Microbial Contamination in Commercial Honey: Insights for Food Safety and Quality Control
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Characterization and Collection
2.2. Characterization of Honey
2.2.1. Physicochemical Analyses
2.2.2. Microbiological Analysis
2.2.3. Amplicon Sequencing and 16S Analysis
2.3. Pangenomic Analysis
2.3.1. Comparative Analysis of Genomes: Phylogenetic Analysis
2.3.2. Complete Reference Genomes for In Silico Analysis and Research of Virulence and Antimicrobial Resistance Genes
3. Results and Discussion
3.1. Physicochemical Profile of Honey
3.2. Microbial Count
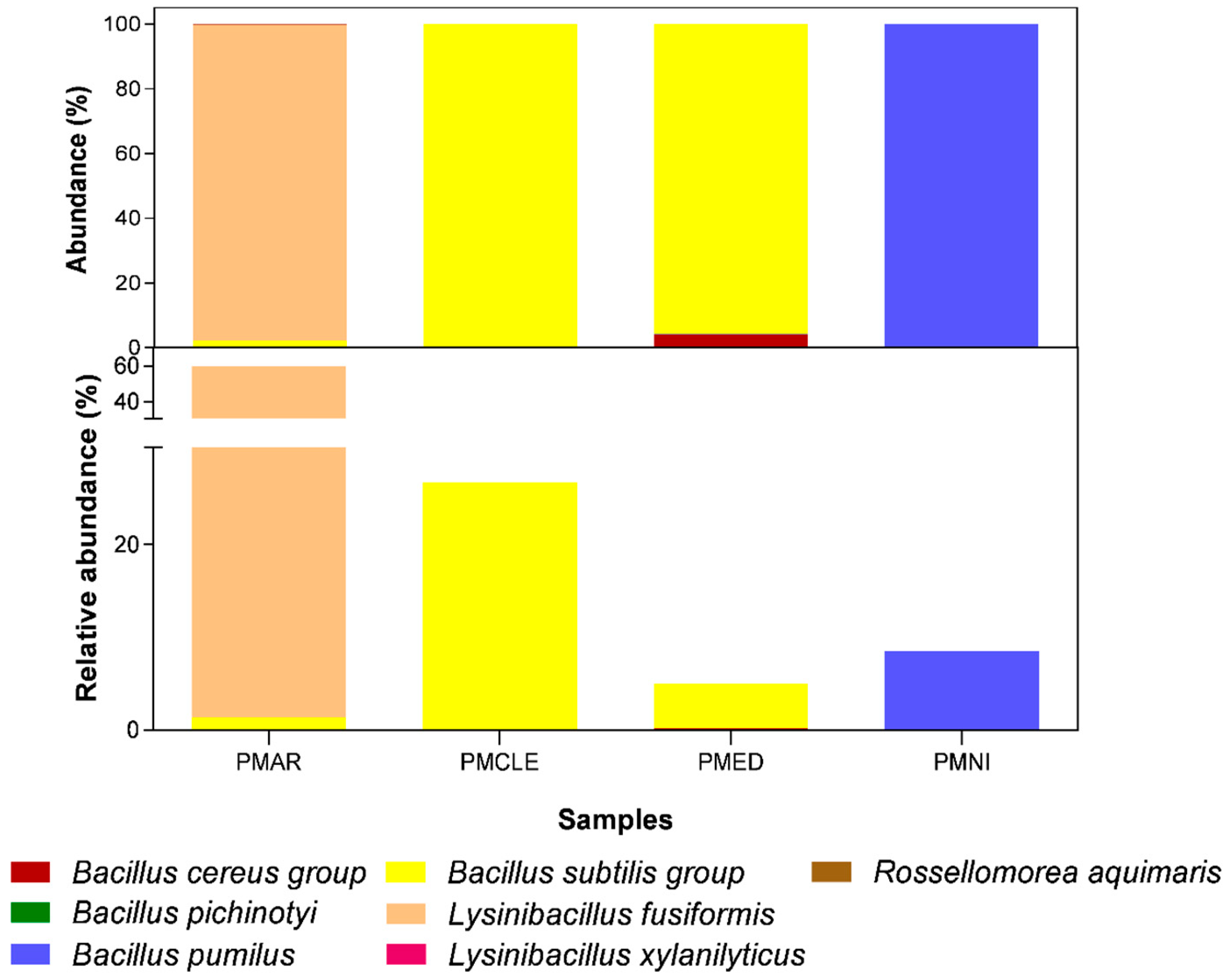
3.3. Bacterial Population Profile in Honey
3.4. Search for Resistance and Virulence Genes in Reference Genomes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO; WHO. Revised Codex Standard for Honey. In Codex Alimentarius; FAO: Rome, Italy, 2001. [Google Scholar]
- Silva, M.S.; Rabadzhiev, Y.; Eller, M.R.; Iliev, I.; Ivanova, I.; Santana, W.C. Microorganisms in Honey. Honey Anal. 2017, 500, 233–257. [Google Scholar] [CrossRef]
- Escuredo, O.; Dobre, I.; Fernández-González, M.; Seijo, M.C. Contribution of Botanical Origin and Sugar Composition of Honeys on the Crystallization Phenomenon. Food Chem. 2014, 149, 84–90. [Google Scholar] [CrossRef] [PubMed]
- FAO. FAOSTAT; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2025. [Google Scholar]
- Brasil Tabela 74: Produção de Origem Animal, Por Tipo de Produto. Available online: https://sidra.ibge.gov.br/tabela/74 (accessed on 15 January 2025).
- Wolff, L.F. Apicultura Sustentável Na Propriedade Familiar de Base Ecológica. In Embrapa Clima Temperado. Circular Técnica; Embrapa Clima Temperado: Pelotas, Brazil, 2007; Volume 64. [Google Scholar]
- Hung, K.-L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The Worldwide Importance of Honey Bees as Pollinators in Natural Habitats. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172140. [Google Scholar] [CrossRef] [PubMed]
- Paxton, R. Conserving Wild Bees. Bee World 1995, 76, 53–55. [Google Scholar] [CrossRef]
- Guimarães, N.P. Apicultura, a Ciência Da Longa Vida; Itatiaia Ltda: Belo Horizonte, Brazil, 1989. [Google Scholar]
- de Freitas Barbosa, W.; Khan, A.; Silva, L. Technological Level and Profitability Honey Production from Bees (Apis mellifera) in Ceará. Rev. De Econ. E Sociologia Rural 2004, 42, 171–188. [Google Scholar] [CrossRef]
- Barbosa, W.d.F.; de Sousa, E.P. Nível Tecnológico e Seus Determinantes Na Apicultura Cearense. Rev. De Política Agrícola 2013, 22, 32–47. [Google Scholar]
- Brasil Agência Nacional de Vigilância Sanitária (ANVISA). Instrução Normativa No 160, de 1 de Julho de 2022. Estabelece Os Limites Máximos Tolerados (LMT) de Contaminantes Em Alimentos; Brasil Agência Nacional de Vigilância Sanitária (ANVISA): Brasília, Brazil, 2022.
- Brasil. Ministério da Agricultura e Pecuária. Mapa do Leite. Available online: https://www.gov.br/agricultura/pt-br/assuntos/producao-animal/mapa-do-leite/mapa-do-leite (accessed on 1 November 2023).
- Brasil Ministério da Agricultura, Pecuária e Abastecimento (MAPA). Instrução Normativa n.° 11, de 20 de Outubro de 2000—Regulamento Técnico de Identidade e Qualidade do Mel; Brasil Ministério da Agricultura, Pecuária e Abastecimento (MAPA): Brasília, Brazil, 2000.
- Lutz, I.A. Métodos Físico-Químicos Para Análise de Alimentos; ANVISA: São Paulo, Brazil, 2008; p. 1020. [Google Scholar]
- Souza, P.; Pacheco, L.; Souza, L.; Silva Filho, M.; Rosário, A.; Moura Júnior, J.; Tavares, Z.; Negrão, C.; Souza, E.; Silva, A. Ciência e Tecnologia de Alimentos: Pesquisa e Práticas Contemporâneas; Científica Digital: São Paulo, Brazil, 2021; pp. 469–476. [Google Scholar]
- White, J.W., Jr.; Subers, M.H.; Schepartz, A.I. Identification of Inhibin, the Antibacterial Factor in Honey, as Hydrogen Peroxide and Its Origin in The Glucose Oxidase System of Honey. Acta Enzym. 1963, 75, 57–70. [Google Scholar]
- Malika, N.; Mohamed, F.; Chakib, E.A. Antimicrobial Activities of Natural Honey from Aromatic and Medicinal Plants on Antibio-Resistant Strains of Bacteria. Int. J. Agric. Biol. 2004, 6, 289–293. [Google Scholar]
- Wen, Y.; Wang, L.; Jin, Y.; Zhang, J.; Su, L.; Zhang, X.; Zhou, J.; Li, Y. The Microbial Community Dynamics During the Vitex Honey Ripening Process in the Honeycomb. Front. Microbiol. 2017, 8, 1649. [Google Scholar] [CrossRef]
- Roxo, I.; Amaral, A.; Portugal, A.; Trovão, J. A Preliminary Metabarcoding Analysis of Portuguese Raw Honeys. Arch. Microbiol. 2023, 205, 386. [Google Scholar] [CrossRef]
- Luca, L.; Pauliuc, D.; Oroian, M. Honey Microbiota, Methods for Determining the Microbiological Composition and the Antimicrobial Effect of Honey—A Review. Food Chem. X 2024, 23, 101524. [Google Scholar] [CrossRef]
- Sinacori, M.; Francesca, N.; Alfonzo, A.; Cruciata, M.; Sannino, C.; Settanni, L.; Moschetti, G. Cultivable Microorganisms Associated with Honeys of Different Geographical and Botanical Origin. Food Microbiol. 2014, 38, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Alippi, A.M.; Abrahamovich, E. HiCrome Bacillus Agar for Presumptive Identification of Bacillus and Related Species Isolated from Honey Samples. Int. J. Food Microbiol. 2019, 305, 108245. [Google Scholar] [CrossRef] [PubMed]
- López, A.C.; Alippi, A.M. Feasibility of Using RFLP of PCR-Amplified 16S rRNA Gene(s) for Rapid Differentiation of Isolates of Aerobic Spore-Forming Bacteria from Honey. J. Microbiol. Methods 2019, 165, 105690. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.R.; Sogin, J.H.; Worobo, R.W. Microbiome Analysis of Raw Honey Reveals Important Factors Influencing the Bacterial and Fungal Communities. Front. Microbiol. 2023, 13, 1099522. [Google Scholar] [CrossRef]
- Mercosul. GMC/RES—Regulamento Técnico MERCOSUL de Identidade e Qualidade do Mel; Mercosul: Montevideo, Uruguay, 1994. [Google Scholar]
- Bo, J.; Qiongqiong, L.; Zhihua, Z.; Jiali, D.; Feng, Q.; Meicheng, Y. Analysis of Contaminated Microorganisms in Commercial Honey. Shanghai J. Prev. Med. 2022, 34, 77–80. [Google Scholar]
- Governo do Estado do Rio Grande do Sul Geografia. Available online: https://estado.rs.gov.br/geografia (accessed on 27 May 2025).
- ISO 6611:2004; Milk and Milk Products—Enumeration of Colony-Forming Units of Yeasts and/or Moulds—Colony-Count Technique at 25 °C. International Organization for Standardization (ISO): Geneva, Switzerland, 2004.
- Salfinger, Y.; Tortorello, M.L. (Eds.) Compendium of Methods for the Microbiological Examination of Foods; American Public Health Association: Washington, DC, USA, 2015. [Google Scholar]
- ISO 4831:2006; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Detection and Enumeration of Coliforms—Most Probable Number Technique. International Organization for Standardization (ISO): Geneva, Switzerland, 2006.
- AOAC International. AOAC Official Method 2011.03—Salmonella in Variety of Food. VIDAS® Salmonella; AOAC International: Rockville, MD, USA, 2011. [Google Scholar]
- ISO 6579-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella Spp. International Organization for Standardization (ISO): Geneva, Switzerland, 2020.
- Association Française de Normalisation (AFNOR). VIDAS Listeria Monocytogenes II (VIDAS LM02). Ref. 30704 Enrichment Stage at 37 °C, 12/11–03/04; AFNOR: La Plaine Saint-Denis, France, 2010. [Google Scholar]
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method. International Organization for Standardization (ISO): Geneva, Switzerland, 2017.
- ISO 16649-3:2015; Microbiology of the Food Chain—Horizontal Method for the Enumeration of Beta-Glucuronidase Positive Escherichia coli—Part 3: Detection and Most Probable Number Technique Using 5-Bromo-4-Chloro-3-Indolyl-β-D-Glucuronide. ISO: Geneva, Switzerland, 2025.
- ISO 6888-1:2021; Microbiology of the Food Chain—Horizontal Method for Enumerating Coagulase-Positive Staphylococci (Staphylococcus aureus and Other Species)—Part 1: Method Using Baird-Parker Agar Medium. International Organization for Standardization (ISO): Geneva, Switzerland, 2021.
- Erhardt, M.M.; de Castro Oliveira, W.; Fröder, H.; Marques, P.H.; Oliveira, M.B.P.P.; dos Santos Richards, N.S.P. Lactic Bacteria in Artisanal Cheese: Characterization through Metagenomics. Fermentation 2023, 9, 41. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Wang, Y.; Qian, P.-Y. Conservative Fragments in Bacterial 16S rRNA Genes and Primer Design for 16S Ribosomal DNA Amplicons in Metagenomic Studies. PLoS ONE 2009, 4, e7401. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Pedroso, M.A.P.; de Castro Oliveira, W.; Felice, A.G.; Cunha, S.C.S.; Oliveira, M.B.P.P.; dos Santos Richards, N.S.P. Pathogenic and Harmful Bacteria in Dairy Technology: Genomic Characterization and Its Correlation with Physicochemical Parameters of Sheep’s Cheese Sold in Southern Brazil. Fermentation 2024, 10, 397. [Google Scholar] [CrossRef]
- Rodrigues, D.L.N.; Ariute, J.C.; Rodrigues da Costa, F.M.; Benko-Iseppon, A.M.; Barh, D.; Azevedo, V.; Aburjaile, F. PanViTa: Pan Virulence and resistance Analysis. Front. Bioinform. 2023, 3, 1070406. [Google Scholar] [CrossRef] [PubMed]
- Bertoldi, F.C.; dos Reis, V.D.A.; Gonzaga, L.V.; Congro, C.R. Caracterização Físico-Química E Sensorial de Amostras de Mel de Abelhas Africanizadas (Apis mellifera L.) produzidas no pantanal. Evidencia 2007, 7, 63–74. [Google Scholar]
- Vieira, T.R.; Noguez, C.S.; dos Santos, M.A.; Wagner, S.A. Caracterização Físico-Química e Botânica Do Mel de Abelhas Sem Ferrão (Meliponini), de Ocorrência No Vale Do Taquari–RS, Objetivando Edição de RTIQ. Res. Soc. Dev. 2023, 12, e29312340846. [Google Scholar] [CrossRef]
- da Cruz Antonio, J.; Tiecher, A. Caracterização Físico-Química de Méis Produzidos no Município de Itaqui—RS. In Proceedings of the 5th Simpósio de Segurança Alimentar: Alimentação e Saúde, Bento Gonçalves, Brazil, 26–29 May 2015; pp. 1–4. [Google Scholar]
- Brasil Ministério da Agricultura, Pecuária e Abastecimento (MAPA). Portaria n.° 795, de 10 de Maio de 2023—Define as Normas Higiênico Sanitárias e Tecnológicas Para os Estabelecimentos que Elaborem Produtos de Abelhas e Seus Derivados; Brasil Ministério da Agricultura, Pecuária e Abastecimento (MAPA): Brasília, Brazil, 2023.
- Cecchi, H.M. Fundamentos Teóricos e Práticos em Análise de Alimentos; Editora da UNICAMP: Campinas, Brazil, 2003; Volume 2, ISBN 85-268-1472-9. [Google Scholar]
- Silva, P.A.M. Qualidade dos Produtos da Abelha. In Seminário Nordestino Pecuário, 7th ed.; Federação da Agricultura e Pecuária do Estado do Ceará: Fortaleza, Brazil, 2003; pp. 7–8. [Google Scholar]
- Venturini, K.S.; Sarcinelli, M.F.; da Silva, L.C. Características do Mel. In Boletim Técnico da Universidade Federal do Espírito Santo; UFES: Vitória, Brazil, 2007. [Google Scholar]
- de Lima da Silva, B. Diagnóstico do Sistema de Producão e Qualidade do Mel de Apis mellifera. Master’s Thesis, Universidade Federal de Viçosa, Viçosa, Brazil, 26 February 2007. [Google Scholar]
- Albuquerque, J.C.G.; Sobrinho, M.E.; de Lima Lins, T.C. Análise da Qualidade do Mel de Abelha Comercializado Com e Sem Inspeção na Região de Brasília-DF, Brasil. Semin. Ciênc. Biól. Saúde 2021, 42, 71–80. [Google Scholar] [CrossRef]
- Gois, G.C.; Lima, C.A.B.; Silva, L.T.; Evangelista-Rodrigues, A. Composição do Mel de Apis mellifera: Requisitos de qualidade. Acta Vet. Bras. 2013, 7, 137–147. [Google Scholar]
- İstanbullugil, F.R.; Taş, N.; Acaröz, U.; Arslan-acaroz, D.; Çakmak, Ö.; Evrenkaya, S.; Gürler, Z. A Review on the Antimicrobial Effect of Honey on Salmonella and Listeria monocytogenes: Recent Studies. Manas J. Agric. Vet. Life Sci. 2023, 13, 210–225. [Google Scholar] [CrossRef]
- Almasaudi, S.B.; Al-Nahari, A.A.M.; Abd El-Ghany, E.S.M.; Barbour, E.; Al Muhayawi, S.M.; Al-Jaouni, S.; Azhar, E.; Qari, M.; Qari, Y.A.; Harakeh, S. Antimicrobial Effect of Different Types of Honey on Staphylococcus aureus. Saudi J. Biol. Sci. 2017, 24, 1255–1261. [Google Scholar] [CrossRef]
- Al-Ghamdi, S.; Alsulami, T.; Alshamri, G.; Ahmed, M.; Ibrahim, M.; Husain, F.M.; Alameen, A.A.; Fulleros, R.; Elamin, W. Physicochemical Properties of Honey and Honey-Like Foods and Their Impact on E. coli Survival: A comparative study. Food Chem. Adv. 2025, 6, 100891. [Google Scholar] [CrossRef]
- Vázquez-Quiñones, C.R.; Moreno-Terrazas, R.; Natividad-Bonifacio, I.; Quiñones-Ramírez, E.I.; Vázquez-Salinas, C. Microbiological Assessment of Honey in México. Rev. Argent. Microbiol. 2018, 50, 75–80. [Google Scholar] [CrossRef]
- Iurlina, M.O.; Fritz, R. Characterization of Microorganisms in Argentinean Honeys from Different Sources. Int. J. Food Microbiol. 2005, 105, 297–304. [Google Scholar] [CrossRef]
- Pereira, A.P.R. Caracterização de Mel Com Vista à Produção de Hidromel. Master’s Thesis, Instituto Politécnico de Bragança, Escola Superior Agrária de Bragança, Bragança, Portugal, 2008. [Google Scholar]
- Pereira, I.A.; Teixeira, F.M.; Finger, R.M.; de Azevedo Souza, M. Microbiological Quality of Honey Market in North-Centre of Rio de Janeiro. Rev. Cienc. Agrovet. 2024, 23. [Google Scholar] [CrossRef]
- Oliveira, E.G.; Nascimento, A.R.; Costa, M.C.P.; Monteiro Neto, V. Qualidade microbiológica do mel de tiúba (Melipona Compressipes Fasciculata) Produzido no ESTADO do Maranhão. Hig. Aliment. 2005, 92–99. [Google Scholar]
- Balzan, S.; Carraro, L.; Merlanti, R.; Lucatello, L.; Capolongo, F.; Fontana, F.; Novelli, E.; Larini, I.; Vitulo, N.; Cardazzo, B. Microbial Metabarcoding Highlights Different Bacterial and Fungal Populations in Honey Samples from Local Beekeepers and Market in North-Eastern Italy. Int. J. Food Microbiol. 2020, 334, 108806. [Google Scholar] [CrossRef]
- Bovo, S.; Utzeri, V.J.; Ribani, A.; Cabbri, R.; Fontanesi, L. Shotgun Sequencing of Honey DNA Can Describe Honey Bee Derived Environmental Signatures and the Honey Bee Hologenome Complexity. Sci. Rep. 2020, 10, 9279. [Google Scholar] [CrossRef]
- Jacinto-Castillo, D.F.; Canto, A.; Medina-Medina, L.A.; O’Connor-Sánchez, A. Living in Honey: Bacterial and Fungal Communities in Honey of Sympatric Populations of Apis mellifera and the Stingless Bee Melipona beecheii, in Yucatan, Mexico. Arch. Microbiol. 2022, 204, 718. [Google Scholar] [CrossRef]
- Økstad, O.A.; Kolstø, A.-B. Genomics of Bacillus Species. In Genomics of Foodborne Bacterial Pathogens; Wiedmann, M., Zhang, W., Eds.; Springer: New York, NY, 2011; pp. 29–53. ISBN 978-1-4419-7686-4. [Google Scholar]
- Jiménez, G.; Urdiain, M.; Cifuentes, A.; López-López, A.; Blanch, A.R.; Tamames, J.; Kämpfer, P.; Kolstø, A.-B.; Ramón, D.; Martínez, J.F.; et al. Description of Bacillus toyonensis sp. nov., a Novel Species of the Bacillus cereus group, and Pairwise Genome Comparisons of the Species of the Group by Means of ANI Calculations. Syst. Appl. Microbiol. 2013, 36, 383–391. [Google Scholar] [CrossRef]
- Østensvik, Ø.; From, C.; Heidenreich, B.; O’sullivan, K.; Granum, P. Cytotoxic Bacillus Spp. Belonging to the B. Cereus and B. Subtilis Groups in Norwegian Surface Waters. J. Appl. Microbiol. 2004, 96, 987–993. [Google Scholar] [CrossRef]
- From, C.; Hormazabal, V.; Granum, P.E. Food Poisoning Associated with Pumilacidin-Producing Bacillus pumilus in Rice. Int. J. Food Microbiol. 2007, 115, 319–324. [Google Scholar] [CrossRef]
- Jay, J.M.; Loessner, M.J.; Golden, D.A. (Eds.) Food Poisoning Caused by Gram-Positive Sporeforming Bacteria. In Modern Food Microbiology; Springer: Boston, MA, USA, 2005; pp. 567–590. ISBN 978-0-387-23413-7. [Google Scholar]
- Bovo, S.; Ribani, A.; Utzeri, V.J.; Schiavo, G.; Bertolini, F.; Fontanesi, L. Shotgun Metagenomics of Honey DNA: Evaluation of a Methodological Approach to Describe a Multi-Kingdom Honey Bee Derived Environmental DNA Signature. PLoS ONE 2018, 13, e0205575. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, Z.; Han, Y.; Wang, Z.; Fan, J.; Xiao, H. Isolation and Identification of Antifungal Peptides from Bacillus BH072, a Novel Bacterium Isolated from Honey. Microbiol. Res. 2013, 168, 598–606. [Google Scholar] [CrossRef]
- Lopez, D.; Vlamakis, H.; Kolter, R. Generation of Multiple Cell Types in Bacillus subtilis. FEMS Microbiol. Rev. 2008, 33, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Kovács, Á.T. Bacillus subtilis . Trends Microbiol. 2019, 27, 724–725. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust Demarcation of 17 Distinct Bacillus species Clades, Proposed as Novel Bacillaceae genera, by Phylogenomics and Comparative Genomic Analyses: Description of Robertmurraya kyonggiensis sp. nov. and Proposal for an Emended Genus Bacillus Limiting it Only to the Members of the Subtilis and Cereus Clades of Species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar] [CrossRef]
- Ahmed, I.; Yokota, A.; Yamazoe, A.; Fujiwara, T. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and Transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 1117–1125. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Kramer, J.M.; Gilbert, R.J. Bacillus cereus and other Bacillus species. Foodborne Bact. Pathog. 1989, 19, 21–70. [Google Scholar]


| Sample | Acidity (mEq/kg) | pH | Humidity (g/100 g) | Ashes (g/100 g) | Water-Insoluble Solids (g/100 g) | Brix (°B) |
|---|---|---|---|---|---|---|
| PMOR | 31.45 ± 0.1 | 4.16 ± 0.0 | 16.73 ± 0.4 | 0.20 ± 0.0 | 0.12 ± 0.0 | 80.0 ± 0.0 |
| PMSA | 28.85 ± 0.0 | 4.28 ± 0.0 | 15.23 ± 0.2 | 0.09 ± 0.0 | 0.06 ± 0.0 | 78.0 ± 0.0 |
| PMED | 29.83 ± 0.0 | 4.11 ± 0.0 | 17.26 ± 0.7 | 0.14 ± 0.0 | 0.11 ± 0.0 | 76.0 ± 0.0 |
| PMCLE | 26.55 ± 0.0 | 4.22 ± 0.0 | 16.69 ± 0.1 | 0.24 ± 0.0 | 0.08 ± 0.0 | 77.0 ± 0.0 |
| PMDE | 33.47 ± 0.0 | 4.11 ± 0.0 | 17.88 ± 0.5 | 0.57 ± 0.0 | 0.08 ± 0.0 | 79.0 ± 0.0 |
| PMNI | 31.15 ± 0.0 | 4.16 ± 0.0 | 16.90 ± 0.2 | 0.15 ± 0.0 | 0.06 ± 0.0 | 76.0 ± 0.0 |
| PMVA | 24.90 ± 0.0 | 4.55 ± 0.0 | 19.22 ± 0.3 | 0.07 ± 0.0 | 0.08 ± 0.0 | 74.0 ± 0.0 |
| PMCL | 26.16 ± 0.0 | 4.28 ± 0.0 | 17.77 ± 0.4 | 0.16 ± 0.0 | 0.05 ± 0.0 | 77.0 ± 0.0 |
| PMBE | 29.32 ± 0.1 | 4.41 ± 0.0 | 18.43 ± 0.4 | 0.35 ± 0.0 | 0.07 ± 0.0 | 76.0 ± 0.0 |
| PMRO | 25.48 ± 0.0 | 4.21 ± 0.0 | 18.45 ± 0.6 | 0.28 ± 0.0 | 0.08 ± 0.0 | 77.3 ± 0.0 |
| PMAR | 26.54 ± 0.0 | 4.39 ± 0.0 | 18.90 ± 0.3 | 0.40 ± 0.0 | 0.06 ± 0.0 | 74.0 ± 0.4 |
| PMRI | 22.44 ± 0.0 | 4.41 ± 0.0 | 15.72 ± 0.7 | 0.33 ± 0.0 | 0.10 ± 0.0 | 80.0 ± 0.0 |
| Standard (Brasil, 2000) | Max. 50 | - | Max. 20 | Max. 0.6 | Max. 0.6 | - |
| Sample | Salmonella spp. (Absence/ Presence) | Listeria (Absence/ Presence) | E. coli (MPN/g) | Molds and Yeasts (CFU/g) | Total Coliforms (MPN/g) | Thermotolerant Coliforms (MPN/g) | S. coag. Positive (CFU/g) |
|---|---|---|---|---|---|---|---|
| PMOR | Absence | Absence | 0 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMSA | Absence | Absence | 0 | 33.3 ± 15.6 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMED | Absence | Absence | 0 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMCLE | Absence | Absence | 0 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMDE | Absence | Absence | 0 | 263.3 ± 31.1 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMNI | Absence | Absence | 0 | 630.0 ± 13.3 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMVA | Absence | Absence | 0 | 96.7 ± 15.6 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMCL | Absence | Absence | 0 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMBE | Absence | Absence | 0 | 43.3 ± 17.8 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMRO | Absence | Absence | 0 | 85.7 ± 2.9 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMAR | Absence | Absence | 0 | 26.3 ± 1.1 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| PMRI | Absence | Absence | 0 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 | <1.0 × 101 |
| Phylum | Class | Order | Family | Genus | Species | Abundance Relative | Sample |
|---|---|---|---|---|---|---|---|
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus | Bacillus cereus group | 0.20% | PMED |
| Bacilluspichinotyi | 0.06% | PMAR | |||||
| Bacillus pumilus | 8.54% | PMED, PMAR, PMNI | |||||
| Bacillus subtilis group | 32.73% | PMED, PMAR, PMCLE | |||||
| Lysinibacillus | Lysinibacillusfusiformis | 58.33% | PMAR | ||||
| Lysinibacillus xylanilyticus | 0.08% | PMAR | |||||
| Rossellomorea | Rossellomorea aquimaris | 0.06% | PMAR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruxel, F.; Geller, A.M.; Felice, A.G.; Ströher, J.A.; Freitas, A.S.d.; Balen, A.; Oliveira, M.B.P.P.; Oliveira, W.d.C. Microbial Contamination in Commercial Honey: Insights for Food Safety and Quality Control. Microbiol. Res. 2025, 16, 128. https://doi.org/10.3390/microbiolres16060128
Bruxel F, Geller AM, Felice AG, Ströher JA, Freitas ASd, Balen A, Oliveira MBPP, Oliveira WdC. Microbial Contamination in Commercial Honey: Insights for Food Safety and Quality Control. Microbiology Research. 2025; 16(6):128. https://doi.org/10.3390/microbiolres16060128
Chicago/Turabian StyleBruxel, Felipe, Ana Maria Geller, Andrei Giacchetto Felice, Jeferson Aloísio Ströher, Anderson Santos de Freitas, Angela Balen, Maria Beatriz Prior Pinto Oliveira, and Wemerson de Castro Oliveira. 2025. "Microbial Contamination in Commercial Honey: Insights for Food Safety and Quality Control" Microbiology Research 16, no. 6: 128. https://doi.org/10.3390/microbiolres16060128
APA StyleBruxel, F., Geller, A. M., Felice, A. G., Ströher, J. A., Freitas, A. S. d., Balen, A., Oliveira, M. B. P. P., & Oliveira, W. d. C. (2025). Microbial Contamination in Commercial Honey: Insights for Food Safety and Quality Control. Microbiology Research, 16(6), 128. https://doi.org/10.3390/microbiolres16060128










