Abstract
Filamentous fungi hold critical industrial value for their ability to produce enzymes, antibiotics, organic acids, and food fermentation. GATA transcription factors (TFs) serve as central regulators of nitrogen metabolism, synthesis of secondary metabolites, stress adaptation, and directly influence fungal development and pathogenicity in filamentous fungi. In this review, we primarily discuss the structural characterization, different types, and phylogenetic analysis of filamentous fungi GATA TFs in filamentous fungi. Subsequently, we systematically summarize the multifunctions of GATA TFs in the mycelial growth, morphological differentiation, and conidial development of filamentous fungi. In addition, we explore their functions in the synthesis of secondary metabolites such as antibiotics (e.g., cephalosporins, penicillins) and organic acids (e.g., ganoderic acid, fumaric acid) in filamentous fungi. Furthermore, we focus on the key roles of GATA TFs AreA and AreB in nitrogen and carbon metabolism in filamentous fungi and their potential synergistic regulatory relationships. Finally, we review the important roles of GATA TFs in the adaptation of filamentous fungi to environmental changes. This review provides research ideas for the development of genetically engineered strains with optimized growth characteristics, increased target metabolites in the fermentation production process, and enhanced environmental adaptability.
1. Introduction
Filamentous fungi, commonly known as molds, are usually capable of forming branched and luxuriant mycelium and are widely distributed in soil, water, air, and the internal and external environments of plants and animals [1]. Filamentous fungi play an important role in nature, mainly by degrading organic wastes to promote ecological cycles, secreting organic acids and anti-microbial compounds, promoting plant growth, and inhibiting the reproduction of pathogenic microorganisms [2]. Filamentous fungi are primarily classified into Ascomycota, Basidiomycota, Mucoromycota, and Chytridiomycota. Common examples of filamentous fungi include Aspergillus spp., Penicillium spp., Fusarium spp., and Mucor spp. [3]. Among them, Aspergillus spp. is widely used in industrial fermentation and enzyme production. For example, Aspergillus oryzae and Aspergillus sojae are widely used for fermentation of soybeans, rice, and wheat, as well as for the production of soy sauce, miso, and sake by hydrolysis of starch and protein [4]. Filamentous fungi can also be used to prepare a variety of enzyme preparations. For example, Aspergillus fumigatus [5], Aspergillus niger [6] and Aspergillus nidulans [7] secrete a variety of cellulose-degrading enzymes and functional enzymes, such as cellulases, hemicellulases, xylanases, glycoside hydrolases, proteases, and peroxidases [8,9]. Some filamentous fungi, such as Aspergillus spp., Fusarium spp., and Penicillium spp., are capable of synthesizing a wide range of secondary metabolites, including organic acids, antibiotics, pigments, and hormones. For example, A. niger [6] and A. nidulans [10] can produce various organic acids, while A. oryzae generates pharmacologically active metabolites such as trichothecenes and aminoclaurine [11]. Penicillin, a broad-spectrum β-lactam antibiotic initially discovered in Penicillium spp., is effective against a variety of infections and enhances therapeutic efficacy when used in combination with other antibiotics [12,13,14]. It can also be synthesized by A. nidulans, whereas cephalosporins are primarily produced by Acremonium chrysogenum [15,16]. Additionally, Penicillium decumbens has been used for the biosynthesis of silver nanoparticles (Ag-NPs), which significantly enhance the antibacterial efficacy of various antibiotics [17]. Certain Aspergillus species, such as A. fumigatus, produce pigments like DHN-melanin and pyomelanin, which help protect the fungus from environmental stressors, including ultraviolet radiation and oxidative stress, while also enhancing its pathogenicity and promoting host infection [18,19]. In terms of plant hormone biosynthesis, Fusarium fujikuroi synthesizes gibberellins (GA), which are crucial for promoting plant growth and inducing flowering in some mutant strains [20,21]. A. flavus is capable of producing growth inhibitors, humic acid, deoxynivalenol (DON), and zearalenone (ZEA) [22]. However, some fungi also produce toxic secondary metabolites, such as aflatoxin B1, a potent carcinogen that can damage liver cells, suppress the immune system, and pose significant health risks to humans and animals [23]. Moreover, some Aspergillus spp. and Mucor spp., like A. oryzae [24], A. niger [25], and Mucor circinelloides [26], belong to the group of oleaginous microorganisms capable of accumulating substantial amounts of lipids and synthesizing saturated fatty acids [27].
The synthesis of secondary metabolites in filamentous fungi is a complex process that involves the regulation of multiple genes. Many studies have shown that GATA TFs play important roles in influencing growth and morphological differentiation, regulating the biosynthesis of secondary metabolites in filamentous fungi, and adapting to different environmental changes. By identifying the GATA TFs, the major GATA TFs including AreA, AreB, SreA, NsdD, LreA, and LreB were found, and their biological functions were revealed in filamentous fungi Aspergillus, Alternaria, and Fusarium. For example, AreA and AreB are key transcription factors involved in the regulation of nitrogen and carbon metabolism and have been identified in various fungi, including A. nidulans [28], A. flavus [22], and A. oryzae [29] from the genus Aspergillus, Alternaria alternata [30] from the genus Alternaria, and F. fujikuroi [31] from the genus Fusarium. GATA TF SreA was identified in Aspergillus spp., including A. nidulans [32,33], A. niger [34], and A. fumigatus [35]. SreA does not directly regulate the synthesis of secondary metabolites but promotes the accumulation of heme intermediates by inhibiting siderophore biosynthesis and enhancing iron uptake [34]. Additionally, GATA TF NsdD was identified in Aspergillus spp. and proved to play a role in growth and conidiophore formation [20,36]. The GATA TFs LreA and LreB, as light-responsive regulators, are present in a variety of Aspergillus spp. fungi, such as A. nidulans [37], A. oryzae [38], and A. fumigatus [39]. Interestingly, LreA and LreB are also involved in the response to temperature and high salt stress as a regulatory complex of the global regulator VeA and resistance to CuSO4 [30,38]. Therefore, this paper focuses on providing a detailed summary of the structural characterizations, types, biological functions, and potential applications of GATA TFs in filamentous fungi for metabolic pathway optimization.
2. Structures and Types of GATA TFs in Filamentous Fungi
2.1. Structural Characterization of Filamentous Fungi GATA TFs
GATA TFs, crucial regulators of gene expression, are ubiquitously present in fungi, plants, and animals. The GATA TFs modulate gene expression by binding to GATA motifs on DNA. They are characterized by one or two highly conserved type IV zinc finger domains (Cys-X2-Cys-X17–20-Cys-X2-Cys) and a DNA-binding domain that recognizes the conserved sequence (A/C/T)-G-A-T-A-(A/G) within the promoter region of target genes [40]. These transcription factors are pivotal in growth and morphological differentiation, biosynthesis of secondary metabolites, and adaptation to environmental changes. Studies have revealed that proteins containing the Cys-X2-Cys-X17-Cys-X2-Cys and Cys-X2-Cys-X18-Cys-X2-Cys motifs represent two forms of type IV zinc finger proteins, termed type IVa and type IVb, respectively [40].
In different species, there are significant differences in the types and numbers of zinc finger structures. In plants, the majority of GATA TFs possess a single zinc finger structural domain with a Cys-X2-Cys-X18-Cys-X2-Cys motif, while a few may have two or more zinc finger loops, some of which may contain up to 20 amino acid residues [41]. In animals, GATA TFs typically consist of two zinc finger domains, each containing the Cys-X2-Cys-X17-Cys-X2-Cys motif, with leucine at the seventh position (alanine in some nematodes), and the physiologically active DNA-binding domain is located at the carboxyl terminus [41]. Conversely, fungal GATA TFs combine characteristics of both plant and animal GATA TFs. They typically feature a single zinc finger domain and are categorized into two main classes based on the number of amino acid residues in the zinc finger loop: animal-type, with a 17-residue structure, and plant-type, with an 18-residue structure. Although less common, zinc finger loops containing 19 to 20 residues also exist in fungi [38,41,42]. In addition to differences in the zinc finger domains, the number of GATA TFs also varies among plants, animals, and fungi. In plants, it ranges from 19 to 87, while animals typically have 6 GATA TFs. In yeast, more than 10 GATA TFs have been identified. In filamentous fungi, the number of GATA TFs ranges from 3 to 20 [38,41,42,43]. To better illustrate this variation, we summarized the number of GATA TFs across different genera of filamentous fungi based on data retrieved from [http://ftfd.snu.ac.kr/tf.php?a=dv_list&id=5] (accessed on 18 March 2025) (see Table 1).

Table 1.
Number of GATA TFs in common filamentous fungi.
2.2. Types of Filamentous Fungal GATA TFs
The GATA family is a class of transcription factors with a conserved type IV zinc finger domain. Current research indicates that GATA TFs in filamentous fungi have been studied most extensively, especially in Aspergillus, Penicillium, Fusarium, Neurospora, Alternaria, Botrytis, and Blastomyces species. Here, to further explore the phylogenetic relationships of GATA TFs in representative species, we performed multiple sequence alignment using MEGA 6.0 and constructed a neighbor-joining (NJ) tree. The robustness of the NJ tree topology was assessed using 1000 bootstrap replications to ensure the reliability of the phylogenetic analysis. The NJ tree grouped the GATA TFs from different genera into six subgroups based on the number of ZnF_GATA domains and the zinc finger motifs within the GATA domain sequences, namely WC1, WC2, NSDD, SRP, ASD4, and NIT2 (see Figure 1).
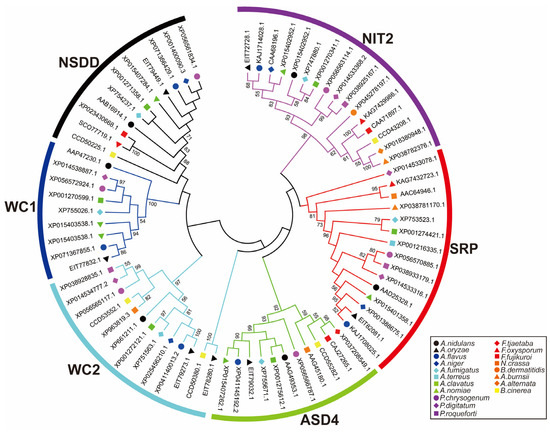
Figure 1.
Phylogenetic analysis of GATA TFs in Aspergillus, Penicillium, Fusarium, Neurospora, Alternaria, Botrytis, and Blastomyces. Comparison of the amino acid sequences of the GATA domains revealed that these GATA TFs can be classified into six subgroups: NIT2, ASD4, SRP, NSDD, WC1, and WC2. The neighbor-joining tree was constructed based on multiple sequence alignment, and the robustness of the tree topology was assessed using 1000 bootstrap replications. Bootstrap support values above 50% are shown at the nodes.
Based on the NJ tree analysis, we have found that the GATA TFs are evolutionarily conserved in Aspergillus, Penicillium, and Fusarium. Therefore, we speculate that the GATA TFs of different species in the same subgroup in the NJ tree might perform the same functions. For example, the NIT2 subgroup primarily regulates nitrogen metabolism, influences hyphal growth and development, and participates in the synthesis of certain secondary metabolites [22,38,44]. The ASD4 subgroup is also involved in the regulation of nitrogen metabolism and is closely associated with organic acid synthesis [45,46,47]. SRP subgroup mainly regulates iron metabolism and is involved in redox reactions and oxidative stress responses [32,33,48]. The NSDD subgroup plays a key role in hyphal growth, development, and spore formation in filamentous fungi [49]. Both WC1 and WC2 are crucial light-sensing transcription factors, responsible for sensing blue light signals and regulating light-responsive reactions, thus influencing fungal growth and development under light conditions [37]. Each subgroup of GATA TFs plays a distinct role in the physiological processes of fungi.
The functional differences between different subgroups are closely related to the differences in their zinc finger structures. For instance, the NIT2 and ASD4 subgroups, involved in nitrogen metabolism regulation, both possess an IVa-type zinc finger structure [38,44,45,46,47,50]. The SRP subgroup, which regulates iron metabolism, contains two IVa-type zinc fingers and is rich in cysteine regions [32,33,48]. The NSDD subgroup, associated with hyphal growth and development, features an IVb-type zinc finger structure, enriched in proline and serine residues [38,47]. Similar to the NSDD subgroup, the WC1 and WC2 subgroups, involved in light response, also contain an IVb-type zinc finger structure [37]. It is noteworthy that in Aspergillus spp., A. oryzae has an additional GATA TF AoSnf5, which is the first time an Aspergillus GATA TF encoding a GATA protein with a 20-residue-spaced Cys-X2-Cys motif has been identified [38].
These factors have been widely identified not only in Aspergillus spp., Penicillium spp., and Fusarium spp., but also in other fungi such as Neurospora crassa, A. alternata, Botrytis cinerea, and Blastomyces dermatitidis. In N. crassa, five GATA TFs have been identified: NIT2, WC-1, WC-2, ASD4, and SRE [51], of which NIT2 is functionally similar to AreA [52], whereas ASD4 shares similarities with the negative regulator AreB [53,54]. Correspondingly, WC-2 contains two type IVb-type zinc finger structural domains and a PAS domain, similar to Aspergillus spp. [55]. Likewise, in A. alternata, researchers identified six GATA TFs, AaAreA, AaAreB, AaLreA, AaLreB, AaNsdD, and AaSreA [30]. The type and number of zinc finger structural domains of AaAreA, AaAreB, AaNsdD, and AaSreA were consistent with those of Aspergillus fungi (particularly similar to AaAreA), whereas AaLreB has one PAS domain in addition to the zinc finger structural domains, and AaLreA contains three PAS domains. In B. cinerea, two GATA TFs have been reported: BcWCL2 and BcLTF1. BcWCL2 is a light-regulated protein associated with circadian rhythm and belongs to the WC-2 group of transcription factors, containing PAS domain(s) [56]. BcLTF1 is homologous to NsdD from the Aspergillus genus [57]. Moreover, in B. dermatitidis, the GATA TF SREB shares structural and functional similarity with SreA from A. nidulans, featuring two typical IVa-type zinc finger domains separated by a cysteine-rich region [58]. To provide a clearer and more detailed presentation of these data, we have summarized the differences in the zinc finger types and numbers of GATA TF subgroups in filamentous fungi mentioned earlier, as shown in Table 2.

Table 2.
Zinc finger structure of GATA transcription factor from filamentous fungi.
Overall, GATA TFs exhibit a high degree of conservation and diversity in filamentous fungi. These factors have been widely identified not only in Aspergillus spp., Penicillium spp., and Fusarium spp., but also in other fungi such as N. crassa and A. alternata. These findings suggest that GATA TFs share similar regulatory functions across different filamentous fungi.
3. Biological Function of GATA TFs in Filamentous Fungi
3.1. The Effect of GATA TFs on the Growth and Development of Filamentous Fungi
In filamentous fungi, GATA TFs are increasingly recognized as key players in developmental processes such as hyphal growth, morphological differentiation, and conidiophore development. Moon et al. demonstrated that the GATA TF NsdD is a key factor in the regulation of conidiophore formation and sexual development in Aspergillus species. Knockdown of NsdD accelerates asexual development, increases spore production, and leads to morphological abnormalities, impaired sexual development, and altered toxin production, while overexpression of NsdD inhibits asexual development and promotes zoospore formation [36]. In addition, NsdD, SfgA, and VosA, as key negative regulators of conidial formation, were found to act at different control points in the developmental-genetic cascade. For example, double knockdown of NsdD and VosA resulted in the rapid formation of large numbers of conidial peduncles in liquid cultures, leading to near-continuous activation of sporulation [36]. NsdD expression is regulated by direct binding to the brlA promoter region, while FlbC and FlbD, key upstream activators of conidial formation, positively regulate brlA activation [59]. Likewise, Szewczyk et al. studied the NsdD gene in A. fumigatus and found that deletion of NsdD prevented the formation of the occlusive capsid and affected mycelial fusion and heterokaryon formation through mating, knockout, and heterokaryon formation experiments [60]. Additionally, the Aspergillus fumigatus NsdD knockout strain exhibited morphological abnormalities when exposed to cell wall disruptors and showed resistance to chitin synthesis inhibitors [60]. Niehaus et al. identified Csm1 as a repressor of conidial formation by functional characterization of Csm1, a homolog of NsdD in F. fujikuroi [49]. Schumacher et al. performed gene deletion and overexpression experiments on BcLTF1 in B. cinerea. The results showed that deletion of BcLTF1 led to conidial overproduction, indicating its inhibitory role in spore production. Additionally, BcLTF1 was found to inhibit mycelial peduncle formation, whereas its overexpression caused the accumulation of undifferentiated aerial mycelium [57]. Furthermore, Shen et al. demonstrated that PaNsdD, the NsdD homolog in P. anserina, plays a crucial role in multiple biological processes, including nutrient growth, senescence, sexual reproduction, stress tolerance, and interspecific antagonism [61]. Fasoyin et al. found that A. flavus strains deficient in AreA exhibited low mycelial density, sparse branching, significantly reduced conidial production, and interference with normal conidiophore formation through AreA knockout experiments [22]. Brenna et al. isolated and characterized the ASD4 gene in N. crassa and showed that ASD4 was not involved in the regulation of nitrogen metabolism but in the process of ascospores and ascospores formation by knockout and backfill experiments [54]. Furthermore, Chen et al. showed that apart from AaLreA, deletion of other AaGATA proteins significantly inhibited mycelial growth and hyphal formation in A. alternata [30]. He et al. used gene silencing to downregulate the Sssre gene in Sclerotinia sclerotiorum, resulting in arrested mycelial growth without nucleus or invasive structure formation [62]. Collectively, these findings amply demonstrate that GATA TFs play diverse regulatory roles in the mycelium growth and sporulation of filamentous fungi.
3.2. Regulatory Roles of GATA TFs in the Secondary Metabolite Biosynthesis of Filamentous Fungi
Filamentous fungi are able to produce many different kinds of secondary metabolites, which are not necessary for maintaining fungal growth, reproduction, and cellular metabolism, but they usually have special biological functions and play an important role in the ecological adaptation process of fungi The secondary metabolites of filamentous fungi, owing to their diverse bioactivities, have significant application value in the fields of medicine, agriculture, and industry. They have been widely developed into antibiotics, anticancer agents, immunosuppressants, and agricultural bioactive compounds [6,10,11,12,13,14,15,16]. Furthermore, some metabolites show great potential for environmental remediation, such as the green synthesis of nanomaterials and the removal of heavy metals. In summary, fungal secondary metabolites are emerging as key resources for advancing biomedicine and sustainable agriculture [17,18,19,20,21]. Therefore, studying the biosynthesis regulation mechanisms of these metabolites, particularly bioactive compounds such as aflatoxin B1, ganoderic acid (GA), fusaric acid, and alcohols, is crucial for enhancing their production.
In the biosynthesis of secondary metabolites in filamentous fungi, GATA TFs serve as pivotal global regulators that orchestrate the activation or repression of secondary metabolic pathways by specifically modulating the transcriptional activity of multiple biosynthetic gene clusters. Aflatoxin B1 is a highly carcinogenic secondary metabolite produced by A. flavus and A. parasiticus, with significant implications for food safety and human health [22]. Studies have shown that GATA TF AreA is a negative regulatory transcription factor for the synthesis of aflatoxin. Aflatoxins are a group of highly carcinogenic mycotoxins produced by A. flavus and related fungi, with potent carcinogenic, mutagenic, and immunosuppressive effects [22]. For example, in A. flavus, knockdown of the AreA gene significantly upregulated the expression of aflK, aflO, aflP, and aflQ genes in the aflatoxin synthesis gene cluster, suggesting that AreA inhibits aflatoxin synthesis [22]. Studies have also found that knocking out the SreA gene significantly inhibits aflatoxin production in A. flavus, with approximately a 70% reduction in toxin levels in the mutant strain. This indicates that SreA also plays a crucial role in aflatoxin biosynthesis in A. flavus [63]. In addition, AreA also plays an important regulatory role in the synthesis of bioactive substances such as organic acids and phenols in filamentous fungi. For example, knockdown of the AcAreA in Acemmonium chrysogenum reduced cephalosporin synthesis but did not affect fungal growth [64]. In F. fujikuroi, AreA acts as a positive regulator to promote GA synthesis, and its gene deletion significantly reduces GA production [65,66]. In A. alternata, AaAreA can directly bind to the aohR promoter sequence to activate transcription, thereby promoting the biosynthesis of alternariol (AOH) and alternariol monomethyl ether (AME) [63]. In Ganoderma lucidum, the ΔAreA mutant strain synthesized significantly more GA than the control strain [67]. Additionally, GATA TF AreB positively regulates the synthesis of fusaric acid [68], as well as the expression of genes in the organic acid gene cluster, particularly under conditions of abundant nitrogen. By sensing changes in nitrogen availability, AreB promotes the expression of nitrogen metabolism-related genes, thereby enhancing the synthesis of organic acids and significantly increasing their yield [53,69]. In A. niger [34] and A. fumigatus [70], SreA is a key regulator of iron metabolism and maintains intracellular iron homeostasis by inhibiting the synthesis of iron carriers under high-iron environments. Meanwhile, Purschwitz et al. found that SreA deletion upregulated heme synthesis genes (e.g., HemA and HemH), indirectly affecting ergosterol and heme synthesis [35]. In Podospora anserina, knockout of the PaNsdD gene inhibits the synthesis of the newly identified secondary metabolite 3-acetyl-4-methylpyrrole in this bacterium [61]. In B. cinerea, deletion of the GATA TF BcWCL2 inhibits the expression of Bcvel1, a regulator of citric acid secretion and tissue acidification, thereby suppressing citric acid secretion [56]. In conclusion, GATA TFs play a crucial role in the biosynthesis of secondary metabolites by regulating the expression of secondary metabolite gene clusters. A deeper understanding of their regulatory mechanisms not only aids in enhancing the production of secondary metabolites but also provides theoretical support for the development of genetically engineered strains with increased yields.
3.3. The Regulatory Roles of GATA TFs in Nitrogen Metabolism and Lipid Synthesis of Filamentous Fungi
Lipids are essential components of cell membranes and play crucial roles in energy storage and signal transduction [71]. Filamentous fungi, due to their rapid growth and metabolic flexibility, have become efficient platforms for lipid production [72]. The type and concentration of nitrogen sources are key factors in regulating lipid biosynthesis. As central regulators of nitrogen metabolism, GATA TFs sense changes in external nitrogen availability, modulating related metabolic pathways and consequently influencing lipid synthesis and accumulation. Nitrogen is a critical component of cellular structure and function, and both eukaryotic and prokaryotic organisms employ fine regulatory mechanisms to ensure stable nitrogen supply, thereby affecting the synthesis of secondary metabolites [73]. Sun et al. found that using sodium nitrate and ammonium molybdate as a mixed nitrogen source in the culture medium significantly increased the production of sphingolipids in oleaginous yeasts through the preparation of sphingolipids from olive oil [74]. Furthermore, Dzurendova et al. increased the biomass, lipid, and fatty acid content of oleaginous Mucoromycota fungi using yeast extract as a nitrogen source and under low phosphorus conditions [75]. Aguilar et al. found that the Ustilago maydis accumulated a large number of triacylglycerol-containing lipid droplets in the absence of nitrate and urea as nitrogen sources, and that the fatty acids in the droplets were palmitic acid, palmitic acid, and urea [76]. The fatty acids abundant in them were palmitic, linoleic, and oleic acids [76]. Microalgae, though not classified as fungi, are microorganisms capable of photosynthesis. Maltsev et al. analyzed the effects of nitrogen and phosphorus concentrations on lipid production in microalgae and found that appropriate nitrogen limitation increased lipid production and promoted microalgal growth, whereas nitrogen enrichment was detrimental to lipid accumulation [77]. Many of their studies suggest that regulating nitrogen metabolism effectively promotes the synthesis and accumulation of intracellular lipids in filamentous fungi. Interestingly, the utilization of nitrogen sources, such as nitrate and urea, and intracellular nitrogen metabolism in filamentous fungi are highly regulated by GATA TFs. For example, in oleaginous yeast, the GATA TF SpGAT1 significantly increased lipid production at low C/N ratios, whereas the absence of SpGAT1 led to a decrease in lipid production at high C/N ratios in the medium. SpGAT1 promotes sterol ester (SE) accumulation by regulating the glyoxylate cycle genes (ICL1, ICL2) and the pyruvate bypass gene (ACS) and by binding to the ACAT2 promoter under low C/N ratio conditions [78]. Similarly, in Yarrowia lipolytica, knockdown of the GATA TFs gzf3 and gzf2 significantly increased lipid accumulation. The expression of enzymes related to malate metabolism, β-oxidation, and ammonia utilization were significantly upregulated in the Δgzf3, Δgzf2, and Δgzf1 strains as compared to the wild-type strain [79]. This shows that GATA TFs regulate the uptake and utilization of nitrogen sources by recognizing and activating genes related to nitrogen metabolism, thereby affecting intracellular lipid synthesis and accumulation in yeast and filamentous fungi.
Studies on nitrogen metabolism in filamentous fungi have focused on glutamate and glutamine. Filamentous fungi assimilate ammonium to glutamate or glutamine via the NADP-dependent glutamate dehydrogenase (NADP-GDH) or glutamine synthetase–glutamine synthase (GS-GOGAT) pathways. Glutamate is involved in protein synthesis as an amino donor, whereas glutamine plays a role in the synthesis of essential metabolites such as nucleic acids, amino sugars, histidine, tyrosine, and asparagine [80,81]. For example, in A. nidulans, GATA TFs AreA and AreB work together to form a complementary regulatory network for nitrogen metabolism. AreA primarily activates the expression of nitrogen assimilation-related genes under nitrogen limitation, while AreB inhibits the expression of these genes under nitrogen abundance, thus maintaining the balance of nitrogen metabolism [35]. Small et al. found that under nitrogen-limiting conditions, the interaction of AreA with TamA led to the dissociation of AreA from the NMRA-AreA complex, activating the expression of nitrogen metabolism genes [82], which encode enzymes involved in nitrogen metabolism, such as arginase (agaA) and ornithine aminotransferase (otaA) [28]. Arginine is degraded by agaA into ornithine and urea, releasing a nitrogen source that can be utilized by the cell. Ornithine then enters the ornithine cycle and is converted to glutamate by otaA, regulating cellular nitrogen metabolism and indirectly promoting ATP synthesis [83]. This process not only optimizes nitrogen metabolism but also indirectly promotes lipid synthesis and accumulation by affecting ATP synthesis, thereby enhancing lipid production under nitrogen-limiting conditions. In contrast, under carbon-rich conditions, AreA changes from an activator to a repressor, inhibiting the expression of agaA and otaA [28], leading to a reduction in lipid synthesis. AreA participates in the process of nitrogen assimilation by directly or indirectly regulating the expression of NADP-GDH, which promotes the conversion of glutamine to glutamate [29]. Lamb et al. found that in the presence of glutamine, the NMRA binds to AreA and thus inhibits its activity. NMRA is also one of the transcription factors involved in the regulation of nitrogen source metabolism, and its transcriptional activity is partially regulated by MeaB, which can either mediate nitrogen repression together with AreA or independently [84].
In contrast, AreB acts as a negative regulator of nitrogen metabolism. Wong et al.’s investigations revealed that ΔAreB did not show significant changes in nitrogen utilization under various nitrogen sources (e.g., ammonium and glutamate), but under nitrogen-limiting conditions, AreB deletion enhanced the activation of AreA [53]. It was also found that the regulatory pathway of AreB involves the regulation of several target genes, including agaA, otaA, as well as the fmdS gene encoding formamidase. When ammonium is present, AreB represses the expression of agaA and otaA, whereas under nitrogen-limited conditions, AreB represses the expression of fmdS [28]. FmdS gene encodes Formamidase (FmdS), which catalyzes the hydrolysis of formamide to produce NH4+, thus providing an available source of nitrogen. Furthermore, the deletion of AreB significantly affects the transcriptional levels of several key regulators involved in carbon and nitrogen metabolism, including TamA, CreA, XprG, and CpcA [85]. TamA functions as a co-activator of AreA and is involved in the regulation of nitrogen metabolism, while CreA acts as a carbon metabolism repressor and mediates Carbon Catabolite Repression (CCR). In filamentous fungi, CCR is a crucial regulatory mechanism that ensures the preferential utilization of favorable carbon sources, such as glucose, while repressing the use of secondary sources like ethanol, acetate, and fatty acids under nutrient-rich conditions [86]. As a zinc finger transcription factor, CreA serves as the central repressor in CCR by inhibiting the expression of genes involved in the metabolism of alternative carbon sources. Through upregulation of CreA, AreB may indirectly enhance CCR, thereby suppressing the utilization of non-preferred carbon sources under specific environmental conditions [82]. Additionally, AreB regulates the expression of XprG, a global regulator involved in carbon starvation responses, further linking AreB to the broader network governing carbon source availability and metabolic regulation [87]. Macios et al. also found that AreB deletion significantly reduces the synthesis of the four tricarboxylic acids in the TCA cycle, namely α-ketoglutarate, succinate, fumarate, and malate, and decreases the efficiency of the TCA cycle [28]. In contrast, CpcA is negatively regulated by AreB, which is activated to promote the synthesis of amino acids and their precursors (e.g., purines, pyrimidines) under conditions of amino acid deficiency or environmental stress, while AreB, as its negative regulator, inhibits the relevant metabolic pathways, thereby reducing the supply of precursors for some secondary metabolites [65]. Svetlov et al. investigated the GATA TFs regulating nitrogen metabolism in Saccharomyces cerevisiae, and found that the yeast regulates the expression of nitrogen-metabolizing enzymes and transport systems mainly through the mechanism of Nitrogen Catabolite Repression (NCR), which is dependent on the activators Gln3p and Gat1p, as well as the repressors Dal80p and Deh1p/Gzf3p. Based on the regulatory model of Uga43/Dal80 competitive repression in S. cerevisiae, it has been hypothesized that AreB may repress the transcriptional activation of AreA through competitive binding to the GATA site [88]. The above findings indicate that AreA and AreB have well defined biological functions in nitrogen metabolism regulation (see Figure 2) and may play roles in regulating lipid synthesis in filamentous fungi by modulating nitrogen source utilization and related metabolic pathways. Specifically, AreA activates the expression of nitrogen metabolism-related genes, such as agaA and otaA, under nitrogen-limiting conditions, enhancing the synthesis of glutamate, which is a key precursor for lipid synthesis, particularly in the production of TAGs and phospholipids. Conversely, AreB suppresses the expression of nitrogen metabolism-related genes under nitrogen-rich conditions, reducing nitrogen metabolism activity and indirectly inhibiting lipid synthesis. Additionally, AreB further suppresses lipid synthesis by competitively inhibiting AreA activity under nitrogen-limiting conditions. However, the specific molecular regulatory mechanisms of AreA and AreB are still unclear and need to be further investigated.
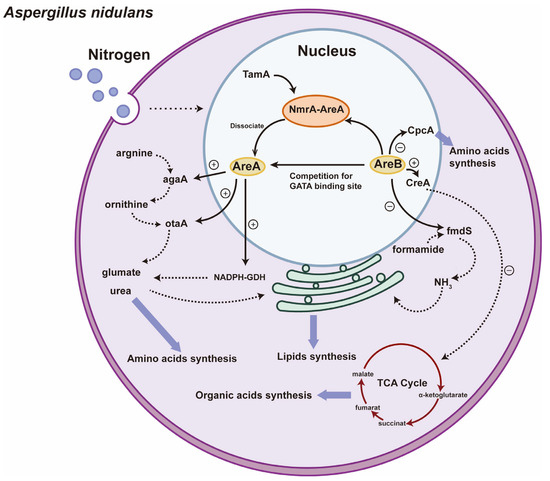
Figure 2.
Regulation of nitrogen metabolism by GATA TFs AreA and AreB in A. nidulans. Under nitrogen-limiting conditions, the NmrA-AreA complex dissociates, releasing AreA, which promotes the expression of agaA and otaA, catalyzing arginine degradation to supply nitrogen and participating in nitrogen assimilation to facilitate ATP synthesis. The generated nitrogen is used for lipid and amino acid synthesis. AreB negatively regulates AreA, fmdS, and CpcA, catalyzing formamide hydrolysis to release ammonia and contribute to the synthesis of amino acids and their precursors. Additionally, AreB activates CreA, inhibiting the synthesis of organic acids in the TCA cycle. Blue circles: nucleus; purple circles: cytoplasm; solid arrows inside nucleus: processes occurring before transfer to cytoplasm; dashed arrows inside cytoplasm: processes occurring within cytoplasm; blue arrows: metabolic flow toward organic acids, lipids, and amino acid synthesis; green circles and irregular ovals: mitochondria; red arrows: TCA cycle; yellow ovals: AreA and AreB genes; orange ovals: NmrA–AreA complex.
A comprehensive review of the literature reveals that most GATA TFs are involved in lipid biosynthesis indirectly by regulating nitrogen metabolism, whereas a few, such as SREB, directly participate in the regulation of lipid metabolism. Harrison et al. found that the GATA TF SREB (similar to SreA in A. nidulans) in Blastomyces dermatitidis is involved in the regulation of iron carrier synthesis, iron uptake, and the maintenance of iron homeostasis, in addition to affecting lipid and amino acid metabolism. Analysis of the ΔSREB mutant strain revealed a significant decrease in the transcript levels of genes related to TAG and ergosterol synthesis, with less effect on the phospholipid synthesis pathway. This suggests that SREB plays an important regulatory role in lipid metabolism, especially triglyceride and ergosterol biosynthesis [58].
3.4. The Roles of GATA TFs in Response to Stress in Filamentous Fungi
Fungi are often affected by changes in the external environment during growth, such as temperature, pH, osmotic pressure, light, and oxidative stress. These environmental changes may regulate their metabolic processes, thus affecting the growth status of the fungus and the production of secondary metabolites. The current studies suggest that GATA TFs are pivotal regulators in filamentous fungi, enabling adaptation to diverse environmental stresses by coordinating gene expression related to survival, metabolism, and virulence. Filamentous fungi are highly sensitive to temperature changes, which significantly affects their growth, morphological development, and pathogenicity. Gauthier et al. found that fungal growth correlates with temperature, and biomass accumulation and carbon utilization efficiency exhibit a nonlinear relationship with temperature [58]. Additionally, some biphasic fungi switch to a pathogenic yeast form with increased temperature, highlighting the key role of temperature in regulating fungal growth and pathogenicity [89,90,91,92]. Gauthier et al. found that in B. dermatitidis, a deletion mutant of the GATA TF SREB, failed to fully transform from yeast to a filamentous morphology when the temperature was lowered from 37 °C to 22 °C. Whereas at 22 °C, SREB was able to regulate lipid metabolism to promote filamentous growth and help the fungus to adapt to the temperature change. This study suggests that SREB plays a temperature-induced morphology transition in temperature-induced morphological transformation [58]. Bugeja et al. found that although the direct response mechanism of AreA to temperature changes in Penicillium marneffei remains unclear, its differential regulatory effects on the utilization of non-preferred nitrogen sources at 25 °C and 37 °C imply that AreA may contribute to the fungus’s adaptation to temperature variation [93]. This leads to the conclusion that GATA TFs play an important regulatory role in the adaptation of fungi to temperature changes.
In the industrial fermentation process, high concentration of salts often causes osmotic stress, affecting cellular water balance and ionic homeostasis, thus negatively affecting the growth of filamentous fungi. A number of studies have confirmed that GATA TFs play an important role in fungal resistance to salt stress. ΔAreA mutant strains exhibit enhanced growth capacity under osmotic stress, likely due to their improved efficiency in utilizing nitrogen sources, a finding also supported by the study of Fasoyin et al. [22]. Crespo et al. found that the GATA TFs GLN3 and GAT1 play an important role under salt stress by co-regulating the expression of ENA1 in S. cerevisiae [94]. This study demonstrated that mutant strains lacking GLN3 and GAT1 exhibit high sensitivity to Li+ and Na+ under high salt concentrations, and the absence of both GATA TFs leads to a significant decrease in the expression of ENA1, which adversely affects cellular salt tolerance [94]. DhGZF3, a gene similar to the GATA TFs Dal80 and Gzf3 in S. cerevisiae, was identified from Debaryomyces hansenii [90]. Overexpression of DhGZF3 in S. cerevisiae wild-type revealed that DhGZF3 enhanced the tolerance of S. cerevisiae to external stresses such as caffeine and rapamycin but reduced its tolerance to Li+ and Na+ [90]. This study suggests that GATA TF DhGZF3 has an important role in regulating yeast to saline and alkaline stresses [95]. Chen et al. evaluated the growth performance of different AaGATA gene mutant strains of A. alternata under various stress conditions by constructing different AaGATA gene mutant strains [30]. It was found that ΔAaAreB and ΔAaSreA mutant strains were sensitive to NaCl, while ΔAaSreA showed higher sensitivity to KCl, and other mutant strains were comparable to the wild-type in terms of tolerance [30]. Moreover, the ΔAaSreA and ΔAaLreA mutant strains showed high sensitivity to 1 mM CuSO4, whereas ΔAaAreB and ΔAaLreB showed greater copper tolerance [30]. These studies suggest that different GATA TFs play different roles in osmotic stress in filamentous fungi.
Similarly, light perception plays a key role in the adaptation of filamentous fungi to environmental changes [96]. Previous studies have shown that LreA and LreB are not only involved in light response but also play a key role in the regulation of sexual development. They sense light signals and regulate the expression of light-responsive genes, thereby influencing processes such as spore formation and hyphal growth [37]. Fuller et al., by knocking out the genes for the GATA TFs LreA and LreB in A. nidulans, found that LreA and LreB promote sexual development in darkness, whereas light prevents this process by repressing their function [39]. Unlike A. nidulans, A. fumigatus can effectively produce conidia in both light and dark environments, indicating that its asexual development is not significantly affected by light. The plant-like, photosensitive pigment FphA acts as a negative regulator, inhibiting the activity of LreA and LreB in the light, thereby influencing the regulation of light responses [39]. Furthermore, it was found that LreA regulates mycelial pigmentation in response to blue light, while FphA does not play a role in this process. More importantly, LreA can help A. fumigatus to better cope with and repair UV-induced cell damage, enhancing its UV resistance [39]. Zhang et al. conducted a systematic analysis of GATA TFs (TgGATAs) in Tolypocladium guangdongense and found that GATA1 (WC-1 homolog) and GATA2 (WC-2 homolog) may not only be early light-responsive genes but also play a role in mycelial color change and fruiting body development [97]. In N. crassa, blue light regulates the expression of genes associated with the differentiation of reproductive structures, as well as the synthesis of secondary metabolites and circadian rhythms. Blue light sensing is mainly regulated by the White Collar Complex (WCC), which consists of the GATA TFs WC-1 and WC-2. WC-1 contains a flavin-binding domain (LOV domain), which binds to the light-sensitive pigment FAD and senses blue light [97], while WC-2 interacts with WC-1 through the PAS domain to form the complexes [98,99]. The WCC binds to gene promoters of the light-inducible elements to regulate the transcription of light-responsive genes. In addition to light perception, WCC regulates circadian rhythms by affecting the biological clock [100] and influences the selection of sexual versus asexual development by regulating DNA methylation status [101].
In filamentous fungi, excessive accumulation of reactive oxygen species (ROS) caused by internal or external factors triggers the activation of antioxidant defense mechanisms to mitigate ROS-induced damage to cellular structures and functions. GATA TFs play a regulatory role in this process. The GATA TF SreA contributes to oxidative stress regulation in A. fumigatus by modulating iron homeostasis. Its deletion leads to increased intracellular iron levels, which in turn induce iron-mediated oxidative stress and the upregulation of antioxidative pathways [70]. In B. cinerea, the GATA TF BcLTF1 plays a pivotal role in regulating oxidative stress by maintaining the balance between ROS production and scavenging. Deletion of bcltf1 results in hypersensitivity to oxidative stress and mitochondrial dysfunction, indicating that BcLTF1 is essential for coping with oxidative challenges induced by light exposure or host infection [57]. Similarly, in Cordyceps militaris, a GATA TF such as CmAreA regulates the transcription of Cmhyd1, facilitating mycelial adaptation to oxidative stress and osmotic pressure, thereby enhancing tolerance to extreme temperatures and oxidative stress [102]. These findings highlight the conserved yet diverse roles of GATA TFs in regulating oxidative stress responses across different filamentous fungi. By modulating key genes involved in iron homeostasis, ROS detoxification, and stress adaptation, GATA TFs such as SreA, BcLTF1, and CmAreA play crucial roles in complex regulatory networks.
In summary, these studies illustrate that GATA TFs in filamentous fungi play a regulatory role under a variety of external environmental factors such as temperature, pH, osmotic pressure, light, and oxidative stress. Therefore, the utilization of GATA TFs can enhance the tolerance of strains to different growth environments, which is of great significance for production practice and application potential.
4. Conclusions and Future Directions
GATA TFs are widely found in fungi, plants, and animals and regulate gene expression by recognizing and binding to GATA motifs on DNA. This paper reviews the structure and classification of GATA TFs in filamentous fungi, discusses their functions in mycobacterial growth and development, nitrogen metabolism and environmental adaptation, focuses on their key roles in affecting lipid synthesis through regulating nitrogen metabolism, and illustrates the potential applications of filamentous fungi in multiple fields.
Filamentous fungi are pivotal in industrial biotechnology for producing organic acids, enzymes, antibiotics, and functional secondary metabolites. GATA TFs, implicated in carbon/nitrogen metabolism and secondary metabolite synthesis, could be engineered to optimize filamentous fungi performance. Important progress has been made in the functional studies of GATA TFs in recent years, but their specific mechanisms of action in different filamentous fungi are still not completely clear. For example, the GATA TFs AreA and AreB are involved in lipid synthesis by regulating nitrogen metabolism, but their specific target genes in the biosynthetic pathway have not yet been clarified, and the regulatory mechanism and molecular mechanism of action still need to be investigated in depth. In addition, most of the studies still remain in laboratory model strains and lack systematic validation in industrial and environmentally relevant fungi.
Therefore, future studies can combine multi-omics (transcriptomics, proteomics, metabolomics) analysis, systems biology, and synthetic biology to further resolve the global regulatory network of GATA TFs in filamentous fungi. Meanwhile, the use of gene-editing technology to precisely regulate the function of GATA TFs is expected to optimize the synthesis pathway of secondary metabolites and improve their production, thus promoting their application in biomedicine, agriculture, and industrial microbial biotechnology. For example, coupling multi-omics with CRISPR-Cas9 could identify GATA TF targets for enhancing yields of high-value compounds like penicillin or organic acids. Furthermore, exploring the roles of GATA TFs in stress responses (e.g., temperature, osmotic stress, oxidative stress, etc.) may improve fungal resilience in industrial fermentation, aligning with global trends in sustainable biomanufacturing.
Author Contributions
D.H., Y.L. and R.Z., Formal Analysis; D.H. and Y.L., Writing—Original Draft Preparation; D.H., Investigation; C.J., Software and Methodology; C.J., Review and Editing; C.J., Supervision; C.J., Funding Acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31900063), Jiangxi Provincial Department of Education Science and Technology Project (GJJ211103), and Doctoral Scientific Research Foundation of Jiangxi Science and Technology Normal University (2018BSQD027).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kerkaert, J.D.; Huberman, L.B. Regulation of nutrient utilization in filamentous fungi. Appl. Microbiol. Biotechnol. 2023, 107, 5873–5898. [Google Scholar] [CrossRef]
- Ghimire, P.S.; Jin, C. Genetics, molecular, and proteomics advances in filamentous fungi. Curr. Microbiol. 2017, 74, 1226–1236. [Google Scholar] [CrossRef]
- Lorenzini, M.; Cappello, M.S.; Logrieco, A.; Zapparoli, G. Polymorphism and phylogenetic species delimitation in filamentous fungi from predominant mycobiota in withered grapes. Int. J. Food Microbiol. 2016, 238, 56–62. [Google Scholar] [CrossRef]
- Orban, A.; Fraatz, M.A.; Rühl, M. Aroma profile analyses of filamentous fungi cultivated on solid substrates. Adv. Biochem. Eng. Biotechnol. 2019, 169, 85–107. [Google Scholar]
- Adav, S.S.; Ravindran, A.; Sze, S.K. Quantitative proteomic study of Aspergillus fumigatus secretome revealed deamidation of secretory enzymes. J. Proteom. 2015, 119, 154–168. [Google Scholar] [CrossRef]
- Adav, S.S.; Li, A.A.; Manavalan, A.; Punt, P.; Sze, S.K. Quantitative iTRAQ secretome analysis of Aspergillus niger reveals novel hydrolytic enzymes. J. Proteome Res. 2010, 9, 3932–3940. [Google Scholar] [CrossRef]
- Kawasaki, L.; Wysong, D.; Diamond, R.; Aguirre, J. Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J. Bacteriol. 1997, 179, 3284–3292. [Google Scholar] [CrossRef]
- Appels, F.V.W.; Dijksterhuis, J.; Lukasiewicz, C.E.; Jansen, K.M.; Wösten, H.A.B.; Krijgsheld, P. Hydrophobin gene deletion and environmental growth conditions impact mechanical properties of mycelium by affecting the density of the material. Sci. Rep. 2018, 8, 4703. [Google Scholar] [CrossRef]
- Wösten, H.A.B. Filamentous fungi for the production of enzymes, chemicals, and materials. Curr. Opin. Biotechnol. 2019, 59, 65–70. [Google Scholar] [CrossRef]
- Saykhedkar, S.; Ray, A.; Ayoubi-Canaan, P.; Hartson, S.D.; Prade, R.; Mort, A.J. A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum stover. Biotechnol. Biofuels 2012, 5, 52. [Google Scholar] [CrossRef]
- He, B.; Tu, Y.; Jiang, C.; Zhang, Z.; Li, Y.; Zeng, B. Functional genomics of Aspergillus oryzae: Strategies and progress. Microorganisms 2019, 7, 103. [Google Scholar] [CrossRef]
- Sonne, M.; Jawetz, E. Comparison of the action of ampicillin and benzylpenicillin on enterococci in vitro. Appl. Microbiol. 1968, 16, 645–648. [Google Scholar] [CrossRef]
- Wu, X.L.; Yang, Y.J.; Li, S.X.; Huang, L.; Liang, F. Clinical effects of different penicillin administration regimens in the treatment of group B Streptococcus infection in pregnant women. Drug Eval. 2023, 20, 1542–1545. [Google Scholar]
- Dutta, A.K.; Phull, P.S. Treatment of Helicobacter pylori infection in the presence of penicillin allergy. World J. Gastroenterol. 2021, 27, 7661–7668. [Google Scholar] [CrossRef]
- Van Den Berg, M.; Gidijala, L.; Kiela, J.; Bovenberg, R.; Vander Keli, I. Biosynthesis of active pharmaceuticals: β-lactam biosynthesis in filamentous fungi. Biotechnol. Genet. Eng. Rev. 2010, 27, 1–32. [Google Scholar]
- Liu, L.; Chen, Z.; Liu, W.; Ke, X.; Tian, X.; Chu, J. Cephalosporin C biosynthesis and fermentation in Acremonium chrysogenum. Appl. Microbiol. Biotechnol. 2022, 106, 6413–6426. [Google Scholar] [CrossRef]
- Majeed, S.; Abdullah, M.S.; Dash, G.K.; Ansari, M.T.; Nanda, A. Biochemical synthesis of silver nanoparticles using filamentous fungi Penicillium decumbens (MTCC-2494) and its efficacy against A-549 lung cancer cell line. J. Nanomater. 2016, 2016, 2184134. [Google Scholar]
- Perez-Cuesta, U.; Aparicio-Fernandez, L.; Guruceaga, X.; Martin-Souto, L.; Abad-Diaz-de-Cerio, A.; Antoran, A.; Buldain, I.; Hernando, F.L.; Ramirez-Garcia, A.; Rementeria, A. Melanin and pyomelanin in Aspergillus fumigatus: From its genetics to host interaction. Int. Microbiol. 2020, 23, 55–63. [Google Scholar] [CrossRef]
- Chamilos, G.; Carvalho, A. Aspergillus fumigatus DHN-Melanin. Curr. Top. Microbiol. Immunol. 2020, 425, 17–28. [Google Scholar]
- Pfannmuller, A.; Leufken, J.; Studt, L.; Michielse, C.B.; Sieber, C.M.K.; Guldener, U.; Hawat, S.; Hippler, M.; Fufezan, C.; Tudzynski, B. Comparative transcriptome and proteome analysis reveals a global impact of the nitrogen regulators AreA and AreB on secondary metabolism in Fusarium fujikuroi. PLoS ONE 2017, 12, e0176194. [Google Scholar] [CrossRef]
- Hedden, P.; Sponsel, V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- Fasoyin, O.E.; Yang, K.; Qiu, M.; Wang, B.; Wang, S.; Wang, S. Regulation of Morphology, Aflatoxin Production, and Virulence of Aspergillus flavus by the Major Nitrogen Regulatory Gene areA. Toxins 2019, 11, 718. [Google Scholar] [CrossRef]
- Caceres, I.; Khoury, A.A.; Khoury, R.E.; Lorber, S.; Oswald, I.P.; Khoury, A.E.; Atoui, A.; Puel, O.; Bailly, J.D. Aflatoxin Biosynthesis and Genetic Regulation: A Review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef]
- Lin, H.; Cheng, W.; Ding, H.T.; Chen, X.J.; Zhou, Q.F.; Zhao, Y.H. Direct Microbial Conversion of Wheat Straw into Lipid by a Cellulolytic Fungus of Aspergillus oryzae A-4 in Solid-State Fermentation. Bioresour. Technol. 2010, 101, 7556–7562. [Google Scholar] [PubMed]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.L.; Molina-Jouve, C.; Nicaud, J.M. Yarrowia lipolytica as a Model for Bio-Oil Production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef]
- Fazili, A.B.A.; Shah, A.M.; Zan, X.; Naz, T.; Nosheen, S.; Nazir, Y.; Ullah, S.; Zhang, H.; Song, Y. Mucor circinelloides: A Model Organism for Oleaginous Fungi and Its Potential Applications in Bioactive Lipid Production. Microb. Cell Factories 2022, 21, 29. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Y.; Zhang, S.; Liu, Q.; Dang, W.; Song, Y. Lipid Accumulation and SNF1 Transcriptional Analysis of Mucor circinelloides on Xylose under Nitrogen Limitation. Antonie Van Leeuwenhoek 2023, 116, 383–391. [Google Scholar] [CrossRef]
- Macios, M.; Caddick, M.X.; Weglenski, P.; Scazzocchio, C.; Dzikowska, A. The GATA Factors AREA and AREB Together with the Co-Repressor NMRA, Negatively Regulate Arginine Catabolism in Aspergillus nidulans in Response to Nitrogen and Carbon Source. Fungal Genet. Biol. 2012, 49, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.; Hynes, M.J.; Davis, M.A. Role of the Regulatory Gene areA of Aspergillus oryzae in Nitrogen Metabolism. Appl. Environ. Microbiol. 1998, 64, 3232–3237. [Google Scholar] [CrossRef]
- Chen, Y.; Cao, Y.; Gai, Y.; Ma, H.; Zhu, Z.; Chung, K.R.; Li, H. Genome-Wide Identification and Functional Characterization of GATA Transcription Factor Gene Family in Alternaria alternata. J. Fungi 2021, 7, 1013. [Google Scholar] [CrossRef]
- Michielse, C.B.; Pfannmüller, A.; Macios, M.; Rengers, P.; Dzikowska, A.; Tudzynski, B. The Interplay between the GATA Transcription Factors AreA, the Global Nitrogen Regulator and AreB in Fusarium fujikuroi. Mol. Microbiol. 2014, 91, 472–493. [Google Scholar] [CrossRef]
- Haas, H.; Zadra, I.; Stöffler, G.; Angermayr, K. The Aspergillus nidulans GATA Factor SREA is Involved in Regulation of Siderophore Biosynthesis and Control of Iron Uptake. J. Biol. Chem. 1999, 274, 4613–4619. [Google Scholar] [CrossRef]
- Oberegger, H.; Schoeser, M.; Zadra, I.; Schrettl, M.; Parson, W.; Haas, H. Regulation of freA, acoA, lysF, and cycA Expression by Iron Availability in Aspergillus nidulans. Appl. Environ. Microbiol. 2002, 68, 5769–5772. [Google Scholar] [CrossRef]
- Franken, A.C.; Lechner, B.E.; Werner, E.R.; Haas, H.; Lokman, B.C.; Ram, A.F.; van den Hondel, C.A.; de Weert, S.; Punt, P.J. Genome Mining and Functional Genomics for Siderophore Production in Aspergillus niger. Brief. Funct. Genom. 2014, 13, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Yap, A.; Volz, R.; Paul, S.; Moye-Rowley, W.S.; Haas, H. Regulation of high-affinity iron acquisition, including acquisition mediated by the iron permease FtrA, is coordinated by AtrR, SrbA, and SreA in Aspergillus fumigatus. mBio 2023, 14, e0075723. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Han, K.H.; Yu, J.H. Upstream Regulation of Development and Secondary Metabolism in Aspergillus Species. Cells 2022, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Purschwitz, J.; Müller, S.; Kastner, C.; Schöser, M.; Haas, H.; Espeso, E.A.; Atoui, A.; Calvo, A.M.; Fischer, R. Functional and Physical Interaction of Blue- and Red-Light Sensors in Aspergillus nidulans. Curr. Biol. 2008, 18, 255–259. [Google Scholar] [CrossRef]
- Jiang, C.; Lv, G.; Ge, J.; He, B.; Zhang, Z.; Hu, Z.; Zeng, B. Genome-Wide Identification of the GATA Transcription Factor Family and Their Expression Patterns under Temperature and Salt Stress in Aspergillus oryzae. AMB Express 2021, 11, 56. [Google Scholar] [CrossRef]
- Fuller, K.K.; Ringelberg, C.S.; Loros, J.J.; Dunlap, J.C. The Fungal Pathogen Aspergillus fumigatus Regulates Growth, Metabolism, and Stress Resistance in Response to Light. mBio 2013, 4, e00142-13. [Google Scholar] [CrossRef]
- Teakle, G.R.; Gilmartin, P.M. Two Forms of Type IV Zinc-Finger Motif and Their Kingdom-Specific Distribution between the Flora, Fauna and Fungi. Trends Biochem. Sci. 1998, 23, 100–102. [Google Scholar] [CrossRef]
- Scazzocchio, C. The fungal GATA factors. Curr. Opin. Microbiol. 2000, 3, 126–131. [Google Scholar] [CrossRef]
- Yu, M.; Yu, J.; Cao, H.; Yong, M.; Liu, Y. Genome-wide identification and analysis of the GATA transcription factor gene family in Ustilaginoidea virens. Genome 2019, 62, 807–816. [Google Scholar] [CrossRef]
- Ronsmans, A.; Wery, M.; Szachnowski, U.; Gautier, C.; Descrimes, M.; Dubois, E.; Morillon, A.; Georis, I. Transcription-dependent spreading of the Dal80 yeast GATA factor across the body of highly expressed genes. PLoS Genet. 2019, 15, e1007999. [Google Scholar] [CrossRef]
- Monahan, B.J.; Askin, M.C.; Hynes, M.J.; Davis, M.A. Differential expression of Aspergillus nidulans ammonium permease genes is regulated by GATA transcription factor AreA. Eukaryot. Cell 2006, 5, 226–237. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, B.; Yin, C.; Guo, Y.; Lin, Y.; Pan, L.; Wang, B. Characterization of natural antisense transcript, sclerotia development and secondary metabolism by strand-specific RNA sequencing of Aspergillus flavus. PLoS ONE 2014, 9, e1007999. [Google Scholar] [CrossRef]
- Conlon, H.; Zadra, I.; Haas, H.; Arst, H.N., Jr.; Jones, M.G.; Caddick, M.X. The Aspergillus nidulans GATA transcription factor gene areB encodes at least three proteins and features three classes of mutation. Mol. Microbiol. 2001, 40, 361–375. [Google Scholar] [CrossRef]
- Haas, H.; Marzluf, G.A. NRE, the major nitrogen regulatory protein of Penicillium chrysogenum, binds specifically to elements in the intergenic promoter regions of nitrate assimilation and penicillin biosynthetic gene clusters. Curr. Genet. 1995, 28, 177–183. [Google Scholar] [CrossRef]
- Haas, H.; Angermayr, K.; Stöffler, G. Molecular analysis of a Penicillium chrysogenum GATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 gene. Gene 1997, 184, 33–37. [Google Scholar] [CrossRef]
- Niehaus, E.M.; Schumacher, J.; Burkhardt, I.; Rabe, P.; Spitzer, E.; Münsterkötter, M.; Güldener, U.; Sieber, C.M.K.; Dickschat, J.S.; Tudzynski, B. The GATA-Type Transcription Factor Csm1 Regulates Conidiation and Secondary Metabolism in Fusarium fujikuroi. Front. Microbiol. 2017, 8, 1175. [Google Scholar] [CrossRef]
- López-Berges, M.S.; Schäfer, K.; Hera, C.; Di Pietro, A. Combinatorial function of velvet and AreA in transcriptional regulation of nitrate utilization and secondary metabolism. Fungal Genet. Biol. 2014, 62, 78–84. [Google Scholar] [CrossRef]
- Harrison, K.A.; Marzluf, G.A. Characterization of DNA binding and the cysteine rich region of SRE, a GATA factor in Neurospora crassa involved in siderophore synthesis. Biochemistry 2002, 41, 15288–15295. [Google Scholar] [CrossRef]
- Fu, Y.H.; Marzluf, G.A. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol. Cell. Biol. 1987, 7, 1691–1696. [Google Scholar] [CrossRef]
- Wong, K.H.; Hynes, M.J.; Todd, R.B.; Davis, M.A. Deletion and Overexpression of the Aspergillus nidulans GATA Factor AreB Reveals Unexpected Pleiotropy. Microbiology 2009, 155, 3868–3880. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Haas, H.; Marzluf, G.A. ASD4, a new GATA factor of Neurospora crassa, displays sequence-specific DNA binding and functions in ascus and ascospore development. Biochemistry 2000, 39, 11065–11073. [Google Scholar] [CrossRef] [PubMed]
- Brenna, A.; Talora, C. WC-1 and the proximal GATA sequence mediate a cis-/trans-acting repressive regulation of light-dependent gene transcription in the dark. Int. J. Mol. Sci. 2019, 20, 2854. [Google Scholar] [CrossRef]
- Ren, W.; Qian, C.; Ren, D.; Cai, Y.; Deng, Z.; Zhang, N.; Wang, C.; Wang, Y.; Zhu, P.; Xu, L. The GATA transcription factor BcWCL2 regulates citric acid secretion to maintain redox homeostasis and full virulence in Botrytis cinerea. mBio 2024, 15, e0013324. [Google Scholar] [CrossRef]
- Schumacher, J.; Simon, A.; Cohrs, K.C.; Viaud, M.; Tudzynski, P. The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet. 2014, 10, e1004040. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, G.M.; Sullivan, T.D.; Gallardo, S.S.; Brandhorst, T.T.; Wymelenberg, A.J.V.; Cuomo, C.A.; Suen, G.; Currie, C.R.; Klein, B.S. SREB, a GATA transcription factor that directs disparate fates in Blastomyces dermatitidis including morphogenesis and siderophore biosynthesis. PLoS Pathog. 2010, 6, e1000846. [Google Scholar] [CrossRef]
- Lee, M.K.; Kwon, N.J.; Lee, I.S.; Jung, S.; Kim, S.C.; Yu, J.H. Negative regulation and developmental competence in Aspergillus. Sci. Rep. 2016, 6, 28874. [Google Scholar] [CrossRef]
- Szewczyk, E.; Krappmann, S. Conserved regulators of mating are essential for Aspergillus fumigatus cleistothecium formation. Eukaryot. Cell 2010, 9, 774–783. [Google Scholar] [CrossRef]
- Shen, L.; Gaslonde, T.; Roullier, C.; Wang, H.; Bodin, J.; Porée, F.H.; Ruprich-Robert, G.; Chapeland-Leclerc, F. Functional Characterization of the GATA-Type Transcription Factor PaNsdD in the Filamentous Fungus Podospora anserina and Its Interplay with the Sterigmatocystin Pathway. Appl. Environ. Microbiol. 2022, 88, e0237821. [Google Scholar] [CrossRef] [PubMed]
- He, X.Y. Functional Study of GATA Transcription Factors SsAREA and SsSRE in Sphaerotheca fuliginea. Ph.D. Thesis, Jilin University, Changchun, China, 2021. [Google Scholar]
- Wei, S.; Zhang, Y.; Wu, M.; Lv, Y.; Zhang, S.; Zhai, H.; Hu, Y. Mechanisms of methyl 2-methylbutyrate suppression on Aspergillus flavus growth and aflatoxin B1 biosynthesis. Int. J. Food Microbiol. 2024, 409, 110462. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pan, Y.; Liu, G. Disruption of the nitrogen regulatory gene AcareA in Acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism. Fungal Genet. Biol. 2013, 61, 69–79. [Google Scholar] [CrossRef]
- Tudzynski, B.; Homann, V.; Feng, B.; Marzluf, G.A. Isolation, characterization and disruption of the areA nitrogen regulatory gene of Gibberella fujikuroi. Mol. Genet. Genom. 1999, 261, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ke, X.; Jia, R.; Huang, L.; Liu, Z.; Zheng, Y. Gibberellic acid overproduction in Fusarium fujikuroi using regulatory modification and transcription analysis. Appl. Microbiol. Biotechnol. 2023, 107, 3071–3084. [Google Scholar] [CrossRef]
- Wang, L.; Wang, C.; Xu, L.; Wang, M. Regulation of nitrogen utilization and mycotoxin biosynthesis by the GATA transcription factor AaAreA in Alternaria alternata. World J. Microbiol. Biotechnol. 2024, 40, 236. [Google Scholar] [CrossRef]
- Zhu, J.; Sun, Z.; Shi, D.; Song, S.; Lian, L.; Shi, L.; Ren, A.; Yu, H.; Zhao, M. Dual functions of AreA, a GATA transcription factor, on influencing ganoderic acid biosynthesis in Ganoderma lucidum. Environ. Microbiol. 2019, 21, 4166–4179. [Google Scholar] [CrossRef]
- Niehaus, E.M.; von Bargen, K.W.; Espino, J.J.; Pfannmüller, A.; Humpf, H.U.; Tudzynski, B. Characterization of the fusaric acid gene cluster in Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2014, 98, 1749–1762. [Google Scholar] [CrossRef]
- Schrettl, M.; Kim, H.S.; Eisendle, M.; Kragl, C.; Nierman, W.C.; Heinekamp, T.; Werner, E.R.; Jacobsen, I.; Illmer, P.; Yi, H.; et al. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 2008, 70, 27–43. [Google Scholar] [CrossRef]
- Yang, Y.; Heidari, F.; Hu, B. Fungi (Mold)-Based Lipid Production. Methods Mol. Biol. 2019, 1995, 51–89. [Google Scholar]
- Hassane, A.M.A.; Eldiehy, K.S.H.; Saha, D.; Mohamed, H.; Mosa, M.A.; Abouelela, M.E.; Abo-Dahab, N.F.; El-Shanawany, A.A. Oleaginous fungi: A promising source of biofuels and nutraceuticals with enhanced lipid production strategies. Arch. Microbiol. 2024, 206, 338. [Google Scholar] [CrossRef] [PubMed]
- Marzluf, G.A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 1997, 61, 17–32. [Google Scholar] [PubMed]
- Sun, X.; Chen, Y.Y.; Wu, Y.N.; Chen, B.; Xi, C.H.; Jiang, L.G. Method for Preparing Sphingolipids Using Olive Oil Fermentation. Chinese Patent CN201610087770.3, 11 June 2021. [Google Scholar]
- Dzurendova, S.; Zimmermann, B.; Tafintseva, V.; Kohler, A.; Ekeberg, D.; Shapaval, V. The influence of phosphorus source and the nature of nitrogen substrate on the biomass production and lipid accumulation in oleaginous Mucoromycota fungi. Appl. Microbiol. Biotechnol. 2020, 104, 8065–8076. [Google Scholar] [CrossRef]
- Aguilar, L.R.; Pardo, J.P.; Lomelí, M.M.; Bocardo, O.I.L.; Juárez Oropeza, M.A.; Guerra Sánchez, G. Lipid droplets accumulation and other biochemical changes induced in the fungal pathogen Ustilago maydis under nitrogen-starvation. Arch. Microbiol. 2017, 199, 1195–1209. [Google Scholar] [CrossRef]
- Maltsev, Y.; Kulikovskiy, M.; Maltseva, S. Nitrogen and phosphorus stress as a tool to induce lipid production in microalgae. Microb. Cell Factories 2023, 22, 239. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Xu, H.; Yang, Q.; Xu, Y.; Yang, H.; Qiao, D.; Cao, Y. GATA-type transcriptional factor SpGAT1 interacts with SpMIG1 and promotes lipid accumulation in the oleaginous yeast Saitozyma podzolica zwy-2-3. Biotechnol. Biofuels Bioprod. 2022, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Pomraning, K.R.; Bredeweg, E.L.; Baker, S.E. Regulation of nitrogen metabolism by GATA zinc finger transcription factors in Yarrowia lipolytica. mSphere 2017, 2, e00038-17. [Google Scholar] [CrossRef]
- Javelle, A.; Morel, M.; Rodríguez-Pastrana, B.R.; Botton, B.; André, B.; Marini, A.M.; Brun, A.; Chalot, M. Molecular characterization, function, and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Mol. Microbiol. 2003, 47, 411–430. [Google Scholar] [CrossRef]
- Breuninger, M.; Trujillo, C.G.; Serrano, E.; Fischer, R.; Requena, N. Different nitrogen sources modulate activity but not expression of glutamine synthetase in arbuscular mycorrhizal fungi. Fungal Genet. Biol. 2004, 41, 542–552. [Google Scholar] [CrossRef]
- Small, A.J.; Hynes, M.J.; Davis, M.A. The TamA protein fused to a DNA-binding domain can recruit AreA, the major nitrogen regulatory protein, to activate gene expression in Aspergillus nidulans. Genetics 1999, 153, 95–105. [Google Scholar] [CrossRef]
- Khandelwal, R.; Srivastava, P.; Bisaria, V.S. Recent advances in the production of malic acid by native fungi and engineered microbes. World J. Microbiol. Biotechnol. 2023, 39, 217. [Google Scholar] [CrossRef] [PubMed]
- Lamb, H.K.; Ren, J.; Park, A.; Johnson, C.; Leslie, K.; Cocklin, S.; Thompson, P.; Mee, C.; Cooper, A.; Stammers, D.K.; et al. Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis. Protein Sci. 2004, 13, 3127–3138. [Google Scholar] [CrossRef] [PubMed]
- Chudzicka-Ormaniec, P.; Macios, M.; Koper, M.; Weedall, G.D.; Caddick, M.X.; Weglenski, P.; Dzikowska, A. The role of the GATA transcription factor AreB in regulation of nitrogen and carbon metabolism in Aspergillus nidulans. FEMS Microbiol. Lett. 2019, 366, fnz066. [Google Scholar] [CrossRef] [PubMed]
- Kuruvilla, F.G.; Shamji, A.F.; Schreiber, S.L. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc. Natl. Acad. Sci. USA 2001, 98, 7283–7288. [Google Scholar] [CrossRef]
- Strauss, J.; Horvath, H.K.; Abdallah, B.M.; Kindermann, J.; Mach, R.L.; Kubicek, C.P. The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol. Microbiol. 1999, 32, 169–178. [Google Scholar] [CrossRef]
- Svetlov, V.V.; Cooper, T.G. The Saccharomyces cerevisiae GATA factors Dal80p and Deh1p can form homo- and heterodimeric complexes. J. Bacteriol. 1998, 180, 5682–5688. [Google Scholar] [CrossRef]
- Dagenais, T.R.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin. Microbiol. Rev. 2009, 22, 447–465. [Google Scholar] [CrossRef]
- Boyce, K.J.; Andrianopoulos, A. Fungal dimorphism: The switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol. Rev. 2015, 39, 797–811. [Google Scholar] [CrossRef]
- Romani, L. Immunity to fungal infections. Nat. Rev. Immunol. 2004, 4, 11–24. [Google Scholar] [CrossRef]
- Marty, A.J.; Broman, A.T.; Zarnowski, R.; Dwyer, T.G.; Bond, L.M.; Lounes-Hadj Sahraoui, A.; Fontaine, J.; Ntambi, J.M.; Keleş, S.; Kendziorski, C.; et al. Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in Blastomyces dermatitidis. PLoS Pathog. 2015, 11, e1004959. [Google Scholar] [CrossRef]
- Bugeja, H.E.; Hynes, M.J.; Andrianopoulos, A. AreA controls nitrogen source utilization during both growth programs of the dimorphic fungus Penicillium marneffei. Fungal Biol. 2012, 116, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Crespo, J.L.; Daicho, K.; Ushimaru, T.; Hall, M.N. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 34441–34444. [Google Scholar] [CrossRef]
- García-Salcedo, R.; Casamayor, A.; Ruiz, A.; González, A.; Prista, C.; Loureiro-Dias, M.C.; Ramos, J.; Ariño, J. Heterologous expression implicates a GATA factor in regulation of nitrogen metabolic genes and ion homeostasis in the halotolerant yeast Debaryomyces hansenii. Eukaryot. Cell 2006, 5, 1388–1398. [Google Scholar] [CrossRef]
- Schumacher, J. How light affects the life of Botrytis. Fungal Genet. Biol. 2017, 106, 26–41. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, G.; Deng, W.; Li, T. Distribution, evolution and expression of GATA-TFs provide new insights into their functions in light response and fruiting body development of Tolypocladium guangdongense. PeerJ 2020, 8, e9784. [Google Scholar] [CrossRef] [PubMed]
- Flores-Ibarra, A.; Maia, R.N.A.; Olasz, B.; Church, J.R.; Gotthard, G.; Schapiro, I.; Heberle, J.; Nogly, P. Light-Oxygen-Voltage (LOV)-sensing Domains: Activation Mechanism and Optogenetic Stimulation. J. Mol. Biol. 2024, 436, 168356. [Google Scholar] [CrossRef]
- Ballario, P.; Talora, C.; Galli, D.; Linden, H.; Macino, G. Roles in dimerization and blue light photoresponse of the PAS and LOV domains of Neurospora crassa white collar proteins. Mol. Microbiol. 1998, 29, 719–729. [Google Scholar] [CrossRef]
- Chen, C.H.; Ringelberg, C.S.; Gross, R.H.; Dunlap, J.C.; Loros, J.J. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009, 28, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Kritsky, M.S.; Russo, V.E.; Filippovich, S.Y.; Afanasieva, T.P.; Bachurina, G.P. The opposed effect of 5-azacytidine and light on the development of reproductive structures in Neurospora crassa. Photochem. Photobiol. 2002, 75, 79–83. [Google Scholar] [CrossRef]
- Li, X.; Wang, F.; Liu, M.; Dong, C. Hydrophobin CmHYD1 Is Involved in Conidiation, Infection and Primordium Formation, and Regulated by GATA Transcription Factor CmAreA in Edible Fungus, Cordyceps militaris. J. Fungi 2021, 7, 674. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).