Abstract
Shiga toxin-producing Escherichia coli (STEC) are zoonotic enteric pathogens linked to human gastroenteritis worldwide. To aid the development of pathogen control efforts, the present study characterized the genotypic diversity and pathogenic potential of STEC recovered from sources near agricultural fields in Northwest Mexico. Samples were collected from irrigation river water and domestic animal feces in farms proximal to agricultural fields and were subjected to enrichment followed by immunomagnetic separation and plating on selective media for the recovery of the STEC isolates. Comparative genomic analyses indicated that the recovered STEC with the clinically relevant serotypes O157:H7, O8:H19, and O113:H21 had virulence genes repertoires associated with host cell adherence, iron uptake and effector protein secretion. Subsequent phenotypic characterization revealed multidrug resistance against aminoglycoside, carbapenem, cephalosporin, fluoroquinolone, penicillin, phenicol, and tetracycline, highlighting the need for improved surveillance on the use of antimicrobials. The present study indicated for the first time that river water in the agricultural Culiacan Valley in Mexico is a relevant key route of transmission for STEC O157 and non-O157 with a virulence potential. In addition, feces from domestic farm animals near surface waterways can act as potential point sources of contamination and transport of diverse STEC with clinically relevant genotypes.
1. Introduction
The increased consumption of fresh produce in the United States (U.S.) has contributed to a high demand for year-round availability, and import trade agreements have helped increase the supply of products that are out of season [1,2]. To meet this demand for fresh produce, Mexico has become a leading supplier of agricultural products ($43.4 billion USD) to the U.S. [3], supplying over 65% of the total value of U.S. fresh produce imports [4,5,6]. The Culiacan Valley in the state of Sinaloa in Northwest Mexico has become the single most important region for various fresh produce commodities to be exported to the U.S. [4,7,8]. For the growth of horticultural products in the Culiacan Valley, the Culiacan River provides most of the irrigation water and is formed at the City of Culiacan by the junction of the Tamazula and Humaya Rivers flowing from the Sierra Madre Occidental mountains [9]. In addition, irrigation water is provided from reservoirs on the Humaya and Tamazula Rivers. The wide bed near the Culiacan River and at the entrance to the Gulf of California reflects massive floodwaters sometimes occurring during heavy rainy seasons, resulting in areas near the river overflowing frequently with loss of crops.
The consumption of fresh fruits and vegetables has steadily increased due to their health benefits; however, the increase in fresh produce consumption has correlated with a higher incidence of outbreaks associated with foodborne enteric pathogens [2,10,11]. As a leading cause of foodborne illness worldwide, the enteric zoonotic pathogen Shiga-toxin-producing Escherichia coli (STEC) has been linked to outbreaks from foodborne and waterborne sources [12,13,14]. STEC causes severe gastroenteritis, hemorrhagic colitis, and in rare cases, the life-threatening hemolytic uremic syndrome (HUS) in humans, which has been attributed to the production of Shiga toxins (Stx) by these bacterial pathogens [15,16]. In particular, the Stx family has been categorized into two major toxin types, Stx1 and Stx2. To date, Stx1 contains four toxin subtypes (Stx1a, Stx1c–Stx1d), and the Stx2 group consists of a heterogeneous and diverse group of fourteen toxin subtypes (Stx2a–Stx2m, Stx2o) [17,18,19]. Epidemiological studies have suggested that STEC strains harboring genes encoding subtypes stx2a, stx2c or stx2d are associated with enhanced virulence and severe human illness [20,21,22,23]. By contrast, the other subtypes of stx1 and stx2 appear to be associated with mild disease or asymptomatic carriage [17,20,24,25,26,27].
Hundreds of different STEC serotypes have been documented; however, serotype O157:H7 has been the one best characterized and reported due to its strong association with the onset of severe disease symptoms in humans [13,14,15]. Subsequent epidemiological data have demonstrated that other non-O157 serogroups are significantly linked with human illnesses [28,29,30,31,32]. In particular, six non-O157 serogroups, O26, O45, O103, O111, O121, and O145, have been reported to contribute significantly to enteric infections in humans worldwide and have been subjected to monitoring and surveillance by regulatory and public health agencies. Moreover, evidence has indicated STECs of serogroups O8, O91, O104, O113, and O128 have been reported to be significant causes of human infections worldwide, and these findings have highlighted the need to recognize non-O157 STECs as emerging pathogens since they are potentially as virulent as O157:H7 [28,29,30,31,32,33].
Given many STEC strains producing Stx do not always cause severe disease symptoms in humans, this evidence demonstrates that additional virulence determinants other than Stx contribute to the clinical significance of STEC [13,14,28,32,34]. The diverse virulence potential among distinct serotypes has been attributed to genes on genomic pathogenicity islands, such as the locus for enterocyte and effacement (LEE) and the non-LEE regions. In particular, the LEE-encoded eae gene is required for attachment to intestinal epithelial cells, and the type III secretion system effectors, Esc-Esp, are implicated in host colonization and disease. Moreover, disease severity in non-O157 STEC has been attributed to nle effectors not encoded by the LEE region, and these effectors contribute to altering the host cell response and have been linked to a variety of functions including inhibition of host cell phagocytosis, invasion, cytotoxicity, and bacterial attachment by altering the signal transduction pathways in the mammalian host cell. In addition to these pathogenicity islands, a collection of chromosomal and plasmid-borne virulence genes, encoding adhesins (saa, csg, fim), cytotoxins (east, subA, toxB), and proteases (espP), are thought to enable the attachment and colonization of the human epithelium and consequently contribute to STEC pathogenic potential [13,14,34,35,36]. Screening for these virulence genes in zoonotic and environmental STEC isolates would thus enable a better characterization of risk factors that could potentially lead to sporadic and outbreak-related human illness.
Recently, published reports have documented a significant increase in antimicrobial resistance in STEC O157 and non-O157, harboring virulence profiles and causing serious problems in healthcare settings worldwide [37,38,39]. The burden of antimicrobial resistance among bacterial pathogens is considered a major health problem contributing to almost 5 million deaths worldwide and 192 million disability-adjusted life years. Moreover, resistance to fluoroquinolones and β-lactams, antibiotics used as first-line treatment for human infections, was responsible for over 70% of these deaths due to increased resistance to these agents in recent years [37,38,39]. Excessive and inappropriate use of antibiotics in treating humans and use of antibiotics as growth promoters in animal feed has created conditions promoting antimicrobial resistance among bacterial populations [40,41,42,43]. Consequently, bacteria of the same or different species are capable of transferring resistance genes via horizontal or lateral gene transfer, thereby contributing to an increase in the resistance to multiple classes of antimicrobials. While usage of antibiotics as a growth promoter is banned in Mexico, a recent survey revealed continued usage of antibiotics in agricultural settings for treating plant diseases and for promoting animal growth [42,43,44,45]. Although some surveillance studies have examined the prevalence of STEC from animal reservoirs and environmental sources [46,47,48], more information is still needed on the potential key routes of transmission of STEC, harboring virulence and antimicrobial resistance genes near agricultural fields for exporting produce from Northwest Mexico. To better identify sources of pathogenic STEC, the present study examined the virulence and antimicrobial resistance profiles of STEC O157 and non-O157 isolates recovered from animal fecal samples and river water used for irrigation in the agricultural Culiacan Valley region to aid the development of pathogen control efforts in a region important for fresh produce production.
2. Materials and Methods
2.1. Fecal and Environmental Sample Collection
The river water and fecal samples (n = 94) were collected from October to December 2016 and June to August 2017 at different sampling sites in the agricultural Culiacan Valley in the Northwest of Mexico. The three sampling sites, Agua Caliente de los Monzon (site A), Jotagua (site B), and San Pedro de Rosales (site C), were selected to be 17–33 km apart and to represent the study area in the Culiacan Valley (Figure 1). Sites A and B were located in the mountain hillsides with a semi-humid climate. Site C was in close proximity to the urban areas of Navolato and Culiacan municipalities and had a semiarid climate.
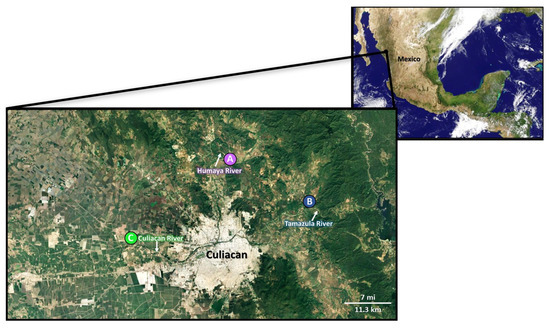
Figure 1.
Sampling sites of STEC strains O157:H7 and non-O157 isolated from river water and animal feces. Enrichment broths were prepared in tryptic soy broth by adding 10–25 g of fecal samples from cattle, chicken, and sheep or from irrigation river water using Moore swabs, collected at sampling sites Agua Caliente de los Monzón (site A), Jotagua (site B), and San Pedro de Rosales (site C) along the Humaya River, Tamazula River, and Culiacan River, respectively, in the agricultural Culiacan Valley in Northwest Mexico.
The fecal samples were collected from available species of animal present at each sampling site. In detail, each sampling site was visited biweekly from October to December of 2016, and the collected samples were 33% (18/54) from river water (Sites A to C) and 67% (36/54) from animal feces. The animal fecal samples corresponded to 50% (18/36) from cattle feces (Sites A to C), 33% (12/36) from chicken feces (Sites B and C), and 17% (6/36) from sheep feces (Site C). During 2017, each sampling site was visited biweekly in June and once in July and August. The collected samples were 30% (12/40) from river water (Sites A to C) and 70% (28/40) from animal feces. The animal fecal samples corresponded to 43% (12/28) from cattle feces (Sites A to C), 43% (12/28) from chicken feces (Sites A to C), and 14% (4/28) from sheep feces (Site C). Approximately 100 g of fecal samples from asymptomatic cattle and sheep and 30 g of fecal samples from chicken were collected from different areas within each sampling site by using a sterile spatula and gloves; they were placed into labeled sterile plastic bags, as documented in previous studies [46]. River water samples were collected with cotton gauze swabs (“Moore swabs”) by anchoring them in a water source to a monofilament line and retrieving them 3–4 days later [49,50]. All collected samples from the various sampling sites were transported immediately under refrigeration to the Microbiology Laboratory (Autonomous University of Sinaloa, Mexico) and then processed within the next 6 h.
2.2. Recovery of Bacterial Isolates from Environmental Samples
For the recovery of bacterial isolates from animal fecal samples from distinct geographical locations in the agricultural Culiacan Valley, a total of 10 g of animal feces were removed with a sterile spatula and placed in a sterile plastic bag [46]. River water samples collected from “Moore swabs” were also placed in a sterile plastic bag [49,50]. The samples in the sterile plastic bags were homogenized manually for 5 to 10 min, followed by addition of 90 mL of tryptic soy broth (TSB) (Becton Dickinson-Bioxon, Nuevo Leon, Mexico). The enrichment cultures were incubated with constant shaking (200 rpm) for 2 h at 25 °C, then at 42 °C for 8 h to enhance the recovery efficiency of stressed STEC isolates from the environmental samples [49,50]; they were held at 4 °C until the following morning without shaking. A 500 µL sample of the enrichment culture and 500 µL of 1× phosphate-buffered solution (PBS) were incubated with 10 µL of Dynabeads® anti-E. coli O157 magnetic beads (Life Technologies, Grand Island, NY, USA) for 30 min with constant mixing. The magnetic beads were then washed twice with PBS containing 0.05% Tween-20 by using a Dynal BeadRetriever Tube Rack (Life Technologies) and finally resuspended in 100 µL of PBS. A 50 µL sample was plated on CHROMagar™ O157 (CHROMagar, Paris, France). Agar plates were subsequently incubated for 18–24 h at 37 °C. Presumptive colonies were selected based on different colony color on CHROMagar, as in previous studies [49].
2.3. Virulence Factor Identification in the Initial Screening of Presumptive STEC Colonies
A total of 21 presumptive STEC isolates were initially screened by PCR amplification for identifying stx1 and stx2 subtypes [51]. All oligonucleotides for the PCR amplifications were purchased from Eurofins Genomics (Louisville, KY, USA). As a template for the PCR reaction, bacterial cultures of the isolates were grown aerobically in TSB broth (Becton Dickinson-Bioxon) for 24 h with constant shaking (200 rpm) at 37 °C, and genomic DNA was extracted using a Wizard Genomic DNA purification kit (Promega Corporation, Madison, WI, USA). The DNA was assessed by fluorometric measurement using a Qubit 4.0 fluorometer (Invitrogen, Carlsbad, CA, USA). PCR amplifications were performed in a 25 µL reaction mixture, each containing 5 µL of DNA template, 0.5 µM of each primer and 12.5 µL of 2× GoTaq® Green Master Mix (Promega Corporation). The reaction mixtures were placed in a Dyad Peltier Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) using the cycling conditions for each targeted gene [51]. Amplified products were analyzed in 1% agarose gels containing 0.04 µL/mL GelRed Nucleic Acid Stain (Phenix Research, Candler, NC, USA), and DNA sequencing analysis (Elim Biopharmaceuticals, Hayward, CA, USA) was performed to verify the amplicon nucleotide sequence for the targeted gene.
2.4. Antimicrobial Susceptibility Testing
The Kirby–Bauer disk diffusion method was performed to test 13 antimicrobials, representing 10 distinct classes, which are commonly used in Mexico for animal food production and human infection treatments [39,52,53,54,55]. Inoculums from each STEC strains were grown aerobically in 5 mL Mueller–Hinton (MH) broth (Becton Dickinson-Bioxon) and incubated at 37 °C to reach a turbidity equal to a McFarland 0.5 standard, according to guidelines provided by the Clinical and Laboratory Standards Institute (CLSI) [56]. MH agar plates were surface inoculated with each STEC culture using sterile cotton swabs, and antimicrobial paper disks (Becton Dickinson-Bioxon) were placed on surface of inoculated MH agar plates. After incubation at 37 °C for 16–18 h, the diameter of the zone of microbial growth inhibition around the antimicrobial disk was measured in millimeters [55]. As a positive control for the antimicrobial susceptibility tests, the E. coli strain ATCC 25,922 (American Type Culture Collection, Manassas, VA, USA) was employed [55]. The minimum inhibitory concentration (MIC) was then determined according to the interpretive criteria established by CLSI to classify the STEC strains as sensitive, intermediate, or resistant to the tested antimicrobial agent [56].
2.5. Phylogenetic and Multilocus Sequence Typing
Multilocus sequence typing (MLST) was performed as previously described [57]. Amplicons of internal fragments of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, recA) were generated by the specific primers with amplification conditions of: 2 min at 95 °C, 30 cycles of 1 min at 95 °C, 1 min at 54 °C (adk, fumC, icd, purA) or 58 °C (recA) or 60 °C (gyrB, mdh), and 2 min at 72 °C, followed by a final extension of 5 min at 72 °C. Sequences for the primers for each target amplification were previously described [57]. Amplicons were analyzed in 1% agarose gels containing 0.04 µL/mL Gel Red Nucleic Acid Stain (Phenix Research). PCR amplicons were purified by using the MinElute® PCR purification kit (QIAGEN, Valencia, CA, USA), and the DNA concentration was quantified using a NanoDrop® ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The sequence of the amplicons was determined on both strands by DNA sequencing analysis (Elim Biopharmaceuticals). Allelic profiles and the corresponding sequence types (STs) were assigned using the E. coli MLST database [58]. Concatenated sequence data with a sequence length of 3423 bp of each distinct representative ST were imported into the Mega X package [59]. An evolutionary phylogeny was constructed using the neighbor-joining method [60], and the topology was validated by bootstrapping (1000 replicates). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches [61]. The evolutionary distances were computed using the maximum composite likelihood method [62].
2.6. Whole-Genome Sequencing and Annotation
The concentration and quality of the genomic DNA extracted from the recovered STEC strains was determined by using a NanoDrop ND-1000 spectrophotometer and a Qubit 4 fluorometer (Thermo Fisher Scientific, Waltham, MA USA). The long-read genome sequencing was performed using a MinION device (Oxford Nanopore Technologies, Oxford, UK). The sequencing libraries were prepared using a total of 400 ng per STEC strain using a Rapid Barcoding Sequencing kit SQK-RBK-004 (Oxford Nanopore Technologies), following the manufacturer’s workflow recommendations and procedures (http://community.nanoporetech.com/protocols/rapid-barcoding-sequencing-sqk-rbk004/checklist_example.pdf (accessed on 1 March 2022)). The prepared libraries were subsequently sequenced on FLO-MIN106 (R9.4.1, active pore number ≥1100) flow cells (Oxford Nanopore Technologies) for a total of 48 h. The raw sequencing reads were base-called, trimmed, and demultiplexed with Guppy version 3.4.4 software using the high-accuracy-mode configuration file entitled “dna_r9.4.1_450bps_hac.cfg”, which was installed on a high-performance desktop workstation (Dell Precision 3630, Dell Technologies, Round Rock, TX, USA) using a Linux Ubuntu version 18.04.6 operating system. Subsequently, the error correction and assembly of the sequencing reads were performed with Flye version 2.9 [63] software by using the recommended preset options for sequencing reads obtained with the Oxford Nanopore Technologies sequencing platform, resulting in assembled genomes with a minimum of 20× depth of coverage for subsequent analysis of gene sequence data [64]. To identify STEC serotypes, the resulting contigs were submitted to the SeroTypeFinder version 2.0 server with the Center for Genomic Epidemiology (available at https://cge.food.dtu.dk/ (accessed on 1 July 2022)) [65].
The assembled genomes of the examined STEC stains, harboring the clinically relevant serotypes O8:H19, O113:H21 and O157:H7 [14,30,32,33], were subsequently examined for the presence of genes encoding putative virulence factors by entering them as input into software ABRicate version 1.0.1 [66] by selecting the parameter cutoffs of 90% coverage and 95% nucleotide identity for screening the matches against entries in the Virulence Factor Database version 2021-03-27 [67]. The ABRicate output for each STEC genome was then represented on a clustered heatmap depicting the presence or absence of the virulence gene profiles, and the heatmap was constructed and displayed with a hierarchical clustering of the genomes based on virulence data using the “Clustermap” function in the python package termed seaborn [68]. The ABRicate output led to an additional subsequent analysis of nucleotide sequences related to the type II secretion system [69], the type III secretion system [70,71], the type VI secretion system [72], and nle effectors [73], and the nucleotide sequences were aligned with the contigs of the reference strains E. coli strain EDL-933 (Accession No. NZ_CP008957), Shigella dysenteriae strain 1617 (Accession No. CP006736), and E. coli O127:H6 strain 2348/69 (Accession No. NC_011601) using the progressiveMauve module [74] in Geneious software version 9.1.8 (Biomatters, Auckland, New Zealand). Additionally, the program MacSyFinder version 2.1.2 with the TXSScan model version 1.1.1 [75] was used to search for components and effectors of the type V secretion system [76] using default parameters and a statistical threshold of significance with a maximal E-value of 0.001 for the analysis.
2.7. Statistical Analyses
Statistical analyses were performed by conducting Fisher’s exact test using R Statistical Software (version 4.2.0; R Foundation for Statistical Computing, Vienna, Austria) [77]. Probability values (p-values) lower than 0.05 were considered significant.
3. Results
3.1. Isolation and Initial Characterization of STEC from River Water and Feces of Farm Animals in the Agricultural Culiacan Valley
The Culiacan Valley in Northwest Mexico is an important region of intensive agriculture (130,000 hectares of crops/42,000 km2 of intensive agriculture) of various fresh produce commodities to be exported to the United States, including tomatoes, peppers, and cucumbers [4,7,8]. The water used for irrigation on these agricultural farms in the Culiacan Valley is provided by a series of canals derived from both the Humaya River and Tamazula River, which merge to become the Culiacan River [78]. These three rivers, Humaya, Tamazula, and Culiacan, are the major rivers in the Culiacan Valley supporting agricultural activities [9]. Given that livestock likely co-mingle near agricultural fields and can contribute as potential point sources of contamination and transport, the present study sampled river water as well as feces from domestic farm animals that were in close proximity to surface waterways serving as a source of irrigation water to agricultural fields. Moreover, the fecal samples were from domestic animals in small farms located in rural communities near rivers used for irrigation, and the primary purpose of raising livestock by these small farms is exclusively for local consumption. The objective of the present study was to identify the primary sources of STEC to enable the development of pathogen control efforts to be prioritized and focused strategically on preventing specific populations from loading the agricultural fields for export produce with foodborne pathogens.
As shown in Figure 1, animal fecal and river water samples were recovered from three sampling sites in small communities at Agua Caliente de los Monzón (site A), Jotagua (site B), and San Pedro de Rosales (site C) along the Humaya River, Tamazula River, and Culiacan River, respectively, in the agricultural Culiacan Valley. The recovery and isolation of presumptive STEC colonies were performed by subjecting the environmental samples to an enrichment step (Figure 2). After an overnight incubation, enrichment broths were subjected to immunomagnetic separation with beads conjugated to anti-O157 antibody [46,50], followed by subsequent plating on selective chromogenic agar (Figure 2), a method developed to enable the recovery of a diverse STEC population with distinctive colonies from environmental samples.
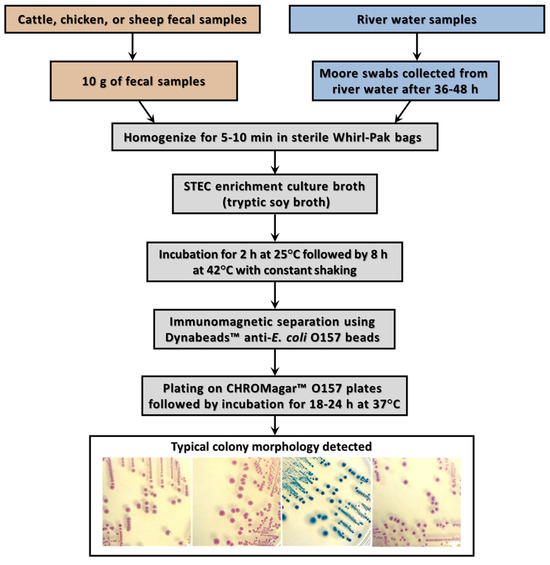
Figure 2.
Culture and isolation methods for efficient STEC recovery. The recovery method for the recovery of STEC isolates from fecal and water samples consisted of an initial enrichment step to resuscitate stressed or injured cells followed by immunomagnetic separation prior to plating on various selective chromogenic media, resulting in the isolation of STEC colonies with a distinctive color morphology.
Presumptive STEC colonies were recovered from 17% (16/94) of the total of the collected samples after plating on indicator solid media. In particular, presumptive STEC colonies were recovered after the enrichment steps in 17% (5/30) of the river samples and 17% (11/64) of animal fecal samples when examining all three sampling sites. Further analysis of the fecal samples indicated that presumptive STEC colonies were predominantly detected in 30% (9/30) of the cattle fecal samples and in 8% (2/24) of the chicken fecal samples. No positive samples were obtained from sheep fecal samples. When analyzing sampling sites (Figure 1), presumptive STEC colonies were recovered predominantly from Jotagua (site B) with 38% (9/24) prevalence, followed by Agua Caliente (site A) and San Pedro (site C), each with a prevalence of 10% for Agua Caliente (3/30) and San Pedro (4/40).
The plating of the selected enrichment broths on the chromogenic and selective solid media resulted in the recovery of 21 presumptive STEC with diverse colony color morphology (Table 1). Most recovered STEC colonies from all sampling sites and sources were 43% (9/21) mauve, while 29% (6/21) of colonies were blue, 24% (5/21) were pink, and 9.5% (2/21) were pink with a blue center (Figure 2). Although the recovery of presumptive STEC isolates from chicken feces predominantly resulted in blue colonies on the selective chromogenic agar, there was no statistically significant correlation between the characteristics of the presumptive colonies and the sampling source for the recovered isolates in the present study.

Table 1.
Bacterial isolates examined in this study and their corresponding multilocus sequence typing (MLST) and Shiga toxin subtypes.
To conduct a preliminary categorization, the presumptive STEC isolates were initially examined for the presence of wxz or wxy genes in the O-antigen gene clusters of common serogroups as well as the flagellin antigen fliC genes. Among the non-O157 isolates, the clinically relevant serotype O8:H19 was detected in 43% (9/21) of the presumptive STEC isolates and was the most predominant among the isolates recovered from cattle (Table 1). A less prevalent detection of isolates harboring serotype O113:H21 was also identified, corresponding to 14% (3/21) of the isolates, and chicken feces were found to be the main source of serotype O113:H19. The present study also identified other non-O157 STEC serogroups, O17 and O185, not previously implicated in human illness [33]. Moreover, only 9.5% (2/21) of the recovered isolates were serotype O157:H7 (Table 1), which has been commonly associated with the development of severe human illness [14,15,79], and river water represented the predominant source of this serotype.
To determine the presence of the key virulence factors in STEC, the Shiga toxin subtypes, including the stx1 subtypes (stx1a, stx1c, stx1d) and stx2 subtypes (stx2a − stx2g), were screened in the recovered isolates (Table 1). Interestingly, the results indicated a high prevalence of the stx1a subtype in 91% (19/22) of the isolates from all sources. Moreover, 69% (9/13) of the cattle STEC isolates were found to harbor the clinically relevant stx2a subtype, while none of the chicken or water isolates were positive for stx2a, indicating a statistically significant correlation between toxin subtype and source (Fisher’s exact test, p-value = 0.0017). The stx2a subtype was not identified in any STEC isolate recovered from river water or chicken fecal samples. By contrast, the STEC strains recovered from river water and animal feces samples in the Culiacan Valley were PCR-negative for the stx subtypes stx1c, stx1d, stx2b, stx2e, stx2f, or stx2g.
3.2. Profiles of Antimicrobial Resistance in the Recovered STEC Isolates
Previous findings have demonstrated that the inappropriate use of antibiotics in the public, private, and agricultural sectors in Mexico has contributed to antimicrobial resistance among enteric bacterial pathogens [39,40,42,80]. Furthermore, surveillance studies in Mexico have provided evidence for an apparent increase in antimicrobial resistance of pathogenic E. coli over the recent years [48,52,55,80]. In the present study, a total of 13 antibacterial agents belonging to various antimicrobial classes (Table S1 in the Supplementary Materials) were selected since these agents are commonly used in Mexico and have been linked to emerging antimicrobial resistance among bacterial pathogens, which are responsible for human illnesses and death [39,52,53,54,55]. Antimicrobial resistance was examined in the recovered STEC isolates by employing the disk diffusion method, a phenotypic method that would enable the relatively unbiased assessment of both the resistance and susceptibility against the tested antimicrobials [81,82]. All of the examined STEC isolates showed susceptibility to two antimicrobials, amoxicillin–clavulanic acid (AMC) and colistin (CST), and the vast majority of the tested STEC strains [95% (20/21)] showed susceptibility to the following antimicrobials: amikacin (AMK), chloramphenicol (CHL), imipenem (IPM), nalidixic acid (NAL), and trimethoprim–sulfamethoxazole (SXT) (Table 2 and Table S2 in the Supplementary Materials). Susceptibility to all tested antimicrobials was observed in the non-O157 isolates BAAL1103 and BAAL1120, recovered from river water and BAAL1112 and BAAL1114, recovered from cattle feces (Table S2).

Table 2.
Antimicrobial non-susceptible profiles identified in the STEC isolates from different sampling sources and sites.
Further analysis of antimicrobial non-susceptibility, representing intermediate and resistant categories [55], was observed when testing six antimicrobials: ampicillin (AMP), cephalothin (CEF), ciprofloxacin (CIP), gentamicin (GEN), kanamycin (KAN), and tetracycline (TET). In particular, the results indicated that 14% (3/21) of the STEC isolates were non-susceptible to either AMP or GEN, while an increased percentage, 24% (5/21) of the isolates were susceptible to TET (Table S2). Interestingly, the highest proportion, 33% (7/21) of the O157 and non-O157 STEC isolates, was found to display non-susceptibility to CEF or KAN (Table S2), antimicrobials belonging to agent classes most prescribed in Mexico [48]. To better characterize multidrug resistance profiles to classes of antimicrobials also commonly used in Mexico, such as aminoglycosides, tetracyclines, cephalosporins, and penicillins [52,53,54,55], statistical analyses were conducted to determine a significant correlation between antimicrobial resistance and either location or source. In the present study, the results indicated that a significant correlation was found between STEC O113:H21 isolates BAAL1005 and BAAL1006, recovered from chicken feces, and resistance to at least one of the tested antimicrobials, AMK, GEN, or KAN, belonging to the aminoglycoside antimicrobial category (Fisher’s exact test, p-value = 0.013) (Table 2). Moreover, the analysis revealed the chicken O113:H21 isolate BAAL1106, the O8:NT isolate BAAL1108, and the O8:H33 isolate BAAL1119 from cattle were found to also display multidrug resistance, indicating non-susceptibility to more than three agents in different classes [55,83]. Interestingly, the cattle isolate BAAL1119 showed multidrug resistance to five different classes of antimicrobials (Table 2), and these findings highlight the need for surveillance of the antimicrobial resistance patterns in foodborne pathogens in a major agricultural region for fresh produce.
3.3. Genetic Relatedness of the Recovered STEC Isolates
To characterize the genetic relatedness among the recovered isolates, MLST was performed to enable a better discrimination among the recovered isolates. To further examine the relationships among the detected STs, the amplified sequences of the housekeeping genes (see Section 2) were concatenated to generate a phylogenetic tree (Figure 3). The results indicated the recovered STEC isolates were classified into nine different STs (Table 1). The MLST analysis revealed that ST-2385 was identified in 43% (9/21) of the recovered STEC isolates, predominantly with serotype O8:H19 (Figure 3), and among those isolates belonging to ST-2385, 78% (7/9) of the isolates were recovered at the Jotagua sampling site. Previous reports and information in the public databases [58] documented that strains belonging to ST-2385 were previously recovered from different sample types, including cattle/manure, irrigation ditch water, river water, and fresh produce [58,84].
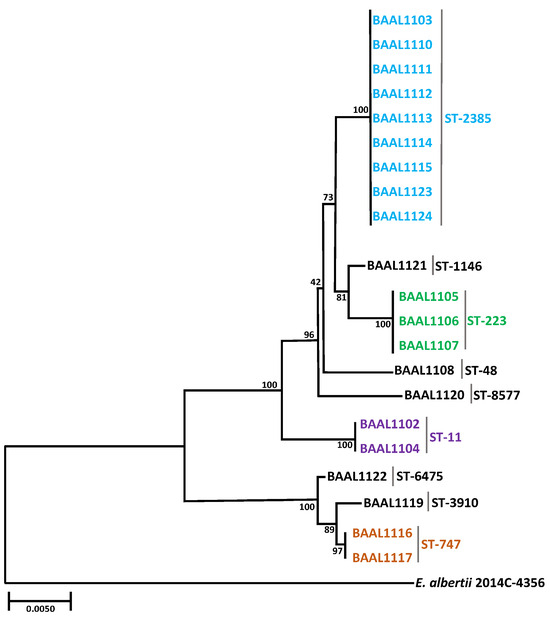
Figure 3.
Phylogenetic tree of sequence types (STs) identified in the recovered STEC isolates. The phylogenetic tree was generated with the neighbor-joining method [60], and the evolutionary distances were computed using the maximum composite likelihood method [62]. STs of isolates recovered from river irrigation water and domestic animal fecal samples are shown in different colors. Bootstrap values, shown at internal notes, were generated from 10,000 replications. The scale bar corresponds to 0.5 nucleotide substitutions per 100 nucleotides.
Interestingly, ST-233 was the next most common ST detected among the recovered STEC O113:H21 isolates and was found to be significantly correlated with chicken isolates from the Jotagua sampling site (Fisher’s exact test, p-value = 0.0006). Moreover, ST-747 was only detected in cattle O17:H45 isolates from the Agua Caliente sampling site and was previously identified in STEC isolates recovered from various leafy greens in the U.S. including spinach, spring mix, and kale [58]. ST-11 was only identified in O157:H7 STEC isolates BAAL1002 and BAAL1004 recovered from river water in the Jotagua sampling site. Previous reports documented ST-11 as a very common ST with over 14,000 entries in public databases, including over 1700 O157:H7 strains [58]. In the present study, ST-11 was only identified in stx2c-positive isolates belonging to serotype O157:H7; however, a significant correlation between ST and virulence profile was not identified among the isolates recovered. Further analysis of branch lengths on the phylogenetic tree indicated isolates with ST-747, ST-6475, and ST-3910 were genetically similar with less than a 0.5% nucleotide difference between them. By contrast, ST-747, ST-6475, and ST-3910, identified in the isolates recovered from Agua Caliente and San Pedro sampling locations, had an almost 2.0% difference to other STs, identified in the isolates recovered from Jotagua (Figure 3).
3.4. Comparative Virulence Profiling of STEC Isolates with Clinically Relevant Serotypes
To conduct more comprehensive analyses of the virulence potential of STEC harboring serotypes O157:H7, O8:H19, and O113:H21 with clinical relevance [14,30,32,33], the present study employed the use of high-resolution sequencing to further characterize the virulence profiles of the examined isolates. As shown in Figure 4, the analysis of the genomic content in all of the examined isolates with clinically relevant serotypes revealed the presence of genes implicated in adhesion (csg, ecpE, fde, fimA, ykgK), invasion (ibe, ompA) and iron uptake (ent, fep, fes). The analysis also revealed that the mandatory effectors of the type V (yadA, pfam03797) and VI (hcp, tss, vgrG) secretion systems were identified in all of the examined STEC isolates with clinically relevant serotypes (Figure 4).
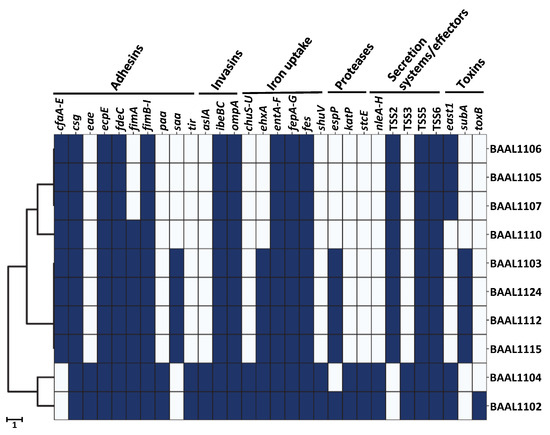
Figure 4.
Virulence determinants in the STEC isolates with the clinically relevant serotypes O157:H7, O8:H19, and O113:H21. A clustered heatmap indicating the presence (blue) or absence (white) of virulence genes in the selected STEC isolates (Table 1) was constructed with a hierarchical clustering of the data based on input from the Virulence Factor Database [67] version 2021-03-27, with the “Clustermap” function in the Python version 3.10 software and the Seaborn package, version 0.12.0 [68]. The scale bar for the hierarchical clustering of the heatmap corresponds to the number of differences between the examined virulence determinants.
Further analysis revealed that the virulence gene profile of the O157:H7 strains differed significantly when compared to the O8:H19 and O113:H21 isolates (Figure 4). In particular, a statistically significant association was observed in the O157:H7 river isolates when examining the presence of the adherence factors, eae and paa genes, as well as the invasion factor, aslA (Fisher’s exact test, p-value = 0.022), adhesins critical for host colonization [13,28,35]. Additionally, the chu operon, encoding for components of the heme utilization system and (along with the katP and stcE genes) encoding proteases [13,36], was significantly correlated with the O157:H7 strains (Fisher’s exact test, p-value = 0.022). Furthermore, numerous genes related to the type III secretion system were exclusively identified in the river O157:H7 strains including the ces genes, encoding chaperones, the esc genes involved in structural components of the secretion system, and the esp genes coding for effector proteins of the type III secretion system [36,70]. The translocated intimin receptor gene tir and the nle genes (Figure 4), as non-LEE effector genes present in the pathogenicity islands of OI-122 and OI-71 [28,73], were also observed to have a significant association (Fisher’s exact test, p-value = 0.022). Virulence factors statistically correlated with the O8:H19 and O113:H21 strains include the cfaA gene, the colonization factor antigen I, which is a well-characterized fimbrial structure implicated in adherence to receptors on human intestinal epithelial cells and with a role in promoting long-term survival in water [85]. Additionally, type II secretion system effectors, gspC–gspL genes forming a general secretion pathway type 2 (Fisher’s Exact Test, p-value = 0.022), were also identified. Finally, enterohemolysin ehxA has been frequently associated with diarrheal symptoms and hemolytic uremic syndrome [86] and was detected in the O113:H21 and most of the O8:H19 isolates examined in the virulence profiling analyses.
Interestingly, the chicken O113:H21 and the river O157:H7 STEC isolates were found only to harbor the astA gene, encoding the enteroaggregative E. coli heat-stable enterotoxin 1 (EAST1) with a potential virulence role, leading to diarrheal disease in humans [87,88]. Although the EAST-1 toxin was originally identified in a subset of enteroaggregative E. coli, additional reports showed that the astA gene was present in other pathotypes of E. coli and bacterial species, indicating that this virulence determinant may be prevalent among enteric bacterial pathogens. Moreover, relevant virulence determinants were exclusively detected in the cattle O8:H19 strains, such as the autoagglutinating adhesion gene saa and the subtilase toxin subunit gene subA. Saa, a plasmid-encoded adhesin, was shown to be involved in the attachment of STEC to eukaryotic cells [35,89]. The subA gene encodes the catalytic subunit A of the toxin, belonging to the family of AB5 toxins, and the internalization of the SubA subunit into the epithelial target cells leads to cell death (apoptosis) in eukaryotic cells [90,91]. Both virulence markers, saa and subA, were originally identified in STEC O113:H21 and were negative for the LEE region [32]. By contrast, the present study only identified saa and subA in serotype O8:H19 isolates, and these results were in agreement with the virulence profile of STEC O8:H19 from an earlier survey [51], potentially indicating a prevalent genotype for isolates with the O8:H19 serotype in this agricultural region in the Northwest of Mexico. In summary, the comparative genomic analyses conducted in the present study indicated that the recovered STEC with the clinically relevant serotypes O157:H7, O8:H19, and O113:H21 had virulence gene repertoires associated with host cell adherence, iron uptake, and effector protein secretion.
4. Discussion
The findings from the present study revealed the presence of diverse STEC recovered from animal feces and agricultural river water within a major agricultural region for export produce. The identification of a high prevalence of positive samples from cattle feces collected at various sites in the Culiacan Valley is consistent with the fact that cattle is a primary animal reservoir for STEC. The colonization of cattle by STEC is typically asymptomatic; however, the levels of colonization can vary significantly among cattle since some animals can have high levels of STEC carriage (“supershedders”) in their gastrointestinal tract for lengthy time periods [14,34]. Interestingly, the present study detected presumptive STEC in the fecal samples from chickens collected at small farms in rural communities in the Culiacan Valley. Although reports have documented isolates of STEC strains previously identified from some retail chicken products and chicken farms in rural communities [92], chickens are not generally considered a prominent animal reservoir for STEC [14,34]. However, animal inoculation experiments have shown that STEC O157 can colonize chickens, and STEC can be shed in chicken feces for up to a year [93]. Although ovine ruminants are considered significant reservoirs of STEC in certain geographical locations such as Australia, New Zealand and Brazil [12,14,94], the present study did not detect any positive samples from sheep fecal samples collected at the small rural farms in Northwestern Mexico. One possible explanation for the lack of STEC detection in sheep in the current survey may be due to a heterogenous STEC distribution in fecal matter as well as greater environmental exposure after excretion, previously shown to contribute to some bacterial decay and an underestimation of the actual STEC prevalence [94]. The results obtained in the present study highlighted that both chickens and cattle could serve as relevant animal reservoirs and should be considered as significant risk factors for STEC in these small rural farms in Northwestern Mexico.
The research findings from the present study demonstrated the recovery of STEC strains with clinically relevant serotypes, O8:H19, O113:H21, and O157:H7, indicating evidence with implications for public health since these relevant serotypes have been previously implicated in disease outbreaks and severe symptoms in humans, such as hemorrhagic colitis and HUS [32,33,95]. In addition, the detection of stx1 and stx2 subtypes demonstrated the presence of important pathogenicity determinants among the recovered STEC isolates from river water and domestic animals. Based on research findings obtained with cultured mammalian cells, the functional activity of the different stx1 and stx2 subtypes has been associated with various levels of virulence [51,96,97]. In particular, the present study identified the stx1a subtype to be highly prevalent among the recovered isolates from all sources. Previous reports have demonstrated that STEC strains expressing Stx1a can potentially result in the development of HUS [17,98,99,100,101]. Furthermore, the stx2 subtyping assays conducted in the present study also identified the subtypes stx2a, stx2c and stx2d, which have been previously linked to severe human illnesses such as bloody diarrhea and HUS [17,98,99,100,101]. The stx subtyping results obtained in the present study indicated that the STEC isolates recovered in the agricultural Culiacan Valley harbored relevant pathogenicity determinants that could potentially lead to the development of severe human illness with significant clinical consequences.
This study also identified effectors associated with the adherence, invasion and colonization of the target host cell, iron uptake, and effector protein secretion. Among the identified virulence determinants, Csg, the curli fimbriae, and the outer membrane protein A, OmpA, have been well characterized in STEC. In particular, curli has been well characterized in pathogenic E. coli and Salmonella enterica and has a role in biofilm formation as well as attachment to the extracellular matrix to enable the successful colonization of the mammalian host cells [102,103]. Recent studies in enterohemorrhagic E. coli have demonstrated OmpA to play an important role in the regulation of multiple pathogenic phenotypes, including attachment on distinct surfaces, biofilm formation, motility, and colonization and invasion of mammalian host cells [104]. Moreover, the presence of the siderophore enterobactin and ferric transport genes highlighted that the examined STEC isolates harbored the determinants for acquisition of iron, an essential nutrient for bacteria growth and increased virulence potential in enteric pathogens [105].
The identification of mandatory effectors of the type V and type VI secretion systems indicated the presence of secretion machineries, which are characteristic virulence pathways required for cell–cell interactions through the adhesion to receptors on the target cell surface, for mediating bacterial competition, and for enabling bacterial colonization and survival by nutrient acquisition [72,76]. Given that these STEC O157:H7 from river water were shown to harbor type III secretion system effectors located on the enterocyte effacement pathogenicity island [36,70,71], these findings indicated these STEC isolates could form attaching and -effacing (A/E) lesions on intestinal mucosa, an essential characteristic of pathogenesis. The detection of the gsp operon as part of the type 2 secretion pathway revealed the encoding of toxins, which includes the heat-labile enterotoxin and a range of hydrolytic enzymes, including proteases, lipases, and carbohydrate-active enzymes, that are translocated across the outer membrane in enteric bacterial pathogens [69].
Moreover, the present study documented multidrug resistance against aminoglycoside, carbapenem, cephalosporin, fluoroquinolone, penicillin, phenicol, and tetracycline among the STEC isolates recovered from domestic animal fecal samples. The observed multidrug resistance to critical agents among the STEC isolates reported in the present study is a matter of serious concern due to the major impact on public health. In particular, the disease outcomes in patients potentially infected with STEC displaying multidrug resistance would be expected to be worse when compared to patients with bacterial infections that are more susceptible to antimicrobials [106]. Additionally, infections with multidrug-resistant bacteria would consequently have much higher medical costs [106], presenting a challenge to the development of effective strategies for both empiric and definitive therapies. Although some antimicrobials (sulfonamides, quinolones and fluoroquinolones) were previously shown to induce Stx production and subsequently encourage the onset of severe disease symptoms, other agents, including macrolides, tetracyclines, aminoglycosides, fosfomycins, ansamycins and cephems, were effective in treating and combating STEC infections when administered early because they failed to induce Stx production [107]. In the present study, multidrug-resistant strains were also found to have non-susceptibility to tetracyclines and aminoglycosides, agents widely used in agriculture at the global level [43], and indicated the necessity of controlling the use of these agents in agricultural settings to prevent the development of resistance among bacterial pathogens.
The analysis of antimicrobial non-susceptibilities to CEF or KAN among the STEC isolates in this study were found to be in agreement with a previous survey, which identified zoonotic STEC in the agricultural Culiacan Valley with 20–30% intermediate and resistant profiles for the antimicrobials KAN or CEF [55]. Based on these observations, the findings obtained in the present study provided compelling evidence in support of the prevalent antimicrobial resistance among STEC isolates to both antimicrobials CEF and KAN in this major produce production region in Mexico. Due to their potency and effectiveness, CEF and KAN continue to be widely prescribed by physicians for use in clinical settings and for veterinarian applications [43,108]. This continued use of CEF and KAN has resulted in a major public health threat due to antibiotic pollution as detected from sampling of various environmental sources, such as surface and ground waters, soil sediment, and animal and plant production sites [43,108]. The environment is thus considered a relevant reservoir of antimicrobial-resistant bacteria. Strains, either from the same species or different species, are capable of transferring resistance determinants via horizontal gene transfer or lateral gene transfer, and this transmission would thus lead to the distribution of and increase in resistance to multiple classes of agents among bacterial populations [37,39,43,108]. In summary, the findings from this study highlighted the imperative need for judicious use as well as for improved surveillance and monitoring programs of the use of antimicrobials for treating human infections and for animal production.
Although previous findings revealed that rural farms in the agricultural Culiacan Valley region could be considered as potential sources of STEC strains [46], the present study documented for the first time that irrigation water in the agricultural Culiacan Valley in Mexico could act as a potential key route of transmission for STEC O157 and non-O157, harboring relevant virulence and antimicrobial resistance genes. In addition, the sampling of animal feces from small rural farms in the present study at strategic locations in the agricultural Culiacan Valley provided supporting evidence that the presence of livestock at sites upstream continued to be potential point sources of contamination since STEC with the clinically relevant serotypes, O157:H7, O8:H19, and O113:H21, were still predominantly recovered, in agreement with previous observations [44]. The results from the present study emphasize the necessity of routine monitoring in this produce production region due to the potential subsequent transport of pathogenic STEC into holding ponds, other conveyances, and eventually into agricultural fields [46,49,50,109]. Future studies will aim to expand the identification of additional point sources of STEC contamination and transport near agricultural fields. This fundamental information will facilitate outreach to growers, processors, and the livestock community about sources of STEC contamination and strategies to improve water quality and implement science-based strategies to prevent any potential on-farm contamination of fresh produce in the agricultural Culiacan Valley in the Northwest of Mexico.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres15010026/s1, Table S1: Antimicrobial agents used in the present study; Table S2. Antimicrobial minimum inhibitory concentration (MIC) values of STEC isolates examined in this study.
Author Contributions
Conceptualization, B.Q., M.S.-B., O.Y.L.-M. and B.A.A.-L.; methodology, B.Q., B.G.L., E.F.B.-H., O.Y.L.-M. and B.A.A.-L.; software, B.G.L.; validation, B.G.L., E.F.B.-H. and B.A.A.-L.; formal analysis, B.Q., B.G.L. and B.A.A.-L.; investigation, B.Q., M.S.-B., B.G.L., E.F.B.-H., O.Y.L.-M. and B.A.A.-L.; resources, B.Q., M.S.-B., O.Y.L.-M. and B.A.A.-L.; data curation, B.Q., B.G.L. and B.A.A.-L.; writing—original draft preparation, B.Q., B.G.L. and B.A.A.-L.; writing—review and editing, B.Q., M.S.-B., B.G.L. and B.A.A.-L.; visualization, B.Q., B.G.L. and B.A.A.-L.; supervision, B.Q., M.S.-B., O.Y.L.-M. and B.A.A.-L.; project administration, B.Q. and B.A.A.-L.; funding acquisition, B.Q. and B.A.A.-L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported in part by The Autonomous University of Sinaloa (Programa de Fomento y Apoyo a Proyectos de Investigación) Grant No. PROFAPI2014/013 and PROFAPI2015/014 and by the United States Department of Agriculture (USDA), Agricultural Research Service (ARS), CRIS Project Number 2030-42000-055-00D.
Institutional Review Board Statement
Ethical review and approval were waived for this study since the fecal samples from the domestic farm animals were collected without having direct contact with the animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete genome sequences of the STEC isolates BAAL1102, BAAL1104, BAAL1103, BAAL1105, BAAL1106, BAAL1107, BAAL1110, BAAL1112, BAAL1115, BAAL1124 are listed with the GenBank accession numbers JAWVHR00000000–JAWVHI000000000 and the BioSample numbers SAMN38166118–SAMN38166127, respectively, under the BioProject number PRJNA1037173 in the National Center for Biotechnology Information (NCBI) BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/ (accessed on 19 November 2023).
Acknowledgments
The authors gratefully thank Jaszemyn Yambao (U.S. Dept. of Agriculture, Albany, CA, USA), Osiris Díaz Torres (CIATEJ, Guadalajara, Jalisco, Mexico) as well as William E. Domínguez-Esquerra, Célica A. Rodríguez-López and Cindy A. Terrazas-Alcaraz (Univ. Auton. Sinaloa, Culiacan, Sinaloa, Mexico) for providing excellent technical assistance. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Callejón, R.M.; Rodríguez-Naranjo, M.I.; Ubeda, C.; Hornedo-Ortega, R.; García-Parrilla, M.C.; Troncoso, A.M. Reported foodborne outbreaks due to fresh produce in the United States and European Union: Trends and causes. Foodborne Pathog. Dis. 2015, 12, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Sivapalasingam, S.; Friedman, C.R.; Cohen, L.; Tauxe, R.V. Fresh produce: A growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 2004, 67, 2342–2353. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Commerce-International Trade Administration. Country Commercial Guides: Mexico—Agriculture. Available online: https://www.trade.gov/country-commercial-guides/mexico-agriculture (accessed on 15 February 2024).
- Huang, S.W. Imports Contribute to Year-Round Fresh Fruit Availability, U.S. Department of Agriculture-Economic Research Service Report FTS-356-01. Available online: http://www.ers.usda.gov/media/1252296/fts-356-01.pdf (accessed on 1 June 2022).
- Lucier, G.; Jerardo, A. Vegetables and Melons Outlook Report. U.S. Department of Agriculture-Economic Research Service Report No. VGS-313. Available online: https://www.ers.usda.gov/webdocs/outlooks/39473/14932_vgs285.pdf?v=981 (accessed on 3 April 2021).
- Karst, T. Mexico’s Dominance in Imports Is Revealed in USDA Statistics. Available online: https://www.thepacker.com/news/produce-crops/mexicos-dominance-imports-revealed-usda-statistics (accessed on 30 January 2023).
- Clemens, R.L. The Expanding U.S. Market for Fresh Produce; Center for Agricultural and Rural Development at Digital Repository Iowa State University: Ames, IA, USA, 2015; pp. 1–3. [Google Scholar]
- Flores, D.U.S. Department of Agriculture-Foreign Agricultural Service: Tomato Annual. GAIN Report MX8025. Available online: https://apps.fas.usda.gov/newgainapi/api/report/downloadreportbyfilename?filename=Tomato%20Annual_Mexico%20City_Mexico_5-30-2018.pdf (accessed on 1 June 2022).
- Rentería-Guevara, S.A.; Rangel-Peraza, J.G.; Rodríguez-Mata, A.E.; Amábilis-Sosa, L.E.; Sanhouse-García, A.J.; Uriarte-Aceves, P.M. Effect of agricultural and urban infrastructure on river basin delineation and surface water availability: Case of the Culiacan River Basin. Hydrology 2019, 6, 58. [Google Scholar] [CrossRef]
- Center for Science in the Public Interest. Outbreak Alert: Closing the Gaps in Our Federal Food Safety Net. Available online: https://cspinet.org/sites/default/files/attachment/outbreak.pdf (accessed on 1 June 2022).
- Sewell, A.M.; Farber, J.M. Foodborne outbreaks in Canada linked to produce. J. Food Prot. 2001, 64, 1863–1877. [Google Scholar] [CrossRef]
- Gyles, C.L. Shiga toxin-producing Escherichia coli: An overview. J. Anim. Sci. 2007, 85, E45–E62. [Google Scholar] [CrossRef] [PubMed]
- Bolton, D.J. Verocytotoxigenic (Shiga toxin-producing) Escherichia coli: Virulence factors and pathogenicity in the farm to fork paradigm. Foodborne Pathog. Dis. 2011, 8, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejek, A.M.; Minnich, S.A.; Hovde, C.J. Escherichia coli 0157:H7 virulence factors and the ruminant reservoir. Curr. Opin. Infect. Dis. 2022, 35, 205–214. [Google Scholar] [CrossRef]
- Karmali, M.A. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 1989, 2, 15–38. [Google Scholar] [CrossRef]
- Karmali, M.A.; Steele, B.T.; Petric, M.; Lim, C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet 1983, 321, 619–620. [Google Scholar] [CrossRef]
- Scheutz, F.; Teel, L.D.; Beutin, L.; Piérard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef]
- Gill, A.; Dussault, F.; McMahon, T.; Petronella, N.; Wang, X.; Cebelinski, E.; Scheutz, F.; Weedmark, K.; Blais, B.; Carrillo, C. Characterization of atypical Shiga toxin gene sequences and description of Stx2j, a new subtype. J. Clin. Microbiol. 2022, 60, e0222921. [Google Scholar] [CrossRef] [PubMed]
- Probert, W.S.; McQuaid, C.; Schrader, K. Isolation and identification of an Enterobacter cloacae strain producing a novel subtype of Shiga toxin type 1. J. Clin. Microbiol. 2014, 52, 2346–2351. [Google Scholar] [CrossRef] [PubMed]
- Bielaszewska, M.; Friedrich, A.W.; Aldick, T.; Schurk-Bulgrin, R.; Karch, H. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: Predictor for a severe clinical outcome. Clin. Infect. Dis. 2006, 43, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Olsen, K.E.; Ethelberg, S.; Scheutz, F. Subtyping method for Escherichia coli Shiga-toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 2007, 45, 2020–2024. [Google Scholar] [CrossRef] [PubMed]
- Boerlin, P.; McEwen, S.A.; Boerlin-Petzold, F.; Wilson, J.B.; Johnson, R.P.; Gyles, C.L. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 1999, 37, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Mariani-Kurkdjian, P.; Bonacorsi, S.; Liguori, S.; Fach, P. Characteristics of emerging human-pathogenic Escherichia coli O26:H11 strains isolated in France between 2010 and 2013 and carrying the stx2d gene only. J. Clin. Microbiol. 2015, 53, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Beutin, L.; Krause, G.; Zimmermann, S.; Kaulfuss, S.; Gleier, K. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 2004, 42, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, A.W.; Bielaszewska, M.; Zhang, W.L.; Pulz, M.; Kuczius, T.; Ammon, A.; Karch, H. Escherichia coli harboring Shiga toxin 2 gene variants: Frequency and association with clinical symptoms. J. Infect. Dis. 2002, 185, 74–84. [Google Scholar] [CrossRef]
- Friedrich, A.W.; Borell, J.; Bielaszewska, M.; Fruth, A.; Tschape, H.; Karch, H. Shiga toxin 1c-producing Escherichia coli strains: Phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 2003, 41, 2448–2453. [Google Scholar] [CrossRef]
- Kawano, K.; Okada, M.; Haga, T.; Maeda, K.; Goto, Y. Relationship between pathogenicity for humans and stx genotype in Shiga toxin-producing Escherichia coli serotype O157. Eur. J. Clin. Microbiol. Infect Dis. 2008, 27, 227–232. [Google Scholar] [CrossRef]
- Coombes, B.K.; Gilmour, M.W.; Goodman, C.D. The evolution of virulence in non-O157 Shiga toxin-producing Escherichia coli. Front. Microbiol. 2011, 2, 90. [Google Scholar] [CrossRef] [PubMed]
- Mathusa, E.C.; Chen, Y.; Enache, E.; Hontz, L. Non-O157 Shiga toxin-producing Escherichia coli in foods. J. Food Prot. 2010, 73, 1721–1736. [Google Scholar] [CrossRef] [PubMed]
- Valilis, E.; Ramsey, A.; Sidiq, S.; DuPont, H.L. Non-O157 Shiga toxin-producing Escherichia coli—A poorly appreciated enteric pathogen: Systematic review. Int. J. Infect. Dis. 2018, 76, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, M.G.; Al-Hindi, R.R.; Esmael, A.; Alotibi, I.A.; Azhari, S.A.; Alseghayer, M.S.; Teklemariam, A.D. The “Big Six”: Hidden emerging foodborne bacterial pathogens. Trop. Med. Infect. Dis. 2022, 7, 356. [Google Scholar] [CrossRef]
- Bettelheim, K.A. The non-O157 Shiga-toxigenic (Verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 2007, 33, 67–87. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.S. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 2007, 85, E63–E72. [Google Scholar] [CrossRef] [PubMed]
- Etcheverría, A.I.; Padola, N.L. Shiga toxin-producing Escherichia coli: Factors involved in virulence and cattle colonization. Virulence 2013, 4, 366–372. [Google Scholar] [CrossRef]
- McWilliams, B.D.; Torres, A.G. Enterohemorrhagic Escherichia coli adhesins. Microbiol. Spectrum 2014, 2. [Google Scholar] [CrossRef]
- Stevens Mark, P.; Frankel Gad, M. The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli. Microbiol. Spectrum 2014, 2. [Google Scholar] [CrossRef]
- Colello, R.; Etcheverría, A.I.; Di Conza, J.A.; Gutkind, G.O.; Padola, N.L. Antibiotic resistance and integrons in Shiga toxin-producing Escherichia coli (STEC). Braz. J. Microbiol. 2015, 46, 1–5. [Google Scholar] [CrossRef]
- Rubab, M.; Oh, D.H. Virulence characteristics and antibiotic resistance profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources. Antibiotics 2020, 9, 587. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Amábile-Cuevas, C.F. Antibiotic resistance in Mexico: A brief overview of the current status and its causes. J. Infect. Dev. Ctries. 2010, 4, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.; Lovari, S.; Cordaro, G.; Di Matteo, P.; Sorbara, L.; Iurescia, M.; Donati, V.; Buccella, C.; Battisti, A. Prevalence and concentration of Verotoxigenic Escherichia coli O157:H7 in adult sheep at slaughter from Italy. Zoonoses Public Health 2009, 56, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Ornelas-Eusebio, E.; García-Espinosa, G.; Laroucau, K.; Zanella, G. Characterization of commercial poultry farms in Mexico: Towards a better understanding of biosecurity practices and antibiotic usage patterns. PLoS ONE 2020, 15, e0242354. [Google Scholar] [CrossRef]
- McManus, P.S.; Stockwell, V.O.; Sundin, G.W.; Jones, A.L. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 2002, 40, 443–465. [Google Scholar] [CrossRef]
- Amézquita-López, B.A.; Quiñones, B.; Cooley, M.B.; León-Félix, J.; Castro-del Campo, N.; Mandrell, R.E.; Jiménez, M.; Chaidez, C. Genotypic analyses of Shiga toxin-producing Escherichia coli O157 and non-O157 recovered from feces of domestic animals on rural farms in Mexico. PLoS ONE 2012, 7, e51565. [Google Scholar] [CrossRef]
- Canizalez-Román, A.; Velázquez-Román, J.; Valdez-Flores, M.A.; Flores-Villaseñor, H.; Vidal, J.E.; Muro-Amador, S.; Guadrón-Llanos, A.M.; González-Nuñez, E.; Medina-Serrano, J.; Tapia-Pastrana, G.; et al. Detection of antimicrobial-resistance diarrheagenic Escherichia coli strains in surface water used to irrigate food products in the Northwest of Mexico. Int. J. Food Microbiol. 2019, 304, 1–10. [Google Scholar] [CrossRef]
- Sánchez-Huesca, R.; Lerma, A.; Guzmán-Saldaña, R.M.E.; Lerma, C. Prevalence of antibiotics prescription and assessment of prescribed daily dose in outpatients from Mexico City. Antibiotics 2020, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Cooley, M.B.; Jay-Russell, M.; Atwill, E.R.; Carychao, D.; Nguyen, K.; Quiñones, B.; Patel, R.; Walker, S.; Swimley, M.; Pierre-Jerome, E.; et al. Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS ONE 2013, 8, e65716. [Google Scholar] [CrossRef] [PubMed]
- Cooley, M.B.; Quiñones, B.; Oryang, D.; Mandrell, R.E.; Gorski, L. Prevalence of Shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front. Cell Infect. Microbiol. 2014, 4, 30. [Google Scholar] [CrossRef] [PubMed]
- Amézquita-López, B.A.; Quiñones, B.; Lee, B.G.; Chaidez, C. Virulence profiling of Shiga toxin-producing Escherichia coli recovered from domestic farm animals in Northwestern Mexico. Front. Cell Infect. Microbiol. 2014, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Canizalez-Román, A.; González-Nuñez, E.; Vidal, J.E.; Flores-Villaseñor, H.; León-Sicairos, N. Prevalence and antibiotic resistance profiles of diarrheagenic Escherichia coli strains isolated from food items in Northwestern Mexico. Int. J. Food Microbiol. 2013, 164, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Homedes, N.; Ugalde, A. Mexican pharmacies and antibiotic consumption at the US-Mexico border. South Med. Rev. 2012, 5, 9–19. [Google Scholar] [PubMed]
- Wirtz, V.J.; Dreser, A.; Gonzales, R. Trends in antibiotic utilization in eight Latin American countries, 1997-2007. Rev. Panam. Salud Publica 2010, 27, 219–225. [Google Scholar] [CrossRef]
- Amézquita-López, B.A.; Quiñones, B.; Soto-Beltrán, M.; Lee, B.G.; Yambao, J.C.; Lugo-Melchor, O.Y.; Chaidez, C. Antimicrobial resistance profiles of Shiga toxin-producing Escherichia coli O157 and non-O157 recovered from domestic farm animals in rural communities in Northwestern Mexico. Antimicrob. Resist. Infect. Control 2016, 5, 1. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: CLSI Document M100-S24; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014; Volume 34, pp. 1–230. [Google Scholar]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M.; Agama Study, G. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, G.V.; Cook, N.M.; Bueno-Sancho, V.; Lewis, C.M.; Persoons, A.; Mitiku, A.D.; Heaton, M.; Davey, P.E.; Abeyo, B.; Alemayehu, Y.; et al. MARPLE, a point-of-care, strain-level disease diagnostics and surveillance tool for complex fungal pathogens. BMC Biol. 2019, 17, 65. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate. Available online: https://github.com/tseemann/abricate (accessed on 1 May 2022).
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Chernyatina, A.A.; Low, H.H. Core architecture of a bacterial type II secretion system. Nat. Commun. 2019, 10, 5437. [Google Scholar] [CrossRef]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Garrido, J.; Ruano-Gallego, D.; Choudhary, J.S.; Frankel, G. The type III secretion system effector network hypothesis. Trends Microbiol. 2022, 30, 524–533. [Google Scholar] [CrossRef]
- Cherrak, Y.; Flaugnatti, N.; Durand, E.; Journet, L.; Cascales, E. Structure and activity of the type VI secretion system. Microbiol. Spectrum 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Coombes, B.K.; Wickham, M.E.; Mascarenhas, M.; Gruenheid, S.; Finlay, B.B.; Karmali, M.A. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 2008, 74, 2153–2160. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Néron, B.; Denise, R.; Coluzzi, C.; Touchon, M.; Rocha, E.P.C.; Abby, S.S. MacSyFinder v2: Improved modelling and search engine to identify molecular systems in genomes. Peer Community J. 2023, 3, e28. [Google Scholar] [CrossRef]
- Meuskens, I.; Saragliadis, A.; Leo, J.C.; Linke, D. Type V secretion systems: An overview of passenger domain functions. Front. Microbiol. 2019, 10, 1163. [Google Scholar] [CrossRef] [PubMed]
- Mehta, C.R.; Patel, N.R. Algorithm 643. FEXACT: A FORTRAN subroutine for Fisher’s exact test on unordered r×c contingency tables. ACM Trans. Math Softw. 1986, 12, 154–161. [Google Scholar] [CrossRef]
- Leyva Morales, J.B.; Valdez, J.B.; Bastidas, P.d.J.B.; Angulo Escalante, M.A.; Sarmiento Sanchez, J.I.; Barraza Lobo, A.L.; Olmeda Rubio, C.; Chaidez Quiroz, C. Monitoring of pesticides in Northwestern Mexico rivers. Acta Univ. 2017, 27, 45–54. [Google Scholar] [CrossRef]
- Amézquita-López, B.A.; Soto-Beltrán, M.; Lee, B.G.; Yambao, J.C.; Quiñones, B. Isolation, genotyping and antimicrobial resistance of Shiga toxin-producing Escherichia coli. J. Microbiol. Immunol. Infect. 2017, 51, 425–434. [Google Scholar] [CrossRef]
- Murillo Llanés, J.; Varón, J.; Félix, J.S.V.; González-Ibarra, F.P. Antimicrobial resistance of Escherichia coli in Mexico: How serious is the problem? J. Infect. Dev. Ctries. 2012, 6, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Burnham, C.-A.D.; Leeds, J.; Nordmann, P.; O’Grady, J.; Patel, J. Diagnosing antimicrobial resistance. Nat. Rev. Microbiol. 2017, 15, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Louie, M.; Cockerill, F.R., 3rd. Susceptibility testing. Phenotypic and genotypic tests for bacteria and mycobacteria. Infect. Dis. Clin. N. Am. 2001, 15, 1205–1226. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Hwang, S.Y.; Kwon, K.H.; Kim, K.Y.; Kim, J.H.; Park, Y.H. Prevalence and characteristics of Shiga toxin-producing Escherichia coli (STEC) from cattle in Korea between 2010 and 2011. J. Vet. Sci. 2014, 15, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Abd El Ghany, M.; Barquist, L.; Clare, S.; Brandt, C.; Mayho, M.; Joffre, E.; Sjöling, Å.; Turner, A.K.; Klena, J.D.; Kingsley, R.A.; et al. Functional analysis of colonization factor antigen I positive enterotoxigenic Escherichia coli identifies genes implicated in survival in water and host colonization. Microb. Genom. 2021, 7, 000554. [Google Scholar] [CrossRef] [PubMed]
- Bielaszewska, M.; Aldick, T.; Bauwens, A.; Karch, H. Hemolysin of enterohemorrhagic Escherichia coli: Structure, transport, biological activity and putative role in virulence. Int. J. Med. Microbiol. 2014, 304, 521–529. [Google Scholar] [CrossRef]
- Dubreuil, J.D. EAST1 toxin: An enigmatic molecule associated with sporadic episodes of diarrhea in humans and animals. J. Microbiol. 2019, 57, 541–549. [Google Scholar] [CrossRef]
- Ménard, L.-P.; Dubreuil, J.D. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): A new toxin with an old twist. Crit. Rev. Microbiol. 2002, 28, 43–60. [Google Scholar] [CrossRef]
- Paton, A.W.; Srimanote, P.; Woodrow, M.C.; Paton, J.C. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 2001, 69, 6999–7009. [Google Scholar] [CrossRef]
- Márquez, L.B.; Velázquez, N.; Repetto, H.A.; Paton, A.W.; Paton, J.C.; Ibarra, C.; Silberstein, C. Effects of Escherichia coli subtilase cytotoxin and Shiga toxin 2 on primary cultures of human renal tubular epithelial cells. PLoS ONE 2014, 9, e87022. [Google Scholar] [CrossRef] [PubMed]
- Paton, A.W.; Srimanote, P.; Talbot, U.M.; Wang, H.; Paton, J.C. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 2004, 200, 35–46. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; van Pelt, W.; van der Voort, M.; Heck, M.; Friesema, I.; Franz, E. Attribution of human infections with Shiga toxin-producing Escherichia coli (STEC) to livestock sources and identification of source-specific risk factors, The Netherlands (2010–2014). Zoonoses Public Health 2018, 65, e8–e22. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-S.; Lee, M.-S.; Kim, J.H. Recent updates on outbreaks of Shiga toxin-producing Escherichia coli and its potential reservoirs. Front. Cell Infect. Microbiol. 2020, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.C.; Burgess, C.M.; Fanning, S.; Duffy, G. An overview of Shiga-toxin producing Escherichia coli carriage and prevalence in the ovine meat production chain. Foodborne Pathog. Dis. 2021, 18, 147–168. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.T.; Sowers, E.G.; Wells, J.G.; Greene, K.D.; Griffin, P.M.; Hoekstra, R.M.; Strockbine, N.A. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 2005, 192, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Fuller, C.A.; Pellino, C.A.; Flagler, M.J.; Strasser, J.E.; Weiss, A.A. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 2011, 79, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, B.; Swimley, M.S. Use of a Vero cell-based fluorescent assay to assess relative toxicities of Shiga toxin 2 subtypes from Escherichia coli. Methods Mol. Biol. 2011, 739, 61–71. [Google Scholar] [CrossRef]
- Byrne, L.; Adams, N.; Jenkins, C. Association between Shiga toxin–producing Escherichia coli O157:H7 stx gene subtype and disease severity, England, 2009–2019. Emerg. Infect. Dis. 2020, 26, 2394. [Google Scholar] [CrossRef]
- Lee, M.S.; Koo, S.; Jeong, D.G.; Tesh, V.L. Shiga toxins as multi-functional proteins: Induction of host cellular stress responses, role in pathogenesis and therapeutic applications. Toxins 2016, 8, 77. [Google Scholar] [CrossRef]
- Griffin, P.M.; Karmali, M.A. Emerging public health challenges of Shiga toxin–producing Escherichia coli related to changes in the pathogen, the population, and the environment. Clin. Infect. Dis. 2017, 64, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Müthing, J.; Schweppe, C.H.; Karch, H.; Friedrich, A.W. Shiga toxins, glycosphingolipid diversity, and endothelial cell injury. Thromb. Haemost. 2009, 101, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Ageorges, V.; Monteiro, R.; Leroy, S.; Burgess, C.M.; Pizza, M.; Chaucheyras-Durand, F.; Desvaux, M. Molecular determinants of surface colonisation in diarrhoeagenic Escherichia coli (DEC): From bacterial adhesion to biofilm formation. FEMS Microbiol. Rev. 2020, 44, 314–350. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Suzue, K.; Takita, A.; Tomita, H. Roles of OmpA in type III secretion system-mediated virulence of enterohemorrhagic Escherichia coli. Pathogens 2021, 10, 1496. [Google Scholar] [CrossRef] [PubMed]
- Kortman, G.A.; Boleij, A.; Swinkels, D.W.; Tjalsma, H. Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS ONE 2012, 7, e29968. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Paterson, D.L. Multidrug-resistant bacteria in the community: Trends and lessons learned. Infect. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef] [PubMed]
- McGannon, C.M.; Fuller, C.A.; Weiss, A.A. Different classes of antibiotics differentially influence Shiga toxin production. Antimicrob. Agents Chemother. 2010, 54, 3790–3798. [Google Scholar] [CrossRef]
- Das, N.; Madhavan, J.; Selvi, A.; Das, D. An overview of cephalosporin antibiotics as emerging contaminants: A serious environmental concern. 3 Biotech 2019, 9, 231. [Google Scholar] [CrossRef]
- Cooley, M.B.; Carychao, D.; Crawford-Miksza, L.; Jay, M.T.; Myers, C.; Rose, C.; Keys, C.; Farrar, J.; Mandrell, R.E. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS ONE 2007, 2, e1159. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).