N-Acyl Homoserine Lactone-Degrading Bacillus sp. Improves the Survival of Penaeus vannamei Larvae Challenged with Vibrio harveyi
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Preparation of Microbial Communities
2.3. Culture Media
2.4. Isolation of Bacillus Strains from the Cultures
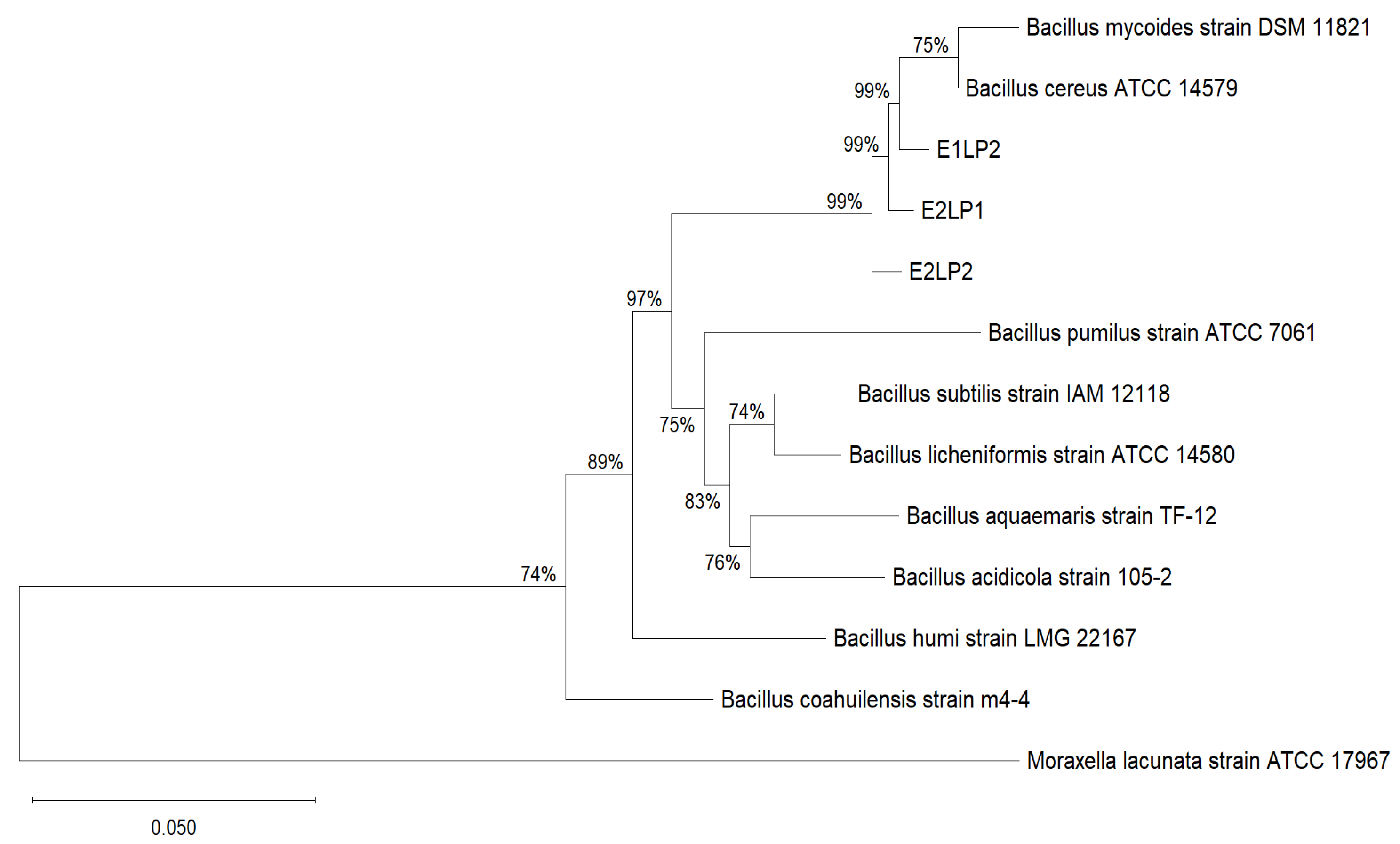
2.5. 16S rRNA Gene Sequencing
2.6. Detection of N-Hexanoyl-L-Homoserine Lactone (HHL)
2.7. Penaeus vannamei Larval Challenge Tests
2.8. Statistical Analysis
3. Results
3.1. Isolation and Identification of Bacillus Strains
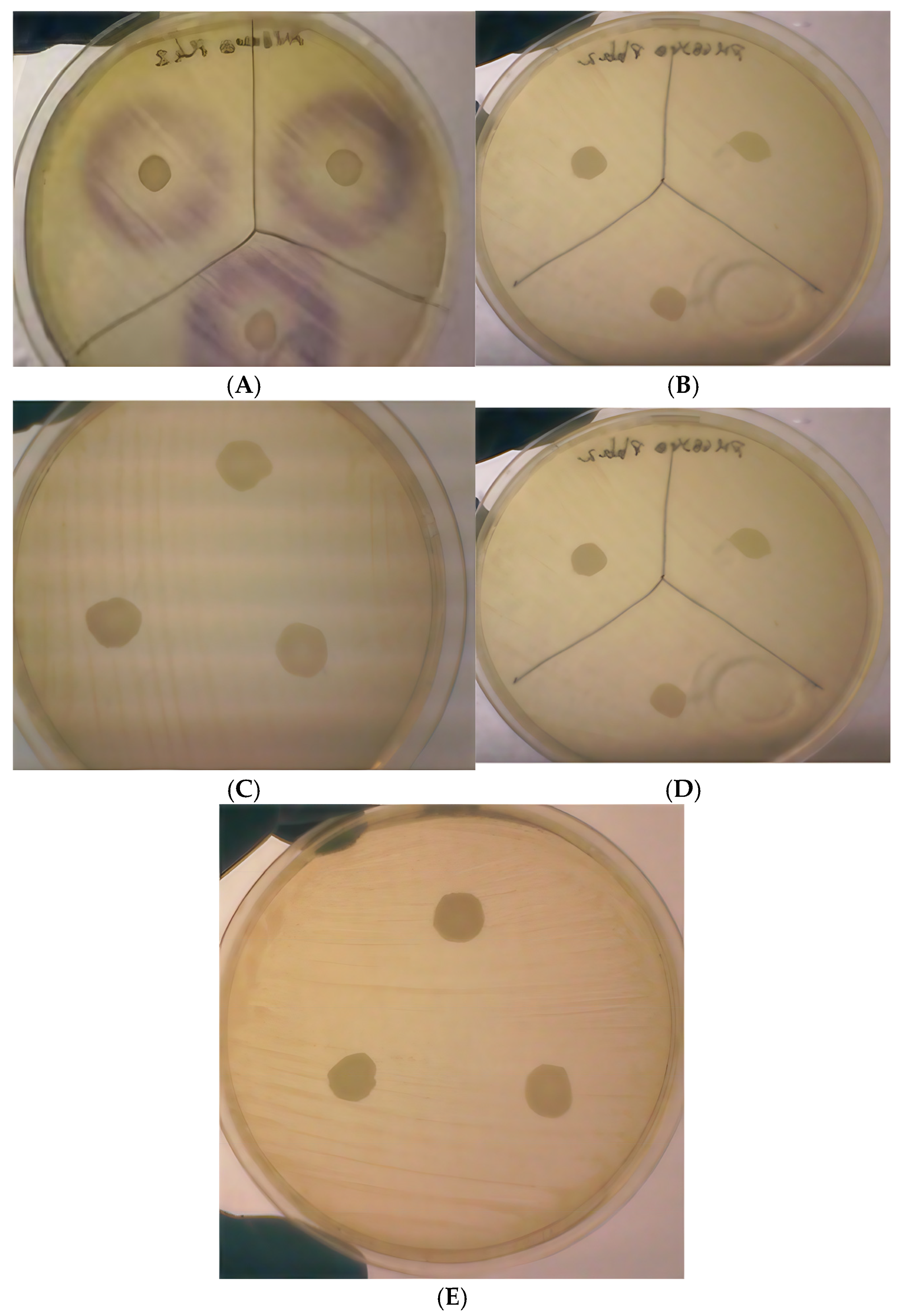
3.2. Acyl Homoserine Lactone (AHL) Inactivation Assay
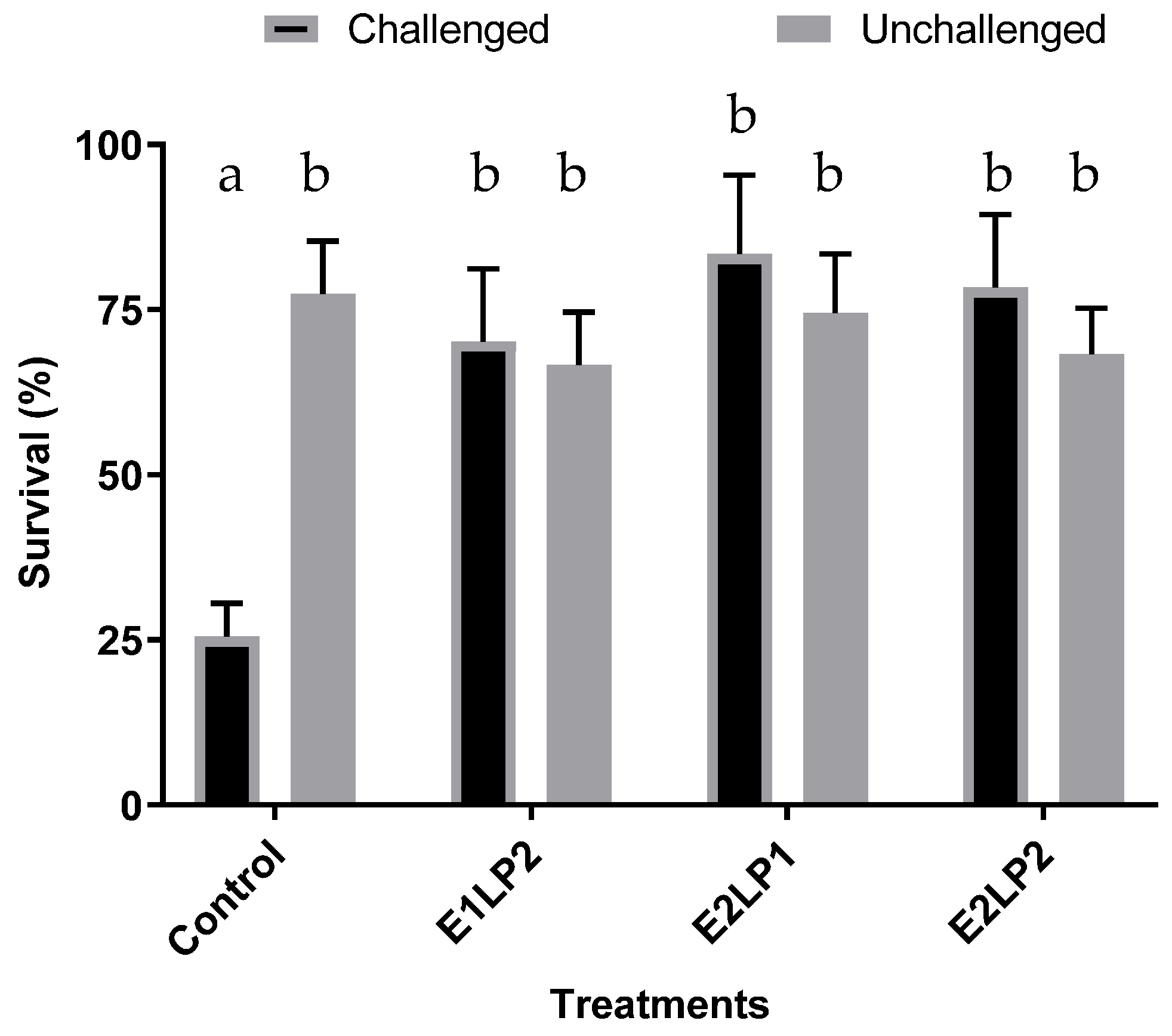
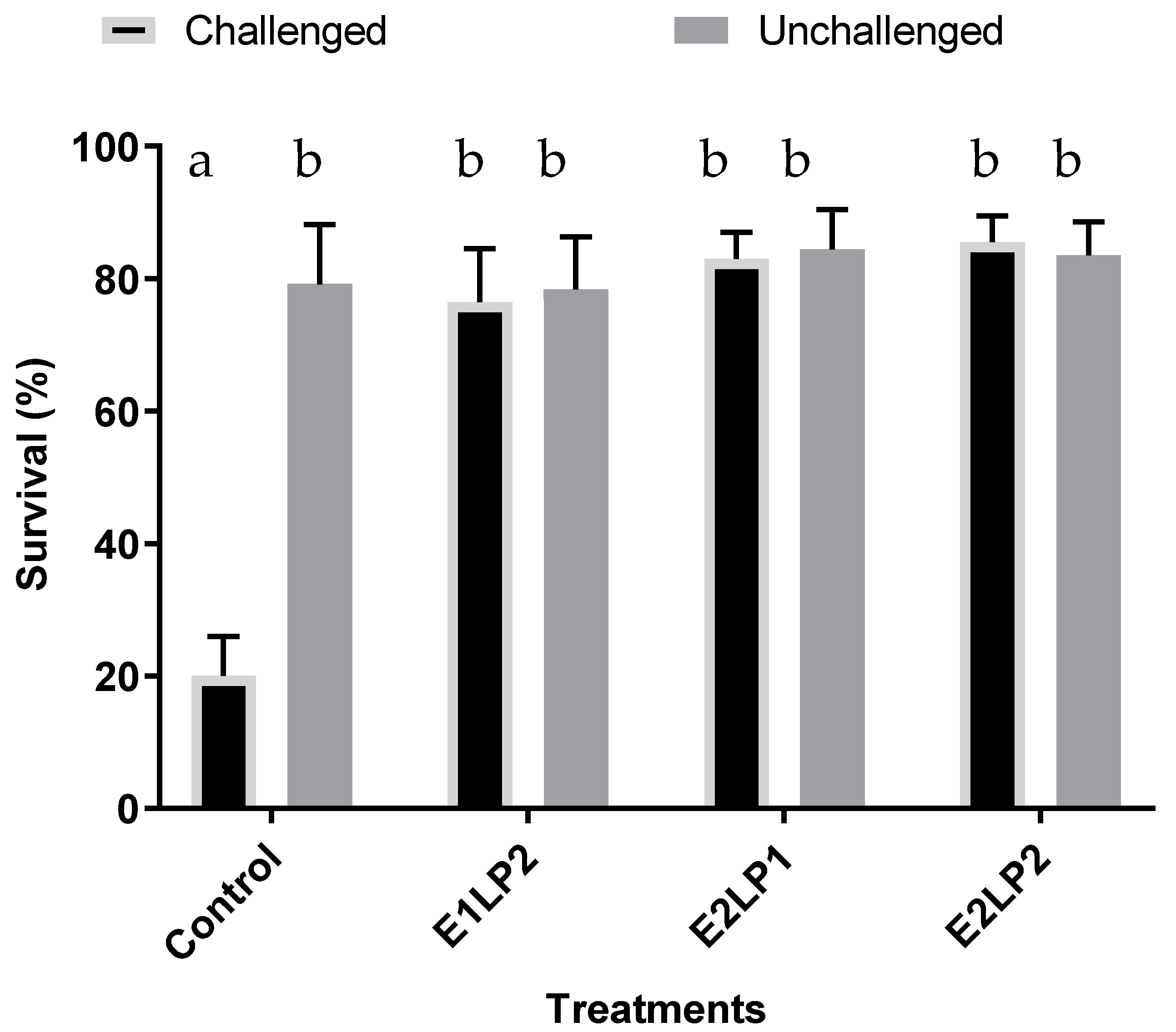
3.3. Bacillus sp. Effect on the Survival of Zoea and Mysis Challenged with Vibrio harveyi
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AHL | Acyl-homoserine lactone |
| MCs | Microbial communities |
| NSS | Nine-salt solution |
| HHL | Hexanoyl-L-homoserine lactone |
References
- Deachamag, P.; Intaraphad, U.; Phongdara, A.; Chotigeat, W. Expression of a phagocytosis activating protein (PAP) gene in immunized black tiger shrimp. Aquaculture 2006, 255, 165–172. [Google Scholar] [CrossRef]
- Kurath, G. Biotechnology and DNA vaccines for aquatic animals. Rev. Sci. Tech.-Off. Int. Épizooties 2008, 27, 175. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, F.; Ding, G.; Li, J.; Guo, X.; Zhu, J.; Zhou, L.; Cai, S.; Liu, X.; Luo, Y.; et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 2012, 6, 1336–1344. [Google Scholar] [CrossRef] [PubMed]
- Saulnier, D.; Haffner, P.; Goarant, C.; Levy, P.; Ansquer, D. Experimental infection models for shrimp vibriosis studies: A review. Aquaculture 2000, 191, 133–144. [Google Scholar] [CrossRef]
- Pruzzo, C.; Gallo, G.; Canesi, L. Persistence of vibrios in marine bivalves: The role of interactions with haemolymph components. Environ. Microbiol. 2005, 7, 761–772. [Google Scholar] [CrossRef]
- Beaz-Hidalgo, R.; Balboa, S.; Romalde, J.L.; Figueras, M.J. Diversity and pathogenecity of Vibrio species in cultured bivalve molluscs. Environ. Microbiol. Rep. 2010, 2, 34–43. [Google Scholar] [CrossRef]
- Mohney, L.L.; Lightner, D.V.; Bell, T.A. An Epizootic of Vibriosis in Ecuadorian Pond-Reared Penaeus vannamei Boone (Crustacea: Decapoda). J. World Aquac. Soc. 1994, 25, 116–125. [Google Scholar] [CrossRef]
- Tyagi, A.; Khushiramani, R.; Karunasagar, I.; Karunasagar, I. Antivibrio activity of recombinant lysozyme expressed from black tiger shrimp, Penaeus monodon. Aquaculture 2007, 272, 246–253. [Google Scholar] [CrossRef]
- Nayak, S.K.; Mukherjee, S.C. Screening of gastrointestinal bacteria of Indian major carps for a candidate probiotic species for aquaculture practices. Aquac. Res. 2011, 42, 1034–1041. [Google Scholar] [CrossRef]
- Gomez-Gil, B.; Roque, A.; Turnbull, J.F. The use and selection of probiotic bacteria for use in the culture of larval aquatic organisms. Aquaculture 2000, 191, 259–270. [Google Scholar] [CrossRef]
- Chythanya, R.; Karunasagar, I.; Karunasagar, I. Inhibition of shrimp pathogenic vibrios by a marine Pseudomonas I-2 strain. Aquaculture 2002, 208, 1–10. [Google Scholar] [CrossRef]
- Wang, Y.B.; XU, Z.R.; Xia, M.S. The effectiveness of commercial probiotics in northern white shrimp Penaeus vannamei ponds. Fish. Sci. 2005, 71, 1036–1041. [Google Scholar] [CrossRef]
- Decamp, O.; Moriarty, D.J.; Lavens, P. Probiotics for shrimp larviculture: Review of field data from Asia and Latin America. Aquac. Res. 2008, 39, 334–338. [Google Scholar] [CrossRef]
- Mihailović, D.T.; Budinčević, M.; Balaž, I.; Mihailović, A. Stability of intercellular exchange of biochemical substances affected by variability of environmental parameters. Mod. Phys. Lett. B 2011, 25, 2407–2417. [Google Scholar] [CrossRef]
- Yu, P.; Ye, H.B.; Shan, H.W.; Ma, S.; Wang, T. Screening, safety evaluation and fermentation conditions optimization of quorum quenching bacteria from Litopenaeus vannamei culture system. Progr. Fish. Sci. 2020, 1–11. [Google Scholar]
- Yu, P.; Wang, T.; Ye, H.B.; Shan, H.W.; Ma, S. Isolation and identification of pathogenic Vibrio spp. retrieved from diseased Litopenaeus vannamei and beneficial role of some functional probiotic bacteria for control. Aquac. Int. 2020, 6, 1403–1420. [Google Scholar] [CrossRef]
- Yu, P.; Ye, H.; Ma, S.; Shan, H. Study of quorum quenching bacteria Cobetia sp. reducing the mortality of Penaeus vannamei infected with Vibrio harveyi: In vitro and in vivo tests. Aquaculture 2021, 534, 736244. [Google Scholar] [CrossRef]
- Natrah, F.M.I.; Kenmegne, M.M.; Wiyoto, W.; Sorgeloos, P.; Bossier, P.; Defoirdt, T. Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquaculture 2011, 317, 53–57. [Google Scholar] [CrossRef]
- Fuqua, C.; Parsek, M.R.; Greenberg, E.P. Regulation of gene expression by cell-to-cell communication: Acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001, 35, 439–468. [Google Scholar] [CrossRef]
- Yousefian, M.; Amiri, M.S. A review of the use of prebiotic in aquaculture for fish and shrimp. Afr. J. Biotechnol. 2009, 8, 7313–7318. [Google Scholar]
- Defoirdt, T.; Thanh, L.D.; Van Delsen, B.; De Schryver, P.; Sorgeloos, P.; Boon, N.; Bossier, P. N-acylhomoserine lactone-degrading Bacillus strains isolated from aquaculture animals. Aquaculture 2011, 311, 258–260. [Google Scholar] [CrossRef]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef]
- Molina, L.; Constantinescu, F.; Michel, L.; Reimmann, C.; Duffy, B.; Défago, G. Degradation of pathogen quorum-sensing molecules by soil bacteria: A preventive and curative biological control mechanism. FEMS Microbiol. Ecol. 2003, 45, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Boon, N.; De Windt, W.; Verstraete, W.; Top, E.M. Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 2002, 39, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406425. [Google Scholar]
- Tinh, N.T.N.; Gunasekara, R.A.; Boon, N.; Dierckens, K.; Sorgeloos, P.; Bossier, P. N-acyl homoserine lactone-degrading microbial enrichment cultures isolated from Penaeus vannamei shrimp gut and their probiotic properties in Brachionus plicatilis cultures. FEMS Microbiol. Ecol. 2007, 62, 45–53. [Google Scholar] [CrossRef]
- Tang, K.H.; Zhang, X.H. Quorum quenching agents: Resources for antivirulence therapy. Mar. Drugs 2014, 12, 3245–3282. [Google Scholar] [CrossRef]
- Dang, W.; Hu, Y.H.M.; Zhang, L. Sun Identification and molecular analysis of a stress-inducible Hsp70 from Sciaenops ocellatus. Fish Shellfish Immunol. 2010, 29, 600–607. [Google Scholar] [CrossRef]
- Gatesoupe, F.J. Probiotic and formaldehyde treatments of Artemia nauplii as food for larval pollack, Pollachius pollachius. Aquaculture 2002, 212, 347–360. [Google Scholar] [CrossRef]
- Nikoskelainen, S.; Salminen, S.; Bylund, G.; Ouwehand, A.C. Characterization of the properties of human-and dairy-derived probiotics for prevention of infectious diseases in fish. Appl. Environ. Microbiol. 2001, 67, 2430–2435. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, K.S.; Turenne, C.Y.; Harmsen, D.; Kabani, A.M. Reassessment of sequence-based targets for identification of Bacillus species. J. Clin. Microbiol. 2004, 42, 1626–1630. [Google Scholar] [CrossRef] [PubMed]
- Hakovirta, J.R.; Preziuso, S.; Hodge, D.; Pillai, S.P.; Weigel, L.M. Identification and analysis of informative single nucleotide polymorphisms in 16S rRNA gene sequences of the Bacillus cereus group. J. Clin. Microbiol. 2016, 54, 2749–2756. [Google Scholar] [CrossRef]
- Liu, B. Uncertain Reliability Analysis. In Uncertainty Theory. Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany; Volume 300.
- Vinoj, G.; Vaseeharan, B.; Thomas, S.; Spiers, A.J.; Shanthi, S. Quorum-quenching activity of the AHL-lactonase from Bacillus licheniformis DAHB1 inhibits Vibrio biofilm formation in vitro and reduces shrimp intestinal colonisation and mortality. Mar. Biotechnol. 2014, 16, 707–715. [Google Scholar] [CrossRef]
- Taga, M.E.; Bassler, B.L. Chemical communication among bacteria. PNAS 2003, 100, 14549–14554. [Google Scholar] [CrossRef]
- Pande, G.S.J.; Scheie, A.A.; Benneche, T.; Wille, M.; Sorgeloos, P.; Bossier, P.; Defoirdt, T. Quorum sensing-disrupting compounds protect larvae of the giant freshwater prawn Macrobrachium rosenbergii from Vibrio harveyi infection. Aquaculture 2013, 406, 121–124. [Google Scholar] [CrossRef]
- Manefield, M.; Harris, L.; Rice, S.A.; de Nys, R.; Kjelleberg, S. Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl. Environ. Microbiol. 2000, 66, 2079–2084. [Google Scholar] [CrossRef]
- Yuniarti, A.; Maftuch, M.; Soemarno, S.; Aulanni’am, A. In vitro and in vivo study of acyl homoserine lactone degrading Bacillus against Vibrio harveyi. Int. J. Biosci. 2015, 6, 338–348. [Google Scholar]
- Monzón-Atienza, L.; Bravo, J.; Torrecillas, S.; Gómez-Mercader, A.; Montero, D.; Ramos-Vivas, J.; Galindo-Villegas, J.; Acosta, F. An In-Depth Study on the Inhibition of Quorum Sensing by Bacillus velezensis D-18: Its Significant Impact on Vibrio Biofilm Formation in Aquaculture. Microorganisms 2024, 12, 890. [Google Scholar] [CrossRef] [PubMed]
- Van Cam, D.T.; Van Hao, N.; Dierckens, K.; Defoirdt, T.; Boon, N.; Sorgeloos, P.; Bossier, P. Novel approach of using homoserine lactone-degrading and poly-β-hydroxybutyrate-accumulating bacteria to protect Artemia from the pathogenic effects of Vibrio harveyi. Aquaculture 2009, 291, 23–30. [Google Scholar] [CrossRef]
- Juhas, M.; Eberl, L.; Tummler, B. Quorum sensing: The power of cooperation in the world of Pseudomonas. Environ. Microbiol. 2005, 7, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Van Cam, D.T.; Nhan, D.T.; Ceuppens, S.; Van Hao, N.; Dierckens, K.; Wille, M.; Sorgeloos, P.; Bossier, P. Effect of N-acyl homoserine lactone-degrading enrichment cultures on Macrobrachium rosenbergii larviculture. Aquaculture 2009, 294, 5–13. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, R.; Cabrera, H.; Niu, Y.; Roncarati, A.; Toledo, A.; Corrales, Y.; Quevedo, Y.; Galosi, L.; Bossier, P.; Arenal, A. N-Acyl Homoserine Lactone-Degrading Bacillus sp. Improves the Survival of Penaeus vannamei Larvae Challenged with Vibrio harveyi. Microbiol. Res. 2025, 16, 56. https://doi.org/10.3390/microbiolres16030056
González R, Cabrera H, Niu Y, Roncarati A, Toledo A, Corrales Y, Quevedo Y, Galosi L, Bossier P, Arenal A. N-Acyl Homoserine Lactone-Degrading Bacillus sp. Improves the Survival of Penaeus vannamei Larvae Challenged with Vibrio harveyi. Microbiology Research. 2025; 16(3):56. https://doi.org/10.3390/microbiolres16030056
Chicago/Turabian StyleGonzález, Reinaldo, Héctor Cabrera, Yufeng Niu, Alessandra Roncarati, Adrian Toledo, Yulaine Corrales, Yanelis Quevedo, Livio Galosi, Peter Bossier, and Amilcar Arenal. 2025. "N-Acyl Homoserine Lactone-Degrading Bacillus sp. Improves the Survival of Penaeus vannamei Larvae Challenged with Vibrio harveyi" Microbiology Research 16, no. 3: 56. https://doi.org/10.3390/microbiolres16030056
APA StyleGonzález, R., Cabrera, H., Niu, Y., Roncarati, A., Toledo, A., Corrales, Y., Quevedo, Y., Galosi, L., Bossier, P., & Arenal, A. (2025). N-Acyl Homoserine Lactone-Degrading Bacillus sp. Improves the Survival of Penaeus vannamei Larvae Challenged with Vibrio harveyi. Microbiology Research, 16(3), 56. https://doi.org/10.3390/microbiolres16030056




