Abstract
Recently, studies suggest that the protective effects of Lactobacillus within the female reproductive tract may be partly due to their ability to form biofilms. This study aims to explore the possibility that Lactobacillus can produce key mediators to further bolster the survival of biofilms in human vaginal microbiomes. Three bacterial species, namely, Lactobacillus gasseri, L. crispatus, and L. jensenii, sourced from human female subjects were used to carry out experiments examining the growth of biofilms using a microfermenter system. The bacteria were used to inoculate a glass rod spatula which was subsequently transferred to the microfermenter system. The resulting biofilm growing on the glass spatula was harvested in media and stored in a −80 °C freezer for gas chromatography–mass spectroscopy analysis. We found that quorum sensing compounds, acyl homoserine lactones (AHLs), were detected in the biofilm of L. crispatus and L. jensenii, but none were detected in L. gasseri. The biofilm produced by L. crispatus and L. jensenii was much higher in quantity than the biofilm produced by L. gasseri. Aside from oligopeptides quorum sensing, lactobacilli were found to also have AHL compounds that may help them produce more biofilms and improve the survival and growth of their bacterial communities in the female genital area.
1. Introduction
Biofilms are clusters of bacteria enclosed in a matrix that attaches to surfaces, be it biological or non-biological, representing a complex community of microbial growth and development governed by a multitude of harmonious cell-to-cell communications [1,2]. These bacteria utilize synthesized peptides or signaling molecules to auto-induce signals via quorum sensing (QS) to then facilitate biofilm formation by altering gene expression, allowing bacteria to evolve in response to external stimuli [3]. This mechanism has been shown to be involved in the attachment, maturation, aggregation, and dispersal of biofilms, promoting antibiotic resistance, pathogenic invasion, and overall survival against competing microorganisms [4]. QS plays a sizable role in the development and maturation process of biofilms and is a topic of interest when humans are involved.
In the context of human health, biofilms play a significant role as many microorganisms can readily adhere and attach to mucosal surfaces. Microbial colonization and subsequent biofilm formation can be mutually beneficial; for example, the microbial community found within the human vaginal tract can be pathogenic, which often leads to infection and chronic conditions [5]. The symbiotic microbial environment found within the human body is critical in maintaining health and protection against invading pathogens [3].
In particular, Lactobacillus crispatus, L. gasseri, L. iners, and L. jensenii, are predominantly in the vagina and contribute to the majority of human vaginal defense against pathogens [5,6]. Lactobacillus attach to the vaginal epithelium, forming a biofilm network that creates a protective barrier against pathogenic microorganisms [5,7]. Historically, these Gram-positive bacilli contributed to vaginal health by primarily producing lactic acid from glycogen metabolism supplied by the vaginal epithelia, thereby lowering the pH and inhibiting the growth of harmful pathogens [5,8]. However, emerging research suggests that the biofilm-forming ability of Lactobacillus mediated by QS may be equally or more important in maintaining a healthy vaginal environment [9].
The QS mechanism in Gram-positive bacteria, such as Lactobacillus, involves autoinducing oligopeptides, which act as messengers in biofilm production [10,11]. In Gram-negative bacilli, acyl-homoserine lactones (AHLs) function as mediators of QS signaling. However, a recent study by Biswa and Doble found a novel Gram-positive marine bacterium capable of producing AHLs [12]. This finding is significant because Gram-positive bacteria could have both oligopeptides and AHLs mediating QS signaling, which could represent a more complex mechanism in biofilm formation that further potentiates its survival.
Given the importance of Lactobacillus spp. in the vaginal microbiome and the scant evidence of AHL production in Gram-positive bacteria, this study aims to explore the possibility that Lactobacillus spp. can produce AHLs for the enhancement and survival of the biofilms they produce, allowing for healthier colonization and longer permanence along the vaginal epithelium, thus preventing pathogenic colonization. By increasing our understanding of the QS mechanisms in Lactobacilli, this research provides new insights into their role in vaginal health and their potential applications in probiotic therapies. The biofilm behavior of three species of Lactobacillus, namely, L. crispatus, L. jensenii, and L. gasseri, will be compared to their biofilm behavior using a microfermenter system, and the subsequent biofilm supernatant will be analyzed using four AHLs: C6-HL, C8-HL, C10-HL, and C12-HL.
2. Materials and Methods
Preparation of the Microfermenter System: The stocks of three bacterial species, L. gasseri (CIP 102991T, Pasteur Institute, Paris, France), L. crispatus (CIP 103603, Pasteur Institute, Paris), and L. jensenii (CIP 69.17T, Pasteur Institute, Paris), were stored in the freezer (−80 °C) in cryovials for use when preparing the microfermenter system. All three Lactobacillus species were stored in the freezer in 10% glycerol–de Mann, Ragosa, and Sharpe (MRS) broth solution. The incubator was set at 37 °C, 5% CO2, with a humidity pan filled with 2 L of autoclaved, distilled water. In advance, 200 mL of MRS broth was freshly prepared and autoclaved to set up the experiment. The microfermenter, spatula, connectors, and Tygon tubes were covered in aluminum foil and autoclaved before use in the microfermenter system. Lactobacillus stock bacteria were removed from the freezer, and an inoculating loop was sterilized via a Bunsen burner and was then used to scrape off the pellet of stock bacteria, which was then placed in a 25 mL Erlenmeyer flask filled with 5 mL of previously autoclaved MRS broth. The flask was then transferred to the incubator and was grown overnight for 24 h. The next day, the media with bacterial growth was removed from the flask, placed into a 15 mL falcon tube, and centrifuged for 15 min at 3200 rpm. The supernatant was discarded, and 5 mL MRS media was added to the falcon tube and vortexed until the pellet was resuspended. The resuspended bacteria were then diluted using MRS media to a set optical density of 0.02 (approximately 104–105 cells/mL of bacteria) using a UV–Visible Spectrophotometer (BioMate 160, Model# GENESYS 1xx, Thermo Fisher Scientific, Waltham, MA, USA). Around 50 mL of MRS media was added into an autoclaved non-bubbling reactor (Figure 1A). The reactor was then inoculated using 1 mL of the OD 0.02 diluted neat bacteria, which approximately contained 104–105 bacteria. Finally, the autoclaved spatula was inserted into a reactor and incubated for 3 h at room temperature.
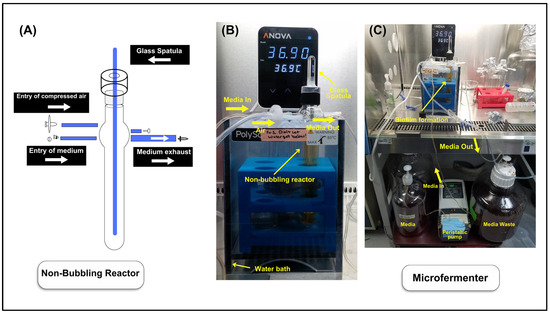
Figure 1.
Illustration of the microfermenter flowthrough culture set up for Lactobacillus species. (A) A non-bubbling reactor with unused capillaries sealed. The flow of MRS broth and airflow in the microfermenter is shown. (B,C) Microfermenter set up is shown.
To complete the system, 2.5 L of MRS broth was prepared and autoclaved. The microfermenter system was set up as shown in Figure 1. The temperature of the water tank was set at 37 °C, and, after three hours of incubation, the non-bubbling reactor was then transferred to the heated water tank. Air was then pressurized (0.04 bar) into the microfermenter system to allow flow in the closed system. The microfermenter system was running for 48 h.
Collecting Biofilm: Around 50 mL of autoclaved MRS broth was added into an autoclaved measuring cylinder to collect the biofilm from a glass spatula. The microfermenter and a glass spatula were placed against a dark surface for photos (Figure 2).

Figure 2.
Biofilm of Lactobacillus species (L. crispatus, L. jensenii, and L. gasseri) formed on the glass surfaces of the microfermenter and spatula.
Under sterile conditions, the glass spatula was placed in the autoclaved measuring cylinder and pulsed in a vortex mixer for a few seconds to separate the biofilm from the spatula. The leftover media from the reactor was transferred into four 15 mL conical tubes. The media in the reactor was centrifuged at 3200 rpm for 15 min at room temperature. After the centrifugation, the media was kept in the freezer for gas chromatography–mass spectrometry analysis.
AHL Extraction: All Lactobacillus species were grown in the microfermenter for 48 h. After the experiment, 15 mL of culture media from the microfermenter chamber was transferred to a 15 mL falcon tube. The media was then centrifuged at 3200 rpm at room temperature for 15 min, after which the first layer was transferred to a fresh tube with the pellet separated. The supernatant was combined with the same amount of ethyl acetate (99.5%, Sigma-Aldrich, St. Louis, MO, USA) in a glass test tube and vortexed for approximately 10 s. The mixture was allowed to separate, and the organic portion was transferred to a small beaker. This extraction was repeated, and both organic portions were combined and allowed to evaporate in a fume hood overnight. The residue was resuspended in 1000 μL of acetonitrile and stored at −20 °C until analyzed by gas chromatography–mass spectrometry.
Acyl-HLS Analysis by Gas Chromatography-Mass Spectrometry: The following standards were purchased from Caymen Chemicals (Ann Arbor, MI, USA): N-butyryl-L-Homoserine Lactone (C4-HL, Cat#10007898), N-hexanoyl-L-Homoserine Lactone (C6-HL, Cat#10011197), N-octonyl-L-Homoserine Lactone (C8-HL, Cat#10011199), N-decanoyl-Homoserine Lactone (C10-HL, Cat#10011201), N-dodecanoyl-Homoserine Lactone (C12-HL, Cat#10011201), and N-tetradecanoyl-Homoserine Lactone (C14-HL, Cat#10011200). Method optimization was performed using 5.0 ppm ethanolic homoserine lactone standards (HLSs) analyzed using a GCMS-TQ8040 gas chromatograph–triple quadrupole mass spectrometer (Shimadzu Scientific Instruments, Columbia, MD, USA). The injector was maintained at 200 °C, and 1.0 μL of the sample was injected with a split ratio of 10.0. Separations were achieved with an RTX-5ms capillary column (30 m, 0.25 mm ID, 0.25 μm film thickness, Restek, Bellefonte, MD, USA) using a He carrier gas at a constant flow of 0.80 mL/min. The column temperature was initially 150 °C, and then it was increased to 275 °C at 15 °C/min and held for 6.00 min. Analytes were ionized by chemical ionization using methane as a reagent gas (flow rate 0.8 mL/min). Following the collection of full scans for each standard, the MRM conditions were established using the Shimadzu Smart MRM program for each of the analytes and can be found in Supplementary Table S1. For each HLS, a quantification and qualification transition were monitored, and identification was based on retention time and ion ratios. The transfer line temperature was 280 °C, and the ion source was maintained at 230 °C. For the quantification of the four most common HLSs, (C6-HL, C8-HL, C10-HL, and C12-HL), a four-point calibration curve from 0.1 to 50 ppm was developed, with all four compounds showing acceptable linearity over this range (R2 > 0.97). Bacterial extracts were analyzed using the optimized MRM method, and compound identification was based on the retention time and both quantification and qualification transitions.
3. Results
Microfermenter: The biofilm produced by three Lactobacillus species is shown in Figure 2. The biofilm produced by L. crispatus and L. jensenii was much higher in quantity than the biofilm produced by L. gasseri. In addition to the quantity, L. crispatus, and L. jensenii adhered more strongly to the spatula when compared to L. gasseri.
Quantification of AHLs: Representative chromatograms from bacterial extracts are shown in Figure 3, and the concentrations for C6-, C8-, C10-, and C12- AHLs are tabulated in Table 1. C8 was typically the most abundant AHL present in all three species. C6 and C12 AHLs were detected in two species, and C10-AHL was detected in only one species.
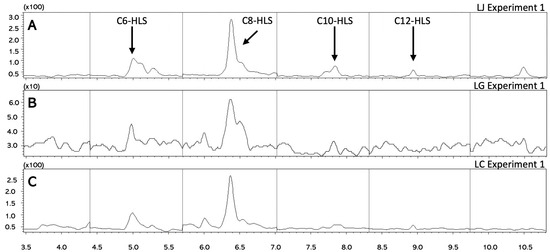
Figure 3.
Chromatograms of AHLs. Representative chromatograms for the analysis of C6-C12 AHLs for (A) Lactobacillus jensenii, (B) Lactobacillus gasseri, and (C) Lactobacillus crispatus.

Table 1.
The levels of AHLs in Lactobacillus species biofilm.
Extracts from a growth experiment for L. jensenii were analyzed (Figure 3A), and all four AHLs were quantified, with concentrations ranging from 140 to 480 ng/mL. Due to difficulties in growing sufficient biofilm from L. gasseri, AHLs from a single sample were extracted and analyzed (Figure 3B). Only C8 was detected; but the concentration of this compound was below the limit of quantification for the method. The lack of QS molecules in this sample may contribute to the difficulties observed in producing a robust biofilm. In L. crispatus (Figure 3C), three AHLs were quantified as follows: 206 ng/mL for C6-AHL, 498 ng/mL for C8-AHL, and 149 ng/mL for C12-AHL.
4. Discussion
The majority of QS-related and AHL-related research studies are focused on Gram-negative bacteria. However, many Gram-positive bacteria play an essential role in human health through QS-related mechanisms, specifically, the Lactobacillus species in the vaginal microbiome [13]. Universally, we know the nature of the signaling molecules (autoinducers) used in Gram-positive QS systems is different from that of Gram-negative bacteria. Miller et al. described in depth the current knowledge of QS mechanisms utilized by these two bacteria classifications [14].
There were several studies published in the past, identifying new QS mechanisms within bacteria [15,16,17], but, as of February 2023, only one study has been documented in the literature by Biswa and Doble that has found the existence of AHL-QS within a marine Gram-positive bacteria [12]. This finding challenged our previously established knowledge and opened up the possibility of Gram-positive bacteria having more than oligopeptide-induced QS that were previously known. Indeed, in our research, we demonstrated that QS compounds, AHLs, were detected in the biofilm of L. crispatus and L. jensenii, but were limited in L. gasseri due to detachment from the spatula, therefore representing an insufficient sample collection. This is a novel finding for lactobacilli in vaginal microbiomes.
As shown in Figure 3, the concentration of the C8-HSLs was dramatically higher than any of the other AHLs in any of the other samples. When extracting the AHLs for GC-MS analysis, all Lactobacillus species were grown in the microfermenter for 48 h since the exact time when these samples made the most AHLs was unknown. In a study conducted by Abbamondi, G.R (2016), their bacterial sample Halomonas smyrnensis (AAD6) started to produce detectable AHLs only after 48 h of incubation [18]. It is possible that the strains were not producing their maximum level of AHLs at the time of extraction. Further analytical studies with a focus on quantifying the concentration of the AHL extracts at different growth intervals could help identify the time when the strains start making the most abundant AHLs.
Interestingly, we also observed that the biofilm concentrations attached to the spatula produced by the L. crispatus and L. jensenii species were much higher than those of L. gasseri. This result is due to the detachment of L. gasseri and the dispersal in to the medium. Supplementary Video S1 shows the growth of L. gasseri over time in the span of 48 h; initial growth can be seen on the spatula and with subsequent detachment as the experiment progresses. Upon detachment, the structural integrity and complex system of a biofilm matrix is lost, and, since the system is continuously removing excess waste media and is replenished with fresh media, our key products of interest would have been flushed out as waste. Unfortunately, upon removing the spatula and upon the interruption of the closed microfermenter system, the dispersed biofilm settled on top of a bed of planktonic growth at the bottom of the reactor, which made the retrieval of the biofilm impossible. This explains why the sample retrieved yielded undetectable traces of AHLs upon GC-MS analysis.
Nonetheless, our quantitative QS data indicate a relatively small level of AHLs and suggest that AHLs might not be the most prominent QS mechanism within these bacterial communities. Although, they might still play a role in cellular communication and, hence, biofilm production. There could very well be more additional communications within these three bacteria communities, which could likely have a more significant influence over the biofilm production. Additionally, a greater variety of AHL compounds should be considered to comprehensively investigate the communication profiles in these Lactobacillus species.
5. Conclusions
According to our research, AHLs were present in the biofilms of L. crispatus and L. jensenii, and, though we detected C8 AHL in L. gasseri, we were not able to quantify the exact concentration. In addition, when compared to L. gasseri, larger concentrations of biofilms were found to be attached to the glass spatula in L. crispatus and L. jensenii due to the detachment of the L. gasseri biofilm from the glass surface. According to our findings, Gram-positive bacteria could have a novel method of QS that facilitates the formation of biofilms that then improve bacterial communities in the vaginal microbiome. The limited size of our study necessitates more research exploring confounding variables in oligopeptide-inducing QS. Further research examining expanded AHL profiles is needed to understand additional influences on biofilm formation.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microbiolres15030100/s1, Supplementary Table S1: MRM method parameters for HLS detection and quantification, Video S1: Timelapse of LG detachment.
Author Contributions
Conceptualization, G.V., K.G., J.G. and A.S.; methodology, K.G., A.S., J.G. and S.D.; validation, F.A.; software, J.G.; formal analysis, C.B.B., A.S. and F.A.; investigation, A.S., F.A. and K.G.; resources, G.V. and K.G.; data curation, S.D., F.A., C.B.B., K.G. and A.S.; writing—original draft preparation, K.G., A.S., S.D., G.V., D.L., T.L. and J.G.—review and editing, A.S., C.F., A.O., D.H., D.L., T.L., S.D., K.G. and J.G.; visualization, S.D. and C.B.B.; supervision, G.V., K.G. and M.G.; project administration, G.V., A.S., K.G. and M.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and was approved by the Institutional Review Board of Texas Tech University Health Sciences Center.
Informed Consent Statement
Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
The data presented in this study are available in the article or Supplementary Materials.
Conflicts of Interest
Author Christopher B. Babayco was employed by the company Kremenak NanoTech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Vestby, L.K.; Gronseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Parsek, M.R.; Greenberg, E. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Younes, J.A.; Lievens, E.; Hummelen, R.; van der Westen, R.; Reid, G.; Petrova, M.I. Women and Their Microbes: The Unexpected Friendship. Trends Microbiol. 2018, 26, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Nicolo, S.; Tanturli, M.; Mattiuz, G.; Antonelli, A.; Baccani, I.; Bonaiuto, C.; Baldi, S.; Nannini, G.; Menicatti, M.; Bartolucci, G.; et al. Vaginal Lactobacilli and Vaginal Dysbiosis-Associated Bacteria Differently Affect Cervical Epithelial and Immune Homeostasis and Anti-Viral Defenses. Int. J. Mol. Sci. 2021, 22, 6487. [Google Scholar] [CrossRef] [PubMed]
- Salas-Jara, M.J.; Ilabaca, A.; Vega, M.; Garcia, A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms 2016, 4, 35. [Google Scholar] [CrossRef]
- Liu, P.; Lu, Y.; Li, R.; Chen, X. Use of probiotic lactobacilli in the treatment of vaginal infections: In vitro and in vivo investigations. Front. Cell. Infect. Microbiol. 2023, 13, 1153894. [Google Scholar] [CrossRef] [PubMed]
- Saxena, P.; Joshi, Y.; Rawat, K.; Bisht, R. Biofilms: Architecture, resistance, quorum sensing and control mechanisms. Indian J. Microbiol. 2019, 59, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, F.; De Craemer, S.; Debunne, N.; Janssens, Y.; Wynendaele, E.; Van de Wiele, C.; De Spiegeleer, B. Peptides as quorum sensing molecules: Measurement techniques and obtained levels in vitro and in vivo. Front. Neurosci. 2017, 11, 183. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Quadri, L.E.; Kuipers, O.P.; De Vos, W.M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 1997, 24, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Biswa, P.; Doble, M. Production of acylated homoserine lactone by Gram-positive bacteria isolated from marine water. FEMS Microbiol. Lett. 2013, 343, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Sturme, M.H. Analysis of Quorum Sensing Regulatory Systems in the Human Isolate Lactobacillus plantarum; Wageningen University and Research: Wageningen, The Netherlands, 2005. [Google Scholar]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, A.O.; Brameyer, S.; Kresovic, D.; Hitkova, I.; Kopp, Y.; Manske, C.; Schubert, K.; Bode, H.B.; Heermann, R. Pyrones as bacterial signaling molecules. Nat. Chem. Biol. 2013, 9, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Brameyer, S.; Kresovic, D.; Bode, H.B.; Heermann, R. Dialkylresorcinols as bacterial signaling molecules. Proc. Natl. Acad. Sci. USA 2015, 112, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Prazdnova, E.V.; Gorovtsov, A.V.; Vasilchenko, N.G.; Kulikov, M.P.; Statsenko, V.N.; Bogdanova, A.A.; Refeld, A.G.; Brislavskiy, Y.A.; Chistyakov, V.A.; Chikindas, M.L. Quorum-sensing inhibition by Gram-positive bacteria. Microorganisms 2022, 10, 350. [Google Scholar] [CrossRef] [PubMed]
- Abbamondi, G.R.; Suner, S.; Cutignano, A.; Grauso, L.; Nicolaus, B.; Toksoy Oner, E.; Tommonaro, G. Identification of N-Hexadecanoyl-L-homoserine lactone (C16-AHL) as signal molecule in halophilic bacterium Halomonas smyrnensis AAD6. Ann. Microbiol. 2016, 66, 1329–1333. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).