Abstract
GeneXpert MTB/RIF is a rapid molecular diagnostic tool capable of simultaneously detecting Mycobacterium tuberculosis and rifampicin resistance. This study aimed to assess the diagnostic precision of GeneXpert MTB/RIF assay to detect pulmonary and extrapulmonary tuberculosis and evaluate the performance for detecting of rifampicin resistance. Of 37,695 samples, 7156 (18.98%) were tuberculosis-positive, and 509 (7.11%) were rifampicin-resistant. The sensitivity, specificity, positive predictive value, negative predictive value, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for pulmonary tuberculosis were 99.87% (95%CI: 99.75–99.94), 99.92% (95%CI: 99.88–99.95), 99.71% (95%CI: 99.54–99.82), 99.97% (95%CI: 99.93–99.98), 21.38% (95%CI: 20.92–21.86), and 99.91% (95%CI: 99.87–99.94), respectively. For extrapulmonary tuberculosis, the sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of GeneXpert MTB/RIF assay accounted for 99.45% (95%CI: 98.73–99.82), 99.84% (95%CI: 99.73–99.92), 98.70% (95%CI: 97.73–99.25), 99.93% (95%CI: 99.84–99.97), 10.64% (95%CI: 9.99–11.31), and 99.80% (95%CI: 99.68–99.88), respectively. Despite its high sensitivity for detecting tuberculosis and rifampicin resistance, GeneXpert MTB/RIF had contradictory results for 20.5% of cases among patients with smear-negative results and 54.9% of cases among patients with a high risk of multidrug-resistant tuberculosis. Of 46% fluoroquinolone-resistant cases, 16.56% (26/157) were multidrug-resistant tuberculosis isolates, and 4.02% (20/498) were isoniazid-resistant, a characteristic distribution leading to about 17.2% of fluoroquinolone-resistance events and relevant marker gyr-A mutations in MDR tuberculosis isolates. Further, our study indicated that increased fluoroquinolone resistance among rifampicin-resistant and isoniazid-resistant tuberculosis endangers the success of newly endorsed MDR-TB regimens.
1. Introduction
Tuberculosis (TB) is a primary reason of morbidity and mortality globally, caused by Mycobacterium tuberculosis (Mtb), which is spread through airborne droplets. Tuberculosis (TB) continues to be endemic in various regions of the world, including in India. It needs surveillance, clinical assessment, testing, contact tracing, and confirmation of diagnosis with supervised or unsupervised treatment regimens for effective eradication [1]. TB was the foremost cause of death from a single contagious agent, ranking above HIV/AIDS until the coronavirus (COVID-19) pandemic emerged. The increase in TB cases from 10.1 million in 2020 to 10.6 million in 2022 reversed many years of slow decline of TB cases worldwide. Similarly, the estimated TB incidence rate (new cases per lakh population per year) increased by 3.9% between 2020 and 2022, following declines in the TB incidence rate by about 2% per year for most of the past two decades [2]. The number of TB patients (new and relapse) has risen 19% from 1,628,161 in 2020 to 1,933,381 in 2021. The main reason for the increase in TB incidence between 2020 and 2021 is the impact of the COVID-19 pandemic on TB detection [3]. Eight countries in the world accounted for more than two-thirds of global TB cases in 2022: India (27%), Indonesia (10%), China (7.1%), the Philippines (7.0%), Pakistan (5.7%), Nigeria (4.6%), Bangladesh (3.6%), and the Democratic Republic of the Congo (3.0%) [2]. Drug-resistant tuberculosis (DR-TB) is a growing issue that threatens TB control programs worldwide, and the incidence rate of MDR/RR-TB cases was 410,000 in 2022. Globally, three countries accounted for the largest share of incident cases of MDR/RR-TB in 2021: India (26% of global cases), the Russian Federation (8.5% of global cases), and Pakistan (7.9% of global cases) [4].
Innovative strategies such as services of rapid molecular diagnostics of TB to people everywhere and high-risk TB patients upfront (accessibility) and an integrated health system approach for the delivery of service with other components, including counselling in the general healthcare system (availability), require exploration and implementation for early diagnosis and decentralized delivery of drug-resistant TB services. Over a half million people have had multidrug-resistant tuberculosis (MDR-TB), and 156,071 were registered in MDR-TB treatment as per the World Health Organization [5] 2020 report. The drug-resistance testing coverage ranges from 46% to 83% among new and formerly treated TB patients [6]. Prompt and ample tuberculosis diagnosis is essential for optimal tuberculosis control strategies, resulting in early treatment of tuberculosis and multidrug-resistant tuberculosis patients. Delays in early diagnosis and appropriate treatment initiation and the high prevalence of HIV in resource-limited settings made tuberculosis and multidrug-resistant-tuberculosis-associated morbidity and mortality relatively high. Molecular tests for tuberculosis and multidrug-resistant tuberculosis have developed remarkably owing to the vital challenges faced by countries with high tuberculosis burden, the emergence of multidrug-resistant tuberculosis, and extremely drug-resistant tuberculosis (XDR-TB) worldwide.
The WHO-endorsed GeneXpert MTB/RIF assay is a fully automated cartridge-based molecular system that revolutionizes tuberculosis (TB) control simultaneously by contributing to the rapid diagnosis of tuberculosis disease and rifampicin drug resistance. Rifampicin-resistant tuberculosis (RR-TB) is a proxy marker for multidrug-resistant tuberculosis (MDR-TB) in more than 90% of cases [7]. Implementing rapid diagnostic tests like GeneXpert and expanding MDR-TB treatment centers at peripheral healthcare facilities were the interventions used to decrease gaps of low case detection and delayed treatment enrolment. The GeneXpert MTB/RIF testing is a rapid molecular diagnostic test performed with an automated cartilage-based GeneXpert machine (Cepheid, Sunnyvale, CA, USA). The GeneXpert tool enables healthcare professionals to improve the speed and quality of TB diagnosis and helps diagnose TB in patients likely to be missed by traditional screening tests [8]. The fact that it allows diagnosis of TB simultaneously as pinpointing resistance to rifampicin treatment is vital in minimizing the transmission of drug-resistant TB in TB-endemic countries. The test function is based on a nucleic-acid amplification assay that detects M. tuberculosis and rifampicin resistance patterns from the sputum and other body fluids [9]. Therefore, this study aimed to assess the wide range of molecular diagnostics for drug-resistant tuberculosis (DR-TB) endorsed by the World Health Organization and assess GeneXpert’s utility for the rapid molecular diagnosis of tuberculosis in a high-burden setting—the southern region of India.
2. Materials and Methods
2.1. Study Setting, Period, and Design
An institution-based retrospective study was conducted from January 2022 to December 2022 in the Intermediate Reference Laboratory, State TB Training Demonstration Centre, Government Hospital for Chest Diseases, Puducherry state, India, to assess the performance utility of the GeneXpert MTB/RIF cartridge in replacement of existing microscopy in the peripheral laboratory. A total of 39,107 TB suspects were enrolled in this study from Puducherry (n = 9819) state and nine adjoining districts (Villupuram n-2912, Kallakuruchi n-3268, Cuddalore n-2442, Thiruchirapalli n-7919, Perambalur n-2326, Thiruvarur n-4019, Nagapattinam n-1048, Tanjavore n-2170, and Thiruvanamalai n-3184) of Tamil Nadu state. All TB patients and rifampicin-resistant TB patients diagnosed by GeneXpert MTB/RIF were the study population for the first-line and second-line line probe assay (LPA) and culture and drug susceptibility testing (DST). Patients with incomplete data and undocumented methods of diagnosis were excluded from the study.
2.2. Sample Processing for Light-Emitting Diode Fluorescent Microscopy
The concentrated samples were smeared on pre-labeled, clean, grease-free microscopic slides and air-dried completely for 15–30 min. The smear was fixed to the slide by passing it over the flame 3 to 5 times for 3 to 4 s each. The slides were placed on a staining rack with the smeared part at the top and the slides not touching each other. The slides were flooded with freshly filtered 0.1% Auramine-phenol and left to stand for 20 min, after which the slides were washed with running water, controlling the water flow to prevent washing away the smear. The excess staining was decolorized entirely with 0.5% acid-alcohol for 2 min and the slides were washed with running water, as before, to wash away the acid alcohol. Then, the slides were counterstained with 0.5% potassium permanganate for 30 s, washed as before with water, and the slides were sloped to air-dry. Stained smears were examined under LED-FM (Primo Star iLED, Carl Zeiss, Gottingen, Germany) with 400× magnification, and 40 fields were examined. LED-FM results were reported for the presence or absence of Acid-Fast Bacilli using the International Union against Tuberculosis and Lung Disease and World Health Organization scale, with a positive result corresponding to ≥1 Acid-Fast Bacilli per 20× for screening and 40× for confirmation [10].
2.3. Expectorated Sputum Sample Processing for GeneXpert MTB/RIF Assay
Using a separate sterile plastic disposable pipette, the GeneXpert sample reagent was added at a 2:1 (v/v) ratio to each specimen in a sputum container with a screw cap. The sputum cup was shaken vigorously 10–20 times using back-and-forth movements in a single shake, and the sample was then incubated in the sputum cup for 15 min at room temperature. As described above, the sputum cup was shaken at least once during incubation. The sputum sample should be liquefied with no visible clumps of sputum after incubation. Then, each GeneXpert MTB/RIF cartridge was labeled with the lab accession number by writing on the sides of the cartridge or using the sterile plastic transfer pipette provided in the GeneXpert/Rif kit; the liquefied sample was drawn into the transfer pipette until the meniscus of the pipette was above the minimum mark and the homogenized, liquefied sample was transferred into the open port of the GeneXpert MTB/RIF cartridge. It was ensured that the correct laboratory number was recorded, matching the cartridge and sputum cup numbers. The pre-labelled barcode in the cartridges was then scanned after switching on the system attached to the GeneXpert instrument. Finally, the cartridge was loaded into the GeneXpert instrument as per the manufacturer’s instructions [11]: The green light stops blinking after clicking to start the test, and the test starts. When the test is finished, the light turns off. Once the run is completed, results are printed automatically. Wait until the system releases the door lock at the run’s end, then open the module door and remove the cartridge. The used cartridges were finally disposed of in the biohazard waste container.
2.4. Lymph Nodes and Other Tissues Sample Processing for GeneXpert MTB/RIF Assay
Lymph nodes and other tissue samples were cut into small pieces in a sterile mortar using a clean, sterile pair of forceps and dissection knives. Approximately 2 mL of sterile phosphate buffer (PBS) was added to a mixer of dissected small pieces of tissue, and sterile PBS solution was ground with a mortar and pestle until a homogeneous suspension was obtained. Next, approximately 0.7 mL of homogenized tissue sample was transferred to a sterile conical screw-capped tube using a transfer pipette. A double volume of Gene- Xpert MTB/RIF Sample Reagent (1.4 mL) was added to 0.7 mL of homogenized tissue using a transfer pipette and vigorously shaken using a vortex for at least 10 s. The suspension was incubated for 10 min at room temperature, and the specimen was shaken vigorously using a vortex for at least 10 s. The processed sample was incubated at room temperature for 5 min and 2 mL was transferred to the GeneXpert MTB/RIF cartridge using a fresh sterile transfer pipette. It was ensured that the correct laboratory number was recorded, matching the cartridge and sputum cup numbers. The pre-labelled barcode was then scanned in the cartridges after switching on the system attached to the GeneXpert instrument. Finally, the cartridge was loaded into the GeneXpert instrument as per the manufacturer’s instructions [12]: The green light stops blinking after clicking to start the test, and the test starts. When the test is finished, the light turns off. Once the run is completed, results are printed automatically. Wait until the system releases the door lock at the run’s end, then open the module door and remove the cartridge. The used cartridges were finally disposed of in the biohazard waste container.
2.5. Processing of Non-Sterile Lymph Nodes and Tissues for GeneXpert MTB/RIF Assay
Lymph nodes and other tissue samples were cut into small pieces in a sterile mortar using a clean, sterile pair of forceps and a sharp, sterile dissection blade. Approximately 2 mL of sterile phosphate buffer was added to ground dissected tissue/PBS solution with a sterile mortar and pestle until a homogeneous suspension was obtained and transferred into a sterile and pre-labeled 50 mL conical tube using a sterile transfer pipette. Sterile 4% Sodium Hydroxide (NaOH) was added equally and the suspension was homogenized using a vortex mixer. The suspension was then incubated for 15 min at room temperature and the tube was filled within 2 cm of the top (e.g., to the 50 mL mark on the tube) with sterile PBS. The whole content was centrifuged at 3000× g for 15 min and the supernatant was discarded into a discard bin containing 5% phenol or other mycobacterial disinfectants. Approximately 1–2 mL sterile PBS was added into deposited pellets using a sterile transfer pipette. About 0.7 mL of homogenized tissue sample was transferred to a sterile conical screw-capped tube using a sterile transfer pipette, and a double volume of GeneXpert MTB/RIF Sample Reagent (1.4 mL) was added to 0.7 mL of homogenized tissue using another sterile transfer pipette. Then, the mixture was vigorously vortexed for at least 10 s and incubated for 10 min at room temperature, and the homogenized specimen was again shaken vigorously and vortexed for at least 10 s. The processed sample was incubated at room temperature for 5 min. Approximately 2 mL of the processed sample was transferred to the GeneXpert MTB/RIF cartridge using a fresh transfer pipette. It was ensured that the correct laboratory number was recorded, matching the cartridge and sputum cup numbers. The pre-labelled barcode was scanned in the cartridges after switching on the system attached to the GeneXpert instrument. Finally, the cartridge was loaded into the GeneXpert instrument as per the manufacturer’s instructions [12]: The green light stops blinking after clicking to start the test, and the test starts. When the test is finished, the light turns off. Once the run is completed, results are printed automatically. Wait until the system releases the door lock at the run’s end, then open the module door and remove the cartridge. The used cartridges were finally disposed of in the biohazard waste container.
2.6. Processing of CSF Samples for GeneXpert MTB/RIF Assay
If the cerebrospinal fluid (CSF) sample volume is less than 2 mL, add an equal volume of the GeneXpert MTB/RIF sample reagent to the CSF sample. Then transfer about 2 mL of the sample mixture directly to the GeneXpert MTB/RIF cartridge. Finally, load the CSF sample cartridge into the GeneXpert instrument as per the manufacturer’s instructions. If the sample volume exceeds 2 mL, transfer all sample content to a sterile conical centrifuge tube and centrifuge for 15 min at 4000 rpm. Carefully discard the supernatant into a discard bin containing 5% phenol or other mycobacterial disinfectants. Then, add a volume of 2 mL of GeneXpert MTB/RIF sample reagent to the deposit using a fresh sterile transfer pipette, and transfer 2 mL of the concentrated CSF sample to the GeneXpert MTB/RIF cartridge. Ensure the correct laboratory number is recorded, matching the cartridge and sputum cup numbers. Scan the pre-labelled barcode in the cartridges after switching on the system attached to the GeneXpert instrument. Finally, load the cartridge into the GeneXpert instrument as per the manufacturer’s instructions [13]. The test starts, and the green light stops blinking after clicking to start the start test. When the test is finished, the light turns off. Once the run is completed, results are printed automatically. Wait until the system releases the door lock at the run’s end, then open the module door and remove the cartridge. The used cartridges are then disposed of in the biohazard waste container.
2.7. DNA Extraction Using GenoLyse for MTBDRplus VER 2.0 Assay
Approximately 1 mL of the liquid culture sample was transferred from each tube to a sterile 1.5 mL microcentrifuge tube with a screw cap using a disposable Pasteur pipette. The samples were centrifuged at 10,000× g for 15 min in a centrifuge. After discarding the supernatant, the pellet was suspended in 100 µL Lysis Buffer (A-LYS) and incubated at 95 °C for 5 min. Approximately 100 µL of Neutralization Buffer (A-NB) was added and the sample was briefly vortexed for 5 s. The samples were centrifuged for 5 min at 10,000× g and 40–80 µL of supernatant was carefully transferred to a separate clean, sterile micro centrifuge tube. The amplification mix (45 µL per PCR tube) was prepared in a room free from contaminating DNA. The amplification Mixer A and B (AM-A and AM-B) have all the reagents required for amplification. After thawing, AM-A and AM-B was mixed carefully. Then, 5 μL of DNA supernatant was added to corresponding PCR tubes except for the contamination control, and 5 µL water was added to one aliquot for the contamination control. All the PCR tubes were placed in the PCR instruments, and the program was run as per the manufacturer’s instructions [14].
2.8. Hybridization for First-Line Drugs
First, 20 µL of pre-warmed Denaturation Solution (DEN, blue) was dispensed in the corner of each of the wells used, and 20 µL of the amplified sample was added, pipetted up and down to mix well, and incubated at room temperature for 5 min. Then, 1 mL of pre-warmed Hybridization Buffer (HYB, green) was carefully added to each well. A strip was placed in each well of the GT Blot tray, ensuring the strips were covered entirely by the solution and that the coated side faces upward. The tray was placed in the GT Blot instrument and incubated at 45 °C for 30 min; the shaking frequency of the GT Blot was to achieve a constant and thorough mixing of the solution. The Hybridization Buffer was aspirated using a sterile Pasteur pipette. Then, 1 mL of Stringent wash solution (STR, red) was added to each strip incubated at 45 °C for 15 min in the GT Blot instrument, and the Stringent wash solution was completely removed using a separate Pasteur pipette. Each strip was washed once with 1 mL of Rinse Solution (RIN) for 1 min in the GT Blot instrument. Next, 1 mL of diluted Conjugate was added to each strip and incubated for 30 min in the GT Blot instrument. The solution was removed using a sterile Pasteur pipette and each strip was washed twice for 1 min with 1 mL of Rinse Solution (RIN) and once for 1 min with approximately 1 mL of distilled water. Then, 1 mL of diluted substrate solution was added to each strip, incubated for 3–20 min, and protected from direct light without shaking. The reaction was stopped as soon as bands were visible by briefly rinsing twice with distilled water. The strips were removed from the tray using tweezers and pasted on an evaluation sheet provided in the kit [15].
2.9. DNA Extraction Using GenoLyse for MTBDRsl VER 2.0 Assay
Approximately 1 mL of culture suspension was transferred into a sterile 1.5 mL screw cap microcentrifuge tube using a disposable Pasteur pipette. The culture suspension was centrifuged at 10,000× g for 15 min in a centrifuge. After discarding the supernatant, the pellet was suspended in 100µL Lysis Buffer (A-LYS) and incubated at 95 °C for 5 min. Approximately 100 µL of Neutralization Buffer (A-NB) was added and the sample was vortexed briefly for 5 s. The liquid suspension was centrifuged at 10,000× g for 5 min and 40–80 µL of supernatant was carefully transferred to a clean, sterile microcentrifuge tube. The amplification mix (45 µL per PCR tube) was prepared in a room free from contaminating DNA. The amplification Mixer A and B (AM-A and AM-B) have all the reagents required for amplification. After thawing, AM-A and AM-B were mixed carefully. Then, 5 μL of DNA supernatant was added to corresponding PCR tubes except for the contamination control, and 5 µL water was added to one aliquot for the contamination control. All the PCR tubes were placed in the PCR instruments, and the program was run as per the manufacturer’s instructions [14].
2.10. Hybridization for Second-Line Drugs
Approximately 20μL of Denaturation Solution was Dispensed (DEN, blue) in the corner of each of the wells in the GT Blot tray, and 20 μL of amplified PCR product was added into each well using a sterile pipette and incubated at room temperature for 5 min. Then, 1 mL of pre-warmed Hybridization Buffer (HYB, green) was carefully added to each well and the tray was gently shaken until the solution was homogenous. A pre-labelled strip was placed into each well using sterile tweezers. The tray was transferred to the GT-Blot instrument and incubated for 30 min at 45 °C. Then, the Hybridization Buffer was completely aspirated using a sterile Pasteur pipette. Next, 1 mL of Stringent wash solution (STR, red) was added to each strip, incubated at 45 °C for 15 min in the GT-Blot instrument, and the Stringent wash solution was completely removed using a separate Pasteur pipette. The strip was washed with 1 mL Rinse Solution (RIN) for 1 min in the GT-Blot instrument, and 1 mL diluted Conjugate was added to each strip and incubated for 30 min in the GT-Blot instrument. The solution was removed using a sterile Pasteur pipette and each strip was washed twice for 1 min with 1 mL of Rinse Solution (RIN) and once for 1 min with approximately 1 mL of distilled water. Then, 1 mL of diluted substrate solution was added to each strip, incubated for 3–20 min, and protected from direct light without shaking. The reaction was stopped as soon as bands were visible by briefly rinsing twice with distilled water. The strips were removed from the tray using tweezers and pasted on an evaluation sheet provided in the kit [16].
2.11. Statistics
The sensitivity, specificity, positive predictive value, negative predictive value, disease prevalence, accuracy, and Kappa Value of GeneXpert MTB/RIF assay were calculated using MedCalc for Windows, version 19.4 (MedCalc Software, Ostend, Belgium) [17].
3. Results
Overall, a total of 39,107 samples of tuberculosis suspects were processed for upfront GeneXpert MTB/RIF testing between January 2022 and December 2022. Of 37,695 (96.4%) samples tested, 18.98% (n = 7156) were positive for tuberculosis and 7.11% (n = 509) were rifampicin-resistant (Figure 1). Among 8581 (22.76%) extrapulmonary samples processed for tuberculosis diagnosis using upfront GeneXpert MTB/RIF assay, 10.72% (n = 920) were positive for tuberculosis, and 3.26% (n = 30) were resistant to the rifampicin drug. Overall, 34,814 samples (92.36%) were referred from public sector facilities, and only 7.64% (n = 2881) were referred from the private sector (Table 1). A total of 1412 (3.61%) GeneXpert MTB/RIF cartridges were wasted in this complete study. A total of 32,825 patients with presumptive TB were enrolled for the study, out of which 15.01% (n = 4927) were positive for M. tuberculosis, and 4.22% (n = 208) were rifampicin-resistant. Among 4870 presumptive DR-TB (Pulmonary) patients, 45.77% (n = 2229) were positive for M. tuberculosis, and 13.50% (n = 301) were rifampicin-resistant (Table 2).
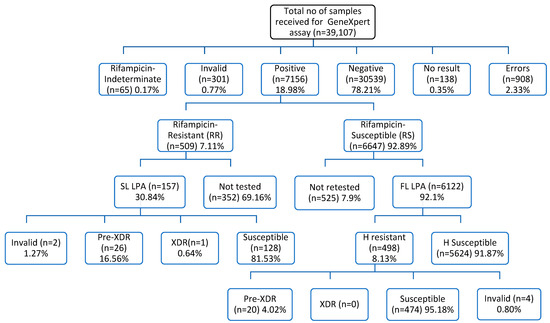
Figure 1.
Samples Flow chart for screening and diagnosis of tuberculosis and drug-resistant tuberculosis cases. SL LPA—Second-Line drug LineProbe Assay; FL LPA—First-Line drug LineProbe Assay; Pre-XDR—Pre-Extremely Drug Resistant; XDR—Extremely Drug Resistant; H—Isoniazid.

Table 1.
Cumulative number of samples received from public and private facilities linked from ten cities in India, 2022.

Table 2.
Laboratory findings of GeneXpert results among presumptive TB and presumptive DR-TB.
Of 2374 samples received from PL-HIV patients, 7.54% (n = 179) were positive for M. tuberculosis, and 5.59% (n = 10) were rifampicin-resistant. Among 2257 pediatric samples processed for diagnosis of TB, 2.22% (n = 50) were positive for M. tuberculosis, 20.53% (n = 2306) were positive for M. tuberculosis among all smear-negative presumptive TB cases—either from previously treated patients or new suspects, and MDR suspect (n = 11,233) samples processed for diagnosis of tuberculosis, and 4.90% (n = 113) were resistant to the rifampicin drug according to the GeneXpert MTB/RIF test. A total of 1820 samples were received from other vulnerable (migratory workers, orphaned, homeless asylum seekers, and displaced people) for diagnosis of tuberculosis using the GeneXpert MTB/RIF assay, out of which 14.45% (n = 263) were positive for M. tuberculosis and 3.80% (n = 10) were rifampicin-resistant. Out of 595 Contacts of TB and DR-TB patient samples tested, 16.97% (n = 101) were positive for M. tuberculosis and 23.76% (n = 24) were rifampicin-resistant. Among 7639 EPTB (extrapulmonary tuberculosis) samples tested, 9.78% (n = 747) were M. tuberculosis-positive and 3.48% (n = 26) were rifampicin-resistant. Of 4026 samples offered for the upfront molecular test, 14.88% (n = 599) were positive for M. tuberculosis and about 1% were rifampicin-resistant (Table 2). Out of the total 3846 presumptive adults (new) notified TB patients who had provided sputum samples for M. tuberculosis diagnosis, 40.90% (n = 1573) were M. tuberculosis-positive and 17.04% (n = 268) were rifampicin-resistant. Of 462 notified pre-treated TB patient samples tested for M. tuberculosis diagnosis, 40.48% (n = 187) were positive for M. tuberculosis and 10.16% (n = 19) were rifampicin-resistant. Among 562 non-responders (drug-sensitive tuberculosis—DSTB and isoniazid mono-resistant tuberculosis—HrTB) tested for M. tuberculosis diagnosis, 83.45% (n = 469) were M. tuberculosis-positive and 2.99% were rifampicin-resistant.
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), disease prevalence, and accuracy of the GeneXpert MTB/RIF assay were calculated from the stratification of patients using the concentrated smear microscopy (Fluorescence) method. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for PTB were found to be 99.87% (95%CI: 0.12–0.07), 99.92% (95%CI: 0.04–0.03), 99.71% (95%CI: 0.17–0.11), 99.97% (95%CI: 0.04–0.01), 21.38% (95%CI: 0.46–0.48), and 99.91% (95%CI: 0.04–0.03), respectively. The Kappa value of 0.997 (95%CI: 0.996-0.998) showed perfect agreement. For EPTB, the sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay counted for 99.45% (95%CI: 0.72–0.37), 99.84% (95%CI: 0.11–0.08), 98.70% (95%CI: 0.97–0.55), 99.93% (95%CI: 0.09–0.04), 10.64% (95%CI: 0.65–0.67), and 99.80% (95%CI: 0.12–0.08), respectively. The Kappa value of 0.990 (95%CI: 0.985–0.995) showed perfect agreement. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for presumptive TB were found to be 99.82% (95%CI: 0.17–0.10), 99.91% (95%CI: 0.04–0.03), 99.51% (95%CI: 0.23–0.16), 99.97% (95%CI: 0.03–0.01), 14.96% (95%CI: 0.38–0.39) and 9.93% (95%CI: 0.04–0.02), respectively (Table 3). The Kappa value of 0.996 (95%CI: 0.995–0.997) showed perfect agreement. For presumptive DR-TB (Pulmonary), the sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay counted for 99.82% (95%CI: 0.28–0.13), 99.77% (95%CI: 0.26–0.15), 99.73% (95%CI: 0.33–0.15), 99.85% (95%CI: 0.25–0.09), 45.73% (95%CI: 1.41–2.41), and 99.92% (95%CI: 0.10–0.05), respectively. The Kappa value 0.996 (95%CI: 0.993–0.998) showed perfect agreement.

Table 3.
Sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the TB screening tool among presumptive TB and presumptive DR-TB.
Among 32,825 presumptive TB enrolled for this study, the sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for PL-HIV out of presumptive TB (n = 2374) were 99.44% (95%CI: 2.53–0.55), 99.91% (95%CI: 0.24–0.08), 98.88% (95%CI: 3.20–0.84), 99.95% (95%CI: 0.27–0.04), 7.50% (95%CI: 1.03–1.13), and 99.87% (95%CI: 0.24–0.10), respectively. The Kappa value of 0.991 (95%CI: 0.981–1.00) showed perfect agreement. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for pediatric cases out of presumptive TB (n = 2257) were found to be 97.96% (95%CI: 8.81–1.99), 99.91% (95%CI: 0.24–0.08), 96.00% (95%CI: 10.28–2.27), 99.95% (95%CI: 0.26–0.04), 2.17% (95%CI: 0.56–0.69), and 99.87% (95%CI: 0.26–0.10), respectively. The Kappa value of 0.969 (95%CI: 0.934–1.00) showed perfect agreement. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for smear-negative, X-ray-suggestive TB (n = 11,233) were found to be 100% (95%CI: 0.16–0.0), 99.99% (95%CI: 0.05–0.01), 99.96% (95%CI: 0.27–0.03) 100% (95%CI: 0.04–0.00), 20.52% (95%CI: 0.74–0.76), and 99.99% (95%CI: 0.04–0.01), respectively. The Kappa value of 0.999 (95%CI: 0.985–0.995) showed perfect agreement. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for other vulnerable group TB (n = 1820) were found to be 99.62% (95%CI: 1.73–0.37), 99.87% (95%CI: 0.33–0.11), 99.24% (95%CI: 2.21–0.57), 99.94% (95%CI: 0.39–0.05), 14.40% (95%CI: 1.59–1.69), and 99.84% (95%CI: 0.32–0.13), respectively(Table 3). The Kappa value of 0.993 (95%CI: 0.986–1.00) showed perfect agreement. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for contacts of TB and DR-TB patients (n = 595) were found to be 100% (95%CI: 3.69–0.00), 99.40% (95%CI: 1.15–0.48), 97.03% (95%CI: 5.67–1.99), 100% (95%CI: 0.74–0.00), 16.47% (95%CI: 2.89–3.23), and 99.50% (95%CI: 0.97–0.40), respectively. The Kappa value of 0.982 (95%CI: 0.961–1.00) showed perfect agreement. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for extrapulmonary TB (n = 7639) were found to be 99.60% (95%CI: 0.78–0.32), 99.90% (95%CI: 0.11–0.06), 99.06% (95%CI: 1.00–0.39), 99.96% (95%CI: 0.09–0.03), 9.73% (95%CI: 0.66–0.68), and 99.87% (95%CI: 0.11–0.07), respectively. The Kappa value of 0.993 (95%CI: 0.988–0.997) showed perfect agreement. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for the upfront molecular test offered(n = 4026) were found to be 99.83% (95%CI: 0.76–0.17), 99.97% (95%CI: 0.13–0.03), 99.83% (95%CI: 1.00–0.15), 99.97% (95%CI: 0.18–0.03), 14.88% (95%CI: 1.09–1.54), and 99.95% (95%CI: 0.13–0.04), respectively. The Kappa value of 0.998 (95%CI: 0.995–1.00) showed perfect agreement.
Among the 4870 presumptive DR-TB (pulmonary) enrolled for this study, the sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for notified TB patients (New)–UDST (n = 3846) were 99.94% (95%CI: 0.34–0.06), 99.87% (95%CI: 0.25–0.10), 99.81% (95%CI: 0.40–0.13), 99.96% (95%CI: 0.27–0.03), 40.85% (95%CI: 1.56–1.57), and 99.90% (95%CI: 0.17–0.07), respectively. The Kappa value of 0.998 (95%CI: 0.996–0.999) showed perfect agreement. The sensitivity, specificity, PPV, NPV, diseases prevalence, and accuracy of the GeneXpert MTB/RIF assay for notified TB patients (previously treated)—UDST (n = 462) were found to be 99.47% (95%CI: 2.41–0.52), 99.64% (95%CI: 1.65–0.35), 99.47% (95%CI: 3.13–0.45), 99.64% (95%CI: 2.15–0.31), 40.48% (95%CI: 4.51–4.63), and 99.57% (95%CI: 1.12–0.38), respectively (Table 3). The Kappa value of 0.991 (95%CI: 0.979–1.00) showed perfect agreement. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for non-responders (n = 562) were found to be 99.57% (95%CI: 1.10–0.38), 97.85% (95%CI: 5.40–1.89), 99.57% (95%CI: 1.23–0.32), 97.85% (95%CI: 5.91–1.60), 83.45% (95%CI: 3.33–2.98), and 99.29% (95%CI: 1.10–0.62), respectively. The Kappa value of 0.974 (95%CI: 0.974–0.999) showed perfect agreement.
The GeneXpert assay test provides semi-quantitative M. tuberculosis detection based on the probe’s Cycle Threshold (Ct)—the number of PCR cycles required for amplifying MTB DNA to detectable levels. GeneXpert assay cycle threshold values (CT) are a semi-quantitative measure of bacillary burden in the specimen. Out of 509 detected RR-TB results, 36 (7.07%) are reported as “High” (Ct < 16), and 98 (19.25%), 163 (32.02%), and 212 (41.65%) are reported as “Medium” (Ct16-22),” Low” (Ct 22-28), and “Very Low” (Ct > 28), respectively (Table 4). Delta Ct (ΔCt) max was calculated as the difference between the earliest and latest Ct across the five molecular beacon (A–E) probes (Table 3). Out of 509 rifampicin-resistant cases, 243 (47.74%) were reported as “dropout” (no hybridization), and 266 (52.25%) were reported as “delayed” (ΔCt > 4), as represented in Table 4. The most common probes for RIF resistance detection were E (n = 68, 13.36%), D (n = 57, 11.20%), and B (n = 55, 10.81%). The probe with the most delayed binding Ct value was categorized as ΔCt 4.1–4.9 (49, 20.16%) and ΔCt > 5 (194, 79.84%).

Table 4.
Distribution of rifampicin-resistance patients in relation to different regions of rpoB gene detected through Probes A, B, C, D, and E.
Among 7156 (18.33%) M. tuberculosis-positive cases, 7.11% (n = 509) were rifampicin-resistant, and 92.89% (n = 6647) were rifampicin-sensitive tuberculosis (Table 1). Of 157 (30.84%) RR tested for MDBDRsl assay, 16.56% (n = 26) were pre-XDR, and 0.64% (n = 1) were XDR tuberculosis. About 69.16% (n = 352) were not tested for MDBDRsl assay due to the non-availability of samples. Of 6647 (92.89%) RS-TB cases, 92.1% (n = 6122) were tested for the MDBDRplus assay, and 525 (7.9%) were not tested for the MDBDRplus assay due to non-availability of samples. Among 6122 RS-TB cases tested for the MDBDRplus assay, 8.13% (n = 498) were HR tuberculosis, and 91.87% (n = 5624) were HS tuberculosis. Of 498 (8.13%) HR tested for the MDBDRsl assay, 4.02% (n = 20) were pre-XDR tuberculosis (Figure 1). Of 46 fluoroquinolone-resistant cases, 16.56% (26/157) were multidrug-resistant tuberculosis isolates and 4.02% (20/498) isoniazid-resistant were fluoroquinolone-resistant, a characteristic distribution leading to about 17.2% of fluoroquinolone-resistance events and relevant marker gyr-A mutations in MDR tuberculosis isolates (Table 5).

Table 5.
The frequency and mutations confer resistance to the drug fluoroquinolone.
4. Discussion
Tuberculosis is the most dreadful infectious disease in the world, with high morbidity and mortality among people. Thus, early detection is of utmost importance for reducing deaths and transmission. The lack of rapid and accurate diagnostic tests hampers global TB control. GeneXpert MTB/RIF assay (Cepheid Inc., Sunnyvale, CA, USA) is the semi-quantitative real-time polymerase chain reaction (PCR) used to rapidly detect the M. tuberculosis complex and rifampicin resistance by amplifying a DNA fragment containing the 81bp hotspot region of the rpoB gene (codons 426–452) that is then hybridized to five molecular beacon probes.
Overall, this study’s MTB and rifampicin-resistant TB frequency was 18.98% and 7.11%, respectively. The incidence rate of M. tuberculosis (18.33%) in this study was higher than the previous study that reported 6.5% [18] and 7.9% [19] in Ethiopia, 13.8% [20] in Nepal, and 12% [21] in India. The incidence rate of M. tuberculosis infection among total suspects (n = 39,107) tested for GeneXpert MTB/RIF was 18.98% (7156/37,695), which is lower than the previous study that reported 38.77% [22] in China, 22.65% [23] and 22.9% [24] in Nigeria, and 23.82% [25] in India. The 3.26% positivity among the 8581 extrapulmonary samples tested for tuberculosis using the GeneXpert MTB/RIF assay in this study is lower than in the previous study (13%) conducted by Anwar et al. [26].
Elbrolosy et al. [27] reported in their study that the sensitivity and specificity of the GeneXpert MTB/RIF assay for PTB were 90.2% and 86.9%, respectively. In contrast, for EPTB, the sensitivity and specificity of GeneXpert MTB/RIF assay counted for 81.6% and 78.9%, respectively. Mulengwa et al. [28] reported the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the GeneXpert MTB/RIF test were 91.6%, 95.3%, 83%, and 97.80%, respectively. However, in this study, the sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for PTB were found to be 99.87%, 99.92%, 99.71%, 99.97%, 21.38%, and 99.91% respectively and for EPTB, the sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay counted for 99.45%, 99.84%, 98.70%, 99.93%, 10.64%, and 99.80%, respectively, compared to concentrated smear (Fluorescence Microscopy) method. Raina et al. [29] reported that the sensitivity, specificity, PPV, and NPV of the GeneXpert MTB/RIF were 100%, 99.5%, 97.5%, and 100%, respectively, compared to the gold standard culture method.
Of 2374 PLHIV samples enrolled for this study, the sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for PLHIV out of presumptive TB were 99.44%, 99.91%, 98.88%, 99.95%, 7.50%, and 99.87%, respectively. In countries with low endemicity rates, the sensitivity and the specificity of GeneXpert MTB/RIF vary between 82%, 95%, and 98% for sensitivity and 96% and 99% for specificity. However, in countries with high endemicity, these rates vary between 80% and 88% for sensitivity and between 95 and 98% for specificity [30,31,32]. In another study, Faria et al. [33] reported that the sensitivity of the GeneXpert MTB/RIF assay ranges from 68% to 100%. Specificity ranged from 91.7% to 100%, the positive predictive value from 79.2% to 96.1%, and the negative predictive value from 84.6% to 99.3%.
In their study, Cox et al. [34] reported 76% sensitivity and 98% specificity for 5717 smear-negative samples processed for GeneXpert MTB/RIF. In our study, the sensitivity of GeneXpert MTB/RIF was also high—100% among smear-negative, and specificity was 99.99% with 99.99% accuracy. Rimal et al. [35] reported the sensitivity, specificity, positive predictive value, and negative predictive values of the GeneXpert MTB/RIF assay for smear-negative sputum samples were 74.3%, 96.6%, 86.7%, and 92%, respectively. In this study, 11,223 sputum samples were collected and processed by microscopy followed by GeneXpert MTB/RIF assay. The sensitivity, specificity, PPV, NPV, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay for smear-negative, X-ray-suggestive TB (n = 11,233) were found to be 100%, 99.99%, 99.96%, 100%, 20.52%, and 99.99%, respectively.
Hebte et al. [36] reported 89.1% (n = 106) positivity and 4.2% (n = 5) rifampicin-resistant TB out of 119 index TB cases enrolled for GeneXpert assay. The positivity rate among the 494 contacts of TB and drug-resistant tuberculosis patients registered for this study was 16.97%. The sensitivity, specificity, positive predictive value, negative predictive value, disease prevalence, and GeneXpert MTB/RIF assay accuracy were 100%, 99.40%, 97.03%, 100.00%, 16.47%, and 99.50, respectively. Our study reported lower positivity than that reported by Hebte et al., and higher than that reported by Gebretsadik et al. [37], which indicated 8.98% (n = 38) positivity and 5.3% (n = 3) rifampicin-resistant TB out of 423 index TB cases enrolled for GeneXpert assay. In their recent study, Gurung et al. [38] reported 4.5% (n = 770) positivity out of 17,114 index TB cases registered for GeneXpert assay. Kalra et al. [39] reported 6.6% positivity (n = 6270) and 8.7% (n = 545) rifampicin-resistant of the total 94,415 presumptive pediatric TB cases diagnosed via GeneXpert MTB/RIF assay. However, the positivity rate among the 2257 pediatric cases enrolled for this study was 2.22%. The sensitivity, specificity, positive predictive value, negative predictive value, disease prevalence, and GeneXpert MTB/RIF assay accuracy were 97.96%, 99.91%, 96.00%, 99.95%, 2.17%, and 99.87%, respectively.
In a previous study, Ibrahim et al. [23] reported the incidence of M. tuberculosis and rifampicin resistance was 22.68% and 4.50%, respectively, out of 2451 samples tested with GeneXpert assay. Of a total of 990 presumptive tuberculosis cases tested [20], the estimated prevalence of M. tuberculosis in presumptive TB patients was 13.8% (95%CI: 11.88–16.16%), and the estimated prevalence of rifampicin resistance in M. tuberculosis-confirmed patients was 10.2% (4.97–15.1%). Of 132 notified new TB cases enrolled in a previous study [40], the positivity on GeneXpert MTB/RIF assay was 78.79%. The sensitivity, specificity, PPV, and NPV of GXP in diagnosing and detecting rifampicin resistance in pulmonary TB were 95%, 93%, 98%, 84%, and 96%, 100%, 100%, and 96%, respectively. The positivity rate among the 3846 notified new TB cases enrolled for this study was 40.90%. The sensitivity, specificity, positive predictive value, negative predictive value, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay were 99.94%, 99.87%, 99.81%, 99.96%, 40.85%, and 99.90%, respectively. Our study reported a higher (40.90%) incidence of M. tuberculosis and rifampicin-resistance (17.04%) rate than the previous studies mentioned here.
In a recent study, Worku et al. [41] reported an 11.9% incidence of M. tuberculosis-positivity rate and 2.5% rifampicin resistance out of 1828 smear-negative and re-treatment cases tested with GeneXpert MTB/RIF assay. Of 462 notified previously treated patients enrolled for this study, the TB-positivity rate was 40.98%. The sensitivity, specificity, positive predictive value, negative predictive value, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay were 99.47%, 99.64%, 99.47%, 99.64%, 40.48%, and 99.57%, respectively. Farra et al. [42,43] reported a 79.1% (488/617) M. tuberculosis-positivity rate and 42.2% rifampicin-resistant in confirmed M. tuberculosis (206/488) in their study out of 617 samples (55.8% relapse; 31.6% failure and 10.2% defaulter) tested using the GeneXpert MTB/RIF assay. Of 562 non-responders (DS-TB and INH-resistant TB) enrolled in this study, the tuberculosis positivity and rifampicin resistance detection rates were 83.45% and 2.99%, respectively, and the sensitivity, specificity, positive predictive value, negative predictive value, disease prevalence, and accuracy of the GeneXpert MTB/RIF assay were 99.57%, 97.85%, 99.57%, 97.85%, 83.45%, and 99.29%, respectively.
In this study, 16.56% (26/157) of multidrug-resistant tuberculosis isolates and 4.02% (20/498) isoniazid-resistant are fluoroquinolone-resistant, a characteristic distribution leading to about 17.2% of fluoroquinolone resistance events and relevant marker gyr-A mutations in MDR tuberculosis isolates. Dreyer et al. [44] reported 69.2% (703/1016) fluoroquinolone resistance among 1016 multidrug-resistant tuberculosis isolates tested for resistance. In India, 36% of the multidrug-resistant tuberculosis isolates are browned to have additional resistance to fluoroquinolone [45,46], and about 3% of MDR-TB isolates are estimated to be extensively drug-resistant (XDR-TB). Sharma et al. [47] reported 3.2% (35/1099) fluoroquinolone resistance among first-line drug-sensitive tuberculosis cases. Furthermore, the increase in fluoroquinolone resistance among isoniazid-resistant tuberculosis suggests that the active adoption of antibiotic stewardship in the community is urgently required.
About 3.6% (n = 1412) of samples were not processed due to challenges in obtaining resamples. Of 1412 samples, 65, 301, 138, and 908 were rifampicin-indeterminate, invalid, no result, and error, respectively. The acceptable rate of rifampicin-indeterminate, invalid, no result, and error calculations need to be defined. This study reported that 69.16% (352/509) of rifampicin-resistant patient samples were not processed to further determine the drug susceptibility pattern of fluoroquinolones and second-line tuberculosis drugs. The control of tuberculosis, once unchecked, is extremely difficult to contain and manage, requiring a multidisciplinary, coordinated set of activities. The cornerstones of classic tuberculosis control approaches include the following:
- Early diagnosis.
- Novel case-finding methods beyond healthcare facilities.
- Shorter and simpler successful treatment regimens for drug-sensitive and drug-resistant tuberculosis.
- A greater focus on prevention strategies.
- Steps to reduce mortality and transmission in adults and children.
The status quo for many rifampicin-resistant patients is a severe systemicness characterized by significant lung damage and high mycobacterial burden. Early detection of rifampicin resistance may facilitate better treatment outcomes and less transmission. Similarly, earlier diagnosis would reduce cumulative immunopathological and structural lung damage (morbidity) and potentially reduce mortality. The molecular characteristics of the disease burden and resistant pattern using the GeneXpert MTB/RIF assay, together with Geographic Information System (GIS) mapping of the location where the specimen was received and tested, could be used as a crude epidemiological tool to identify hot spots of tuberculosis transmission and changes in patterns of circulating rifampicin resistance strains.
5. Conclusions
Our study unveiled a high positivity rate of M. tuberculosis in presumptive TB and presumptive drug-resistant tuberculosis patients. Our analysis also demonstrated a high rifampicin-resistant (RR-TB) tuberculosis rate among M. tuberculosis-confirmed patients. The performance of GeneXpert MTB/RIF detected in M. tuberculosis and rifampicin-resistant patients agrees with that of other researchers who established the diagnosis in a significant proportion of cases. A high rate was observed in both previously treated and treatment-naive patients. In conclusion, GeneXpert can be a valuable diagnostic tool in patients of suspected pulmonary tuberculosis, either Acid-Fast Bacilli smear-negative or -positive, due to its rapidity and synchronized detection of rifampicin resistance, especially advantageous in a patient with MDR and HIV-associated tuberculosis. This study confirms that GeneXpert remains a rapid diagnostic tool for diagnosing TB and confirming sensitivity/resistance to RIF in pulmonary and extrapulmonary samples. The high sensitivity and specificity of GeneXpert MTB/RIF allows ruling out the disease with a high degree of confidence. Molecular epidemiological studies to understand the genetic diversity of M. tuberculosis and link the index cases with secondary infection among close contacts would be valuable. Our study indicated that increased fluoroquinolone resistance among rifampicin-resistant and isoniazid-resistant tuberculosis endangers the success of newly endorsed MDR-TB regimens. Furthermore, fluoroquinolone resistance among isoniazid-resistant tuberculosis suggests that the active adoption of antibiotic stewardship in the community is urgently required. This study recommends that the GeneXpert MTB/RIF assay be used as a replacement for smear microscopy in high-burden peripheral laboratories for the screening of tuberculosis and rifampicin-resistant tuberculosis as early as possible for treatment management.
Author Contributions
Methodology—U.B.; Formal Analysis and Supervision—M.M., G.S. and A.F.; Investigation, Writing and Data Curation—V.R., A.M. (Aaina Muralidhar), A.M. (Anbazhagi Muthukumar) and R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jain, V.K.; Iyengar, K.P.; Samy, D.A.; Vaishya, R. Tuberculosis in the era of COVID-19 in India. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2023; World Health Organization: Geneva, Switzerland, 2023; Available online: https://www.who.int/publications/m/item/global-tuberculosis-report-2023 (accessed on 7 November 2023).
- World Health Organization. Global Tuberculosis Report 2022; World Health Organization: Geneva, Switzerland, 2022; Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 27 October 2022).
- Dean, A.S.; Auguet, O.T.; Glaziou, P.; Zignol, M.; Ismail, N.; Kasaeva, T.; Floyd, K. 25 years of surveillance of drug-resistant tuberculosis: Achievements, challenges, and way forward. Lancet Infect. Dis. 2022, 22, e191–e196. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020; Available online: https://www.who.int/publications/i/item/9789240013131 (accessed on 14 October 2022).
- Nandlal, L.; Perumal, R.; Naidoo, K. Rapid Molecular Assays for the Diagnosis of Drug-Resistant Tuberculosis. Infect. Drug Resist. 2022, 15, 4971–4984. [Google Scholar] [CrossRef] [PubMed]
- Mulu, W.; Abera, B.; Yimer, M.; Hailu, T.; Ayele, H.; Abate, D. Rifampicin—Resistance pattern of Mycobacterium tuberculosis and associated factors among presumptive tuberculosis patients referred to Debre Markos Referral Hospital, Ethiopia: A cross-sectional study. BMC Res. Notes 2017, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Tamirat, K.S.; Kebede, F.B.; Baraki, A.G.; Akalu, T.Y. The Role of GeneXpert MTB/RIF in Reducing Treatment Delay Among Multidrug Resistance Tuberculosis Patients: A Propensity Score Matched Analysis. Infect. Drug Resist. 2022, 15, 285–294. [Google Scholar] [CrossRef]
- Pongpeeradech, N.; Kasetchareo, Y.; Chuchottaworn, C.; Lawpoolsri, S.; Silachamroon, U.; Kaewkungwal, J. Evaluation of the use of GeneXpert MTB/RIF in a zone with high burden of tuberculosis in Thailand. PLoS ONE 2022, 17, e0271130. [Google Scholar] [CrossRef]
- International Union against Tuberculosis and Lung Disease. The Public Health Service National Tuberculosis Reference Laboratory and the National Laboratory Network; International Union against Tuberculosis and Lung Disease: Paris, France, 1998. [Google Scholar]
- Kabir, S.; Parash, M.T.H.; Emran, N.A.; Hossain, A.B.M.T.; Shimmi, S.C. Diagnostic challenges and Gene-Xpert utility in detecting Mycobacterium tuberculosis among suspected cases of Pulmonary tuberculosis. PLoS ONE 2021, 16, e0251858. [Google Scholar] [CrossRef]
- Mukhida, S.; Vyawahare, C.R.; Mirza, S.B.; Gandham, N.R.; Khan, S.; Kannuri, S. Role of GeneXpert MTB/RIF assay for the diagnosis of cervical lymph node tuberculosis and rifampicin resistance. Tzu Chi Med. J. 2022, 34, 418–422. [Google Scholar]
- Xpert MTB/RIF System for the Diagnosis of Pulmonary and Extra-Pulmonary TB and Rifampicin Resistance in Adults and Children. A Pre-Publication Version of the Policy Guidance. Available online: http://www.stoptb.org/wg/gli/assets/documents/WHOPolicyStatementonXpertMTB-RIF2013pre-publication22102013.pdf (accessed on 1 March 2017).
- Smita, S.S.; Venkatesh, K.; Usharani, B.; Anbazhagi, S.; Vidya Raj, C.K.; Chitra, A.; Muthuraj, M. Prevalence and factors associated with multidrug-resistant tuberculosis in South India. Sci. Rep. 2020, 10, 17552. [Google Scholar]
- Vidyaraj, C.K.; Chitra, A.; Smita, S.; Muthuraj, M.; Govindarajan, S.; Usharani, B.; Anbazhagi, A. Prevalence of rifampicin-resistant Mycobacterium tuberculosis among human-immunodeficiency-virus-seropositive patients and their treatment outcomes. J. Epidemiol. Glob. Health 2017, 7, 289–294. [Google Scholar] [CrossRef]
- Aaina, M.; Venkatesh, K.; Usharani, B.; Anbazhagi, M.; Rakesh, G.; Muthuraj, M. Risk Factors and Treatment Outcome Analysis Associated with Second-Line Drug-Resistant Tuberculosis. J. Respir. 2022, 2, 1–12. [Google Scholar] [CrossRef]
- MedCalc for Windows, version 19.4; MedCalc Software: Ostend, Belgium, 2021. Available online: www.medcalc.org/calc(accessed on 14 October 2022).
- Sinshaw, W.; Kebede, A.; Bitew, A.; Tesfaye, E.; Tadesse, M.; Mehamed, Z. Prevalence of tuberculosis, multidrug resistant tuberculosis and associated risk factors among smear negative presumptive pulmonary tuberculosis patients in Addis. BMC Infect. Dis. 2019, 19, 641. [Google Scholar] [CrossRef] [PubMed]
- Wasihun, A.G.; Dejene, T.A.; Hailu, G.G. Frequency of MTB and rifampicin resistance MTB using Xpert-MTB/RIF assay among adult presumptive tuberculosis patients in Tigray, Northern Ethiopia: A cross sectional study. PLoS ONE 2020, 15, e0240361. [Google Scholar] [CrossRef] [PubMed]
- Shiv, K.S.; Pramod, R.B.; Anjana, S.; Deepak, D.; Deepa, G.; Renu, S. Rifampicin-resistant Mycobacterium tuberculosis by GeneXpert MTB/RIF and Associated Factors among Presumptive Pulmonary Tuberculosis Patients in Nepal. Infect. Drug Resist. 2020, 13, 2011–2019. [Google Scholar]
- William, A.; Yogita Rai, Y.; Ravinder Kaur, R. Evaluation of Rifampicin-resistant Tuberculosis in Pediatric Patients by GeneXpert MTB/RIF. J. Microbiol. Infect. Dis. 2021, 11, 81–87. [Google Scholar] [CrossRef]
- Zhua, W.; Wanga, Y.; Lic, T.; Chenc, W.; Wang, W. Gap to End-TB targets in eastern China: A joinpoint analysis from population-based notification data in Zhejiang Province, China, 2005–2018. Int. J. Infect. Dis. 2021, 104, 407–414. [Google Scholar] [CrossRef]
- Ibrahim, M.M.; Tom, M.I.; Umoru, M.A.; Jidda, B.U.; Mustafa, A.I.; Adam, M.; Akbar, S. Trends in the incidence of Rifampicin resistant Mycobacterium tuberculosis infection in northeastern Nigeria. Sci. Afr. 2022, 17, e01341. [Google Scholar] [CrossRef]
- Peter, O.; Ikuabe, I.D.E. Assay in patients with pulmonary tuberculosis in Yenagoa, Nigeria. Pan Afr. Med. J. 2018, 29, 1–4. [Google Scholar]
- Nemagouda, S.K. A three-year experience with genexpert MTB/RIF assay in tuberculosis control programme (RNTCP)—A clinical study. J. Evol. Med. Dent. Sci. 2019, 8, 3080–3083. [Google Scholar] [CrossRef]
- Khan, A.S.; Ali, S.; Khan, M.T.; Ahmed, S.; Khattak, Y.; Irfan, M.; Sajjad, W. Comparison of GeneXpert MTB/RIF assay and LED-FM microscopy for the diagnosis of extra pulmonary tuberculosis in Khyber Pakhtunkhwa, Pakistan. Braz. J. Microbiol. 2018, 4, 909–913. [Google Scholar] [CrossRef]
- Elbrolosy, A.M.; Helbawy, R.H.E.; Mansour, O.M.; Latif, R.A. Diagnostic utility of GeneXpert MTB/RIF assay versus conventional methods for diagnosis of pulmonary and extrapulmonary tuberculosis. BMC Microbiol. 2021, 21, 144. [Google Scholar] [CrossRef] [PubMed]
- Mulengwa, D.L.; Maropeng Charles Monyama, M.C.; Lebelo, S.L. Evaluation of the GeneXpert MTB/RIF assay performance in sputum samples with various characteristics from presumed pulmonary tuberculosis patients in Shiselweni region, Eswatini. Infect. Dis. 2022, 54, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Raina, C.; Sabita, B.; Alina, S.; Manoj, P.; Brajendra, S.; Yengkokpam, S.; Ranjit, S.; Zareena, F.; Rachana, M.; Ali, A.R.; et al. Diagnostic performance of GeneXpert MTB/RIF assay compared to conventional Mycobacterium tuberculosis culture for diagnosis of pulmonary and extra pulmonary tuberculosis, Nepal. Narra J. 2021, 1, e33. [Google Scholar] [CrossRef]
- Li, S.; Liu, B.; Peng, M.; Chen, M.; Yin, W.; Tang, H. Diagnostic accuracy of Xpert MTB/RIF for tuberculosis detection in different regions with different endemic burden: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0180725. [Google Scholar] [CrossRef] [PubMed]
- Opota, O.; Zakham, F.; Mazza-Stalder, J.; Nicod, L.; Greub, G.; Jaton, K. Added Value of Xpert MTB/RIF Ultra for Diagnosis of Pulmonary Tuberculosis in a Low-Prevalence Setting. J. Clin. Microbiol. 2019, 57, e01717-18. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Liu, F.; Lu, X.; Huang, Q. Evaluation of GeneXpert MTB/RIF for detecting mycobacterium tuberculosis in a hospital in China. J. Int. Med. Res. 2017, 45, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Faria, M.G.B.F.; Andrade, R.L.P.; Camillo, A.J.G.; Leite, K.F.S.; Saita, N.M.; Bollela, V.R. Effectiveness of GeneXpert in the diagnosis of tuberculosis in people living with HIV/AIDS. Rev. Saude Publica 2021, 55, 89. [Google Scholar] [CrossRef]
- Cox, H.; Dickson, H.; Ndjeka, N.; Hoog, A.G.; Cobelens, A.F.; Stevens, W. Delays and loss to follow-up before treatment of drug-resistant tuberculosis following implementation of Xpert MTB/RIF in South Africa: A retrospective cohort study. PLoS Med. 2017, 14, e1002238. [Google Scholar] [CrossRef]
- Rimal, R.; Shrestha, D.; Pyakurel, S.; Poudel, R.; Shrestha, P.; Rai, K.R.; Ghimire, G.R.; Rai, G.; Rai, S.K. Diagnostic performance of GeneXpert MTB/RIF in detecting MTB in smear-negative presumptive TB patients. BMC Infect. Dis. 2022, 22, 321. [Google Scholar] [CrossRef]
- Habte, D.; Melese, M.; Hiruy, N.; Gashu, Z.; Jerene, D.; Moges, F.; Yifru, S.; Girma, B.; Kassie, Y.; Haile, Y.K.; et al. The additional yield of GeneXpert MTB/RIF test in the diagnosis of pulmonary tuberculosis among household contacts of smear positive TB cases. Int. J. Infect. Dis. 2016, 49, 179–184. [Google Scholar] [CrossRef]
- Gebretsadik, D.; Ahmed, N.; Kebede, E.; Mohammed, E.; Belete, M.A. Prevalence of Tuberculosis by Automated GeneXpert Rifampicin Assay and Associated Risk Factors among Presumptive Pulmonary Tuberculosis Patients at Ataye District Hospital, North East Ethiopia. Infect. Drug Resist. 2020, 13, 1507–1516. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.C.; Dixit, K.; Paudel, R.; Sah, M.K.; Pandit, R.N.; Aryal, T.P.; Khatiwada, S.U.; Majhi, G.; Dhital, R.; Paudel, P.R.; et al. Comparing Additionality of Tuberculosis Cases Using GeneXpert or Smear-Based Active TB CaseFinding Strategies among Social Contacts of Index Cases in Nepal. Trop. Med. Infect. Dis. 2023, 8, 369. [Google Scholar] [CrossRef] [PubMed]
- Kalra, A.; Parija, D.; Raizada, N.; Sachdeva, K.S.; Rao, R.; Swaminathan, S. Upfront Xpert MTB/RIF for diagnosis of pediatric TB—Does it work? Experience from India. PLoS ONE 2020, 15, e0236057. [Google Scholar] [CrossRef] [PubMed]
- Sasikumar, C.; Utpat, K.; Desai, U.; Joshi, J. The role of Genexpert in the diagnosis of Mycobacterium tuberculosis. Adv. Respir. Med. 2020, 88, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Worku, M.; Agonafir, M.; Yassin, M.A.; Yassin, M.A.; Datiko, D.G.; Theobald, S.; Cuevas, L.E. Use of Xpert MTB/RIF for the Identification of TB and Drug Resistance among Smear-Negative and Re-Treatment Cases in Rural Areas of Ethiopia. Open Microbiol. J. 2019, 13, 188–192. [Google Scholar] [CrossRef]
- Farra, A.; Manirakiza, A.; Yambiyo, B.M.; Zandanga, G.; Lokoti, B.; Arthaud, A.B.; Ngaya, G.; Hermana, G.; Ourandji, L.M.; Ignaleamoko, A.; et al. Surveillance of Rifampicin Resistance with GeneXpert MTB/RIF in the National Reference Laboratory for Tuberculosis at the Institut Pasteur in Bangui, 2015–2017. In Open Forum Infectious Diseases; Oxford University Press: Oxford, UK, 2023. [Google Scholar] [CrossRef]
- Zhou, R.; Zheng, T.; Luo, D.; Zhu, M.; Li, Q.; Xu, Y.; Wang, D.; Luo, J.; Zeng, C.; Wei, G.; et al. Drug resistance characteristics of Mycobacterium tuberculosis isolates obtained between 2018 and 2020 in Sichuan, China. Epidemiol. Infect. 2022, 150, e27. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, V.; Mandal, A.; Dev, P.; Merker, M.; Barilar, I.; Utpatel, C.; Nilgiriwala, K.; Rodrigues, C.; Crook, D.W.; Rasigade, J.P. High fluoroquinolone resistance proportions among multidrug-resistant tuberculosis driven by dominant L2M.tuberculosis clones in the Mumbai Metropolitan region. Genome Med. 2022, 14, 95. [Google Scholar] [CrossRef]
- WHO-Global Tuberculosis Report 2020 [Internet]. WHO. World Health Organization. Available online: http://www.eho.int/tb/publications/global_report/en/ (accessed on 4 November 2020).
- Lee, H.W.; Yim, J.J. Fluoroquinolone resistance in multidrug-resistant tuberculosis patients. Korean J. Intern. Med. 2019, 34, 286–287. [Google Scholar] [CrossRef]
- Rohini Sharma, R.; Singha, B.K.; Kumar, K.; Ramachandran, R.; Jorwa, P. Presence of Fluoroquinolone mono-resistance among drug-sensitive Mycobacterium tuberculosis isolates: An alarming trend and implications. Clin. Epidemiol. Glob. Health 2019, 7, 363–366. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).