona, H. Psychiatrist attitudes towards pharmacogenetic testing, direct-to-consumer genetic testing, and integrating genetic counseling into psychiatric patient care. Psychiatry Res. 2015, 226, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.Y.; Chua, B.Y.; Subramaniam, M.; Suen, E.L.; Lee, J. Clinicians’ perceptions of pharmacogenomics use in psychiatry. Pharmacogenomics 2017, 18, 531–538. [Google Scholar] [CrossRef] [Green Version]
- Gregornik, D.; Salyakina, D.; Brown, M.; Roiko, S.; Ramos, K. Pediatric pharmacogenomics: Challenges and opportunities: On behalf of the Sanford Children’s Genomic Medicine Consortium. Pharm. J. 2021, 21, 8–19. [Google Scholar] [CrossRef]
- Eichmeyer, J.; Rogers, S.; Formea, C.M.; Giri, J.; Jones, J.S.; Schnettler, E.; Schmidlen, T.; Glogowski, E.; Kurz, R.N. Parc Report: A Perspective on the State of Clinical Pharmacogenomics Testing. Pharmacogenomics 2020, 21, 809–820. [Google Scholar] [CrossRef]
- Empey, P.E.; Pratt, V.M.; Hoffman, J.M.; Caudle, K.E.; Klein, T.E. Expanding Evidence Leads to New Pharmacogenomics Payer Coverage. Genet. Med. 2021, 23, 830–832. [Google Scholar] [CrossRef] [PubMed]
- United Healthcare. Pharmacogenetic Testing. Available online: https://www.uhcprovider.com/content/dam/provider/docs/public/policies/medicaid-comm-plan/pharmacogenetic-testing-cs.pdf (accessed on 1 July 2021).
- Tanner, J.A.; Davies, P.E.; Overall, C.C.; Grima, D.; Nam, J.; Dechairo, B.M. Cost-Effectiveness of Combinatorial Pharmacogenomic Testing for Depression from the Canadian Public Payer Perspective. Pharmacogenomics 2020, 21, 521–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitropoulou, C.; Litinski, V.; Kabakchiev, B.; Rogers, S.; Patrinos, G. Parc Report: Health Outcomes and Value of Personalized Medicine Interventions: Impact on Patient Care. Pharmacogenomics 2020, 21, 797–807. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).