Abstract
The microbial biosynthesis of proteins, primary metabolites, and chemicals is gaining extraordinary momentum and is presently viewed as an advancing approach in the industrial research sector. Increased threats to the environment and the possibility of declining petroleum assets have switched the spotlight to microbial cell factories (MCFs). Aside from possessing various advantages over chemical synthesis, such as less toxicity, cheaper methodologies, and an environmentally benign nature, microbes can be cultivated in fermenters, resulting in an effective bioprocessing approach in terms of industrial relevance. As the overwhelming majority of biodiversity is microbial, this review first highlights the microbial biodiversity of industrially vital microorganisms. Then, the paper delineates the production pathways for generating valuable bioproducts via microbial workhorses. Many host cells synthesize bio-compounds as a part of their natural mechanism; however, several techniques have also been developed to attain the desired end product from non-native microbes with selected properties. The microbial biosynthetic pathways can be categorized as native-existing pathways, heterologous pathways, and artificial de novo pathways. Systems metabolic engineering, which integrates metabolic engineering with evolutionary engineering, synthetic biology, and systems biology, has further revolutionized the field of engineering robust phenotypes. The employment of these strategies improves the performance of the strain, eventually achieving high titer and productivity rates of bio-chemicals. Modern trends and tools for exploiting native pathways and designing non-native-created pathways are also briefly discussed in this paper. Finally, the review discusses the use of microbial workhorses for producing a myriad of materials and chemicals, including carboxylic acids, amino acids, plant natural products (PNPs), carotenoids, flavors, and fragrances, unveiling the efficacy of utilizing microbial species to generate sustainable bio-based products.
1. Introduction
Presently, fossil fuel energy, including natural gas, coal, and petroleum, accounts for 85% of the total energy use. The combustion by-product of fossil fuels (i.e., carbon dioxide) is a chief source of global warming and accounts for 52% of the total global warming factors, delineating the utilization of fossil energy as a highly unsustainable approach [1]. Rising concerns, regarding global climate change and fossil fuel depletion, have diverted attention toward developing sustainable alternatives to petroleum-based conventional chemical processes. Advancements in bio-refinery practices have promoted the production of multiple renewable bioproducts, such as polymers, fuels, and chemicals, from microbial sources [2]. Fermentation methodologies that employ microbial host strains induce the formation of several compounds, comprising vitamins, carboxylic acids, biofuels, enzymes, etc. (Figure 1). Thus, these microbial cell factories hold considerable value for generating high-value chemicals and other bio-compounds. Natural products derived from MCFs display multiple advantages, for instance, they have fewer land and water requirements in addition to being ecologically benign. Moreover, products obtained from microbial cells are cheaper and the processes are economically efficient [3]. A recent analysis of environmental biodiversity revealed that 99% of microbial existence was in the consortia form and further highlighted the applications of microbes as functional ingredients in the food sector and in biofuel production [4,5,6]. Generally, there are one million types of naturally derived products. Among them, 25% are biologically active compounds, 60% are derived from plant origins, and the rest are acquired from microbial sources. Microorganisms produce approximately 23,000 known secondary metabolites, of which 42% are derived from fungi and 32% are made by actinomycetes (filamentous bacteria) [7], depicting the high biodiversity and utilization of potential microorganisms as cell factories to attain industrially vital products.
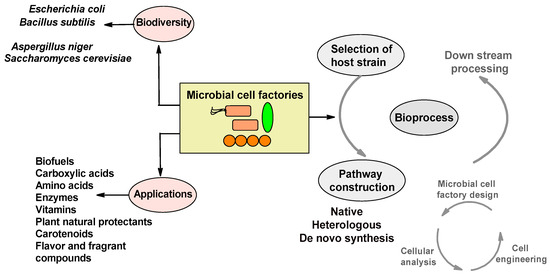
Figure 1.
Overview of microbial cell factories: biodiversity, applications, and industrial processing.
A wide range of microorganisms naturally synthesize various compounds, for example, the production of lactic acid from Gram-positive lactic acid bacteria (LAB), including strains belonging to the genera Lactobacillus sp., Lactococcus sp., Aerococcus sp., and Vagococcus sp. [8], and the formation of propionic acid from Propionibacterium sp., such as P. acidipropionici, P. thoenii, P. freudenreichii, and P. jensenii, under anaerobic fermentation conditions [9]. Technological advancements, which are accelerating the isolation and genetic alterations of production strains, have driven the concept of looking beyond conventional cell factories. At times, naturally occurring cell factories remain ineffective in achieving the desired yield and productivity rates of the end product [10]. The development of metabolic engineering and synthetic biology systems addresses this issue very well. By employing these modern approaches, desired productivity rates can be attained by utilizing microbial strains with ideal properties. The exploitation of the desired characteristics of microbial workhorses, especially those that are generally regarded as safe (GRAS) by the FDA, has led to a considerable progress in synthesizing various chemical compounds [6].
In particular, designing microbial cell factories begins with the selection of the product and microbial strain, followed by biosynthetic pathway identification, which can be native, heterologous, or de novo designs. In order to achieve the formation of new compounds and to ensure the optimal utilization of the selected pathway, enzyme and metabolic engineering techniques have been utilized [11]. With the implementation of modern strategies, several microbial-derived products have also been commercialized. For example, Novo Nordisk [12] and Avansya (DSM/Cargill) [13,14] carried out, respectively, the production of human insulin and steviol glycosides from Saccharomyces cerevisiae. Another company, Biosynthia, is also generating a variety of vitamins by using strains of Escherichia coli [15]. Several strategies have been studied and implemented to date for achieving the desired products from microbial strains, but there exist only a number of microbial-based natural products in the industrial sector, which highlights the importance of creating a commercial market for microbial-based bioproducts. The review aims to discuss the biodiversity and potentiality of microbial cell factories for generating industrially vital bio-compounds by critically summarizing the research and data available in the scientific literature. It also provides a comprehensive overview of biosynthetic pathways and modern bioengineering tools to generate robustness in microbial cells for the efficient production of desired bio-chemicals.
2. Biodiversity of Microbial Cell Factories
Microbial cell factories are employed for the bioconversion of substrates into value-added products (Table 1). These cell factories can overproduce native metabolites, heterologous products, and combinations of heterologous proteins. Microbial genomes express a number of metabolic pathways for the production of several novel chemicals. For millennia, lactic acid bacteria and baker’s yeast have been utilized to produce beverages and fermented foods [15]. Approximately 104 bacterial species have been cultured to date in the laboratory, leaving the majority of natural diversity still to be revealed [16]. The consideration of biodiversity is viewed as a major key factor for enhancing the yield of desired products, in addition to disclosing the potential of microorganisms to generate several valuable compounds. One example is lactic acid bacteria that include Lactococcus sp., Lactobacillus sp., Streptococcus sp., Pediococcus sp., and Leuconostoc sp. These genera fall under the category of the fundamental LAB group, while the peripheral LAB group includes Oenococcus sp., Tetragenococcus sp., Enterococcus sp., Weissella sp., and Sporolactobacillus sp. [17]. The fermentation of LAB strains gives rise to various industrial compounds, including lactic acid, amines, antibacterial and antifungal peptides, vitamins, γ-aminobutyric acids, antioxidant substances, acetone hydrogen peroxides, organic acids (such as butyric acid, succinic acid, acetic acid, and propionic acid), polyols, diacetyls, and flavor precursors [18,19]. Moreover, several yeast genera are also associated with foods and beverages, which include Saccharomyces sp., Cryptococcus sp., Brettanomyces sp., Galactomyces sp., Hansenula sp., Hyphopichia sp., Metschnikowia sp., Rhodotorula sp., Saccharomycopsis sp., Saccharomycodes sp., Schizosaccharomyces sp., Yarrowia sp., Candida sp., Debaryomyces sp., Geotrichum sp., Kluyveromyces sp., and Trichosporo sp. [20]. Aspergillus species are also in the spotlight of fungal research practices, and a large sector of industrial and academic research communities is dedicated to exploring this genus due to its economic importance. Several Aspergillus species are widely utilized in the production of drinks, foods, and organic acids, comprising A. aculeatus, A. oryzae, and A. niger [21]. Moreover, advancements in metabolic engineering and molecular biology tools have resulted in the production of several useful products from non-native microbes as well. The utilization of such tools has evolved the design of cell factories for the production of the desired compounds from microbes having the desired phenotypes, for example, the production of artemisinin (an antimalarial drug) and the sweetener stevia from S. cerevisiae strains and insulin synthesis from Pichia pastoris [22,23].

Table 1.
Biodiversity of microbial cell factories in forming different bio-compounds.
3. Designing Microbial Cell Factories
Microbial cell factories produce various valuable compounds by utilizing inexpensive, renewable feedstock in an eco-friendly and economic manner. For bio-based formations, microorganisms are considered ethically sound and can be cultured securely in fermenters. A recent report by Market Research Future (Market Forecast till 2030) stated that the global market size of bio-based chemicals is expected to reach USD 163.92 billion by 2030 with a compound annual growth rate (CAGR) of 8.30% [56]. Several microorganisms naturally synthesize bioproducts by different mechanisms. These workhorses include E. coli, B. subtilis, Lactic acid bacteria, S. cerevisiae, and A. niger, which naturally form lysine, vitamins, enzymes (glucose oxidase, catalase, phytase, etc.), bioethanol, and proteins, respectively [57,58,59,60,61]. Over the past two decades, this field has experienced tremendous advancements. In addition to upgraded and routine DNA recombinant processes, several other techniques have also been developed to achieve the desired yield of products with ideal properties. These advanced strategies include genome sequencing, in silico metabolic modeling, gene expression profiling, omics techniques, evolutionary approaches, and metabolic engineering strategies [57].
The development of microbial cell factories by utilizing metabolic engineering involves the rewiring of cell metabolism in order to augment native metabolite production or enable cells to generate new products. Traditional strategies are based on eliminating metabolic shunt-related enzymes, modifying metabolic pathways, and balancing the reducing power and ATP to shift the metabolic flux for attaining desired end products [62]. Despite the potential manufacturing of several bioproducts, these metabolic cell factories face several limitations during industrial implementations, such as the production of toxic inhibitors, a low pH environment, and extreme temperature conditions, which leads to a relatively low industrial productivity compared to that acquired at the lab scale [63]. Considering the limitations associated with traditional metabolic approaches, the phenomenon of systems metabolic engineering emerges. Systems metabolic engineering is a more recent approach that includes the integration of traditional metabolic engineering approaches with the tools and strategies of synthetic biology, system biology, and evolutionary engineering. The advent of systems metabolic engineering stimulated the production of high-performance microbial strains, fine and bulk chemicals, biofuels, natural products, and polymers [64]. At first, the development of MCFs begins with the selection of the desired end product and feedstock (carbon source). Before constructing MCFs, there should be a careful evaluation of the market demands of the respective product. Secondly, the carbon substrate or raw materials should be selected pragmatically, concerning the inclusive economics of bio-refineries. Several factors should be taken into account while selecting a substrate, including lignin and sugar composition, overall abundance, seasonal dependency, and costs of transport. Evidently, a substrate with a low cost and stable supply should be used [62]. Then, the host microbial strain to be engineered and the construction pathway of the desired product are selected, followed by evolutionary engineering and pathway optimization to achieve higher yields of the end product.
3.1. Microbial Strain Selection
After the selection of a valuable compound and substrate, a cell factory is selected, i.e., the host microorganism. The criteria for selecting the starting strain comprise the following steps: (i) easier manipulation of the respective organism; (ii) whether the selected product is native, partially native, or non-native; (iii) suitability and sustainability for large-scale production; (iv) optimum growth and desired product formation on simple culture mediums; (v) metabolic capacity towards the end product; (vi) bioprocess compatibility; (vii) cost-effective recovery; (viii) and purification procedures [65]. To date, various studies have reported the engineering of model organisms as microbial cell factories. E. coli and S. cerevisiae have been extensively investigated tools for genetic manipulation. Several other strains have also been widely studied, including Clostridium sp., Bacillus sp., and Pseudomonas sp. Furthermore, a prompt genome-scale metabolic model construction, economical genome-sequencing techniques, and newer genomic and genetic manipulation tools, such as the metabolic engineering of microorganisms through clustered, regularly interspersed short palindromic repeats (CRISPRs), have made the engineering of microbial strains far easier and inexpensive than in the past [66,67,68]. One example includes succinic acid production through the metabolic engineering of Mannheimia succiniproducens. The organism was isolated from cow rumen based on the hypothesis that the CO2-rich environment could contain a bacterium with the capability of performing phosphoenolpyruvate carboxylation. In silico genome-scale metabolic simulations were carried out in order to identify the gene targets, followed by the construction of PALK strain (ΔldhA and Δpta-ackA). Fed-batch cultures of this strain on glycerol and glucose carbon sources provided higher yields of succinic acid, indicating the effectiveness of systems metabolic engineering and genetic manipulation tools for achieving the high productivity of the desired end product [69]. In silico genome-scale simulation and modeling is an effective tool for the selection of suitable production organisms. The systematic computational assessment and extensive evaluation of E. coli capacities for producing varied chemical compounds demonstrated the production of 1777 non-native products by the introduction of heterologous enzymes. Among these products, 279 compounds were reported to have vast commercial applications [70].
3.2. Pathway Construction: Native, Heterologous, and Artificial De Novo
3.2.1. Native Pathway
As discussed earlier, the development of microbial cell factories begins with the selection of a product of interest. It can be a native metabolite, generated by the wild-type host (chassis organism), and the chosen chassis can be made to produce the desired product through supplementary metabolic capabilities. Natural overproducers are favored host strains for the effective production of bioproducts. This is because these microorganisms already exhibit robust metabolic fluxes and a high tolerance toward the target products. Examples include the production of succinate by wild-type S. cerevisiae [71] and butanol and isobutanol production through Clostridium and yeast species, respectively (Figure 1). Well-known Clostridia spp. that are natural producers of biobutanol include C. beijerinckii, C. pasteurianum, C. acetobutylicum, C. cadaveris, C. saccharoperbutylacetonicum, C. tetanomorphum, C. sporogenes, and C. aurantibutyricum [72]. Glutamate and L-lycine are also naturally produced by C. glutamicum [73]. Moreover, these microbial overproducers can also be metabolically engineered for the production of other relevant chemicals, for example, the utilization of the L-lysine-producing C. glutamicum PKC strain as a chassis strain for cadaverine (polyamide monomer) production. L-lysine decarboxylase (LDC) catalyzes a direct decarboxylation reaction to convert L-lysine into cadaverine. The integration of the ldcC gene of E. coli into the genome of the C. glutamicum strain with the concomitant L-lysine exporter (LysE) leads to the production of an engineered C. glutamicum strain, producing greater yields of cadaverine under fed-batch glucose fermentation. Furthermore, after the purification of cadaverine, it can be chemically polymerized with sebacic acid, which can be effectively obtained from plant oil for synthesizing nylon 510 (a commodity plastic exhibiting higher-temperature resistance). Consequently, entirely bio-based 510, along with several other bio-based nylons, can be obtained from similar pathways [74].
It is important to emphasize that the presence of the biosynthesis pathway simply does not provide the assurance of selecting a particular chassis. Even in the presence of an already-existent biosynthetic pathway, often enzymatic or engineering strategies can be prerequisites for escalating the metabolic flux. A recent example showed the overproduction of succinate by creating MCFs by utilizing S. cerevisiae with a computationally derived metabolic engineering strategy. Although S. cerevisiae is a natural producer of succinate, it is consumed during the TCA cycle. Metabolically engineered S. cerevisiae strains can generate industrially desired succinate quantities by removing succinate dehydrogenase and 3-phosphoglycerate dehydrogenase isoenzymes. As a result, the resulting mutant further upregulates the conversion of iso-citrate to succinate and glyoxylate, ultimately counteracting the deficiencies of serine and glycine [71].
3.2.2. Heterologous Pathway
Another approach to create MCFs is installing heterologous pathways into chassis organisms to generate new products. The strategy is usually utilized if the desired product is (i) relevant to higher organisms (such as plants) with costly industrial production rates, (ii) a product of artificial metabolism and non-native to microbes, or (iii) native to a microorganism that is challenging to culture [65]. An example of the establishment of a heterologous pathway is artemisinic acid production, which is an antimalarial drug precursor in S. cerevisiae. For this purpose, firstly, Artemisia annua genes were discovered that were involved in the biosynthesis of artemisinic acid. The pathway constituted the conversion of amorphadiene into artemisinic acid in three steps, catalyzed by amorphadiene oxidase (CPR1, CYB5, and CYP71AV1), artemisinic aldehyde dehydrogenase (ALDH1), and alcohol dehydrogenase (ADH1). Then, the engineering of the S. cerevisiae strain that involves the overproduction of amorphadiene was carried out to downregulate the expression of squalene synthase (ERG9). The later step was carried out under the copper-regulated CTR3 promoter and led to increased amorphadiene concentrations [75].
Another example of utilizing heterologous enzymes for their natural function, involving natural substrates, is the production of resveratrol by engineered E. coli holding phenylpropanoid biosynthetic pathways. The biosynthetic pathway for resveratrol formation in E. coli is built via heterologous enzymes from plants and bacteria. Resveratrol is a natural secondary metabolite and is gaining a lot of attention due to its inhibitory activity and carcinogenesis potential. The production of plant-specific metabolite resveratrol by engineered E. coli has been reported by utilizing tyrosine as an initial precursor. The strategy consisted of designing and expressing the bacterial tyrosine ammonia-lyase (TAL) with artificial phenylpropanoid pathways. This was accomplished by assembling TAL, cinnamate/4-coumarate:CoA ligase (ScCCL), and stilbene synthases (STSs) from the actinomycete Saccharothrix espanaensis, S. coelicolor, and Arachis hypogaea, respectively, on a single plasmid in E. coli. Stilbenes are formed by tyrosine and then converted into 4-coumaric acid, followed by acid activation to form CoA esters. These CoA esters act as a substrate for STSs, condensing with three malonyl-CoA molecules, and results in the production of resveratrol [76]. Moreover, the production of astaxanthin through a metabolically engineered E. coli strain was also demonstrated. Astaxanthin has vast applications in both the pharmaceutical as well as cosmetic and food industries. It is produced by the introduction of a series of heterologous genes, comprising trCrBKT and crtEYIBZ from Chlamydomonas reinhardtii and Pantoea ananatis, respectively, in the E. coli strain, resulting in the establishment of a biosynthetic pathway for astaxanthin production. A higher astaxanthin yield from glycerol was observed after the fed-batch fermentation of the engineered strain. Furthermore, for enhancing its production, additional engineering strategies were also taken into account, such as the fusion of solubility-enhancing tags and E. coli signal peptides to trCrBKT and overexpressing the native genes (ispDF), centered on an in silico metabolic flux analysis [77].
3.2.3. Artificial De Novo Pathway
The design, construction, and establishment of heterologous pathways can facilitate the effective production of desired chemical compounds. However, there is an unavailability of metabolic routes and enzymes for several target chemicals, which shifts the focus toward de novo artificial synthesis for developing new enzymes that exhibit the potential to catalyze similar reactions through directed evolution and protein engineering. The artificial de novo biosynthetic pathway comprises re-arranging characterized enzymes, along with a dependency on enzyme promiscuity, for assisting the production of new products. This approach is also an area of several ongoing research activities [11]. Several efforts have been made to generate industrially vital compounds through artificial de novo pathways. For instance, there is a de novo metabolic pathway for the production of four, five, and six carbon lactams, also referred to as butyrolactam, valerolactam, and caprolactam, respectively. The pathway involves the utilization of ω-amino acids as precursors. The C. propionicum-derived β-alanine CoA transferase (Act) enzyme catalyzed the activation of ω-amino acids, followed by the spontaneous cyclization to lactams. Three metabolically engineered strains of E. coli sp. were developed by presenting this newly constructed pathway, followed by a system-level optimization. This strategy can be applied to attain higher production rates of these industrially vital lactams by fed-batch fermentation in the presence of renewable carbon sources [78]. Another example is the production of 2-pyrrolidone (a butyrolactam), which is a valuable chemical with vast applications as a polymer precursor, solvent, and as an active intermediate in pharmaceuticals. A novel butyrolactam synthase, ORF27, derived from S. aizunensis, catalyzed the ring-closing dehydration of γ-aminobutyrate to butyrolactam by an ATP-dependent mechanism. The recombinant E. coli strain, when metabolically engineered to express the genes encoding mutant glutamate decarboxylase and ORF27, led to the production of higher butyrolactam yields from L-glutamate (Figure 2) [79]. Moreover, the synthesis of resveratrol glucoside derivatives, including resveratrol-4′-O-glucoside and resveratrol-3-O-glucoside, also referred to as resveratroloside and piceid, respectively, can be achieved from simpler carbon sources by initiating de novo pathways in an E. coli system [80].
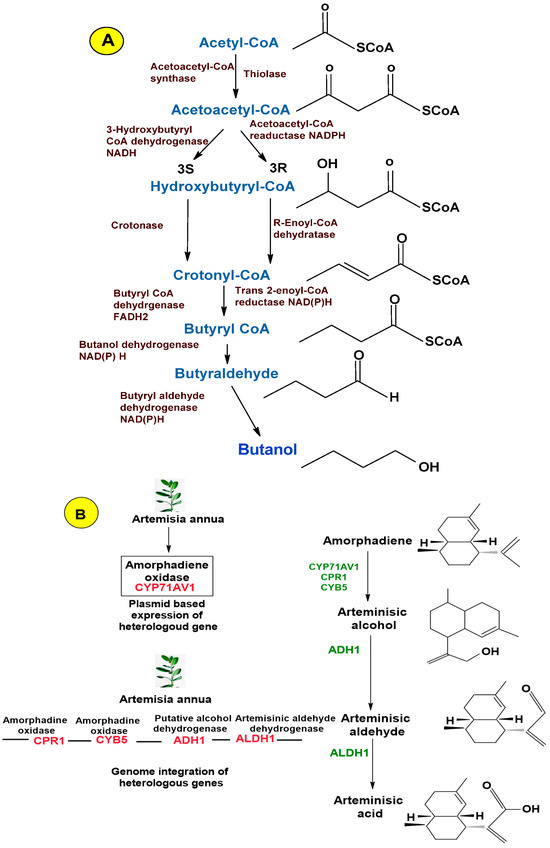
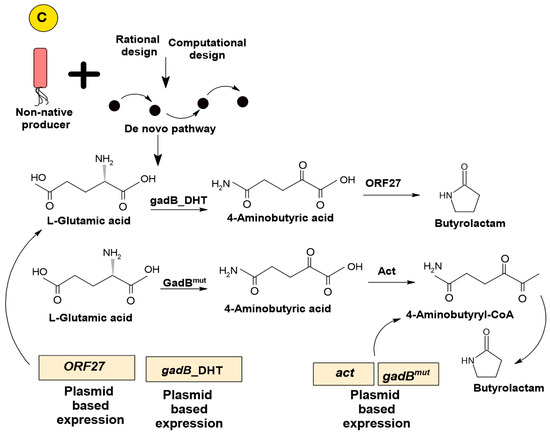
Figure 2.
Biosynthetic pathway construction of microbial cell factories: native, homologous, and artificial de novo. (A) Biosynthetic native mechanism of Clostridium sp. for the production of butanol. (B) Heterologous mechanism for the production of artemisinic acid by engineered S. cerevisiae. (C) Artificial de novo synthesis of butyrolactam by utilizing engineered E. coli as a host strain [57,72,75,79].
Heterologous pathways do not always lead to the production of desired products and also deliver non-economical routes; thus, the exploration of artificial de novo biosynthetic pathways in accordance with enzyme promiscuity is a preferable approach. The rational identification of appropriate enzymes is a daunting task; therefore, computer-aided pathway prediction tools are utilized for devising novel biosynthetic pathways. Several pathway prediction programs have been developed to date, including Biochemical Network Integrated Computational Explorer (BNICE), BRENDA, and Metabolic Tinker [11]. The Genomatica’s in-house SimPheny BioPathway Predictor software (http://www.genomatica.com/, accessed on 4 February 2024) has been applied for the production of 1,4-butanediol (BDO) from central metabolites, such as succinyl-CoA, α-ketoglutarate, L-glutamate, and acetyl-CoA. 1,4-BDO has vast applications in industries, especially for the manufacturing of polyurethanes. The results reveal the suitability of a-ketoglutarate and succinylCoA-based pathways for 1, 4-BDO production [81].
4. Robustness of Microbial Cell Factories
As described earlier, the advancements in engineering strategies and synthetic biology display a great potential for the manufacturing of several bioproducts. However, the implementation of these strategies at the industrial scale faces harsh conditions, such as high temperatures, a low pH, and the presence of toxic inhibitors, ultimately yielding a low productivity compared to that at the lab scale. The robustness of MCFs is their ability to sustain the phenotypes of high efficiencies in the presence of diverse perturbations [63]. The implementation of engineering systems can generate robustness in microbial strains for creating tolerance against toxin compounds and other harsh conditions. The strain tolerance can be increased by the engineering functions of regulation/transcription factors. As an example, the improved tolerance of E. coli is achieved against high-ethanol-stress conditions and different chemicals by engineered sigma factor δ70 that controls the transcription of several genes. Robustness in a microbial cell can also be created by transporter and membrane engineering. Moreover, it can also be achieved by engineering protection, repairing, or competition pathways [82]. A graphical description of the mechanisms behind generating robustness in cell strains is presented in Figure 3.
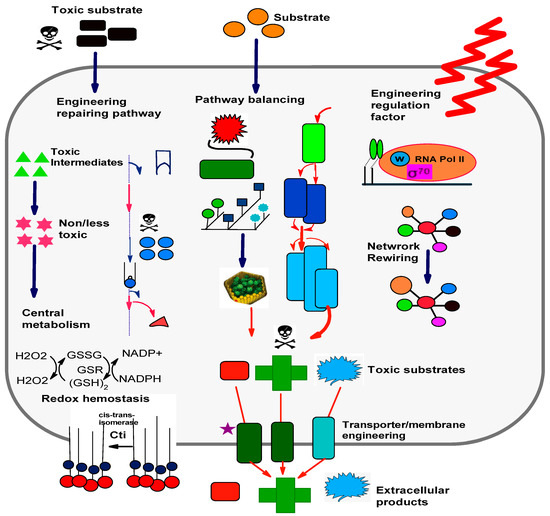
Figure 3.
Engineering of robustness in microbial cell factories: engineering of regulation factors in cells causes the cell to achieve its fitness state for tolerating unfavorable conditions. (1) Introducing synthetic pathways and engineering repairing pathways sustain redox and cellular metabolic hemostases and transform toxic substrate into less/nontoxic compounds, respectively. (2) Pathway balancing evades the buildup of toxic intermediates in addition to refining the biosynthesis rate. (3) Transporter engineering relieves toxicity while membrane engineering generates tolerance in membrane against stress and toxic molecules [82].
Systems metabolic engineering that mixes traditional metabolic engineering methods with synthetic biology, systems biology, and evolutionary engineering is empowering the improvement of MCFs, which are adept at proficiently generating a myriad of materials and chemicals comprising bulk and fine chemicals, biofuels, amino acids, polymers, drugs, and several other natural products [82,83,84]. With the onset of the bio-big data era, artificial intelligence (AI)-centered data-driven methods have been dynamically applied for retrosynthetic algorithms. One recent tactic, RetroPath RL, utilized reinforcement learning as the focal engine for exploring the reaction rules and chemical space compiled in RetroRules [83], thus having a major role in improving the pathway design. Several systems metabolic engineering tools were employed for this purpose, as presented in Table 2.

Table 2.
An overview of the strategies and design tools for the systems metabolic engineering of microorganisms for the efficient production of biochemicals [57,83].
5. Application of Microbial Cell Factories
Microbial cell factories are becoming a fundamental technology for pharmaceutical, food, and chemical industries to satisfy the welfare of an increasing global population and socio-economic development. Microorganisms are used for the production of various products, including carboxylic acids, amino acids, vitamins, enzymes, plant natural products, carotenoids, biogas, and other biofuels. About 52% of FDA-approved chemical entities were naturally derived products during the period of 1981–2006 [85]. The production of varied value-added macromolecules and metabolites was witnessed in the last decade by MCFs, with titers changing from μg/L to mg/L. Moreover, the introduction of metabolic engineering approaches improved the rate, titer, and yield of industrially vital compounds by manipulating the host metabolism, physiology, stress response, carbon–energy balance, and the annihilation of an undesirable ATP sink [10]. Due to MCFs, the industrial biotechnology sector is increasing expeditiously, and numerous biocommodities are also in production. The vast applications of microbial workhorses for generating various industrially relevant chemicals and products are presented in the section below.
5.1. Carboxylic Acids
Carboxylic acids are considered as appealing bio-renewable chemicals due to their flexibility and utilization as precursors for varied industrial chemicals. Currently, conventional chemical processes are used to achieve carboxylic acid synthesis from petroleum resources. However, an excessive reliance on petrochemical-based raw materials and the requirement of toxic metals, including organic solvents and heavy metal catalysts, have questioned the proficiency of chemical processes in terms of exacerbating the environmental burden. As a result of these concerns, deriving carboxylic acids from renewable microbial sources becomes apparent [86]. Carboxylic acids can be produced by several microbial groups, for instance, acetic acid production from A. cerevisiae, A. aceti, Gluconacetobacter liquefaciens, G. entanii, Komagataeibacter intermedius, and K. hansenii [87], and the synthesis of succinic acid from Anaerobiospirillum succiniciproducens, M. succiniciproducens, and Actinobacillus succinogenes [88]. Acetic acid bacteria (AAB) play a major role in the production of food and beverages, such as kombucha and vinegar [74]. Butyric acid also has vast applications in animal feed, food additives, perfumes, and varnishes. In nature, several Clostridium species synthesize butyric acid, particularly, C. thermobutyricum, C. butyricum, C. pasteurianum., and C. acetobutylicum [89].
With the advancements in metabolic engineering, the production of carboxylic acids from metabolically engineered microbial systems has gained much attention to achieve the desired yield and titer. Several non-conventionally engineered microbes have been demonstrated as superior hosts for carboxylic acid biosynthesis. For instance, the enhanced production of propionic acid, which is widely used as a food additive, antimicrobial preservative, and an intermediate for producing polymers, pesticides, and flavorings, can be achieved by engineering P. acidipropionici sp. CGMCC 1.2232 [90], P. freudenreichii subsp. Shermanii DSM 4902 [91], and P. jensenii sp. ATCC 4868 [92]. The FDA has designated several Propionibacterium species (derived from dairy goods) as generally regarded as safe (GRAS), electing them as a suitable approach for producing propionic acid in the food industry [93]. Lactic acid (2-hydroxypropanoic acid) serves as an additive in the food, pharmaceutical, and textile industries. Moreover, it is also used as a synthetic intermediate to manufacture biodegradable polymers [94]. Lactic acid bacteria (LAB), including Lactobacilli and Lactococci sp., are highly favored microbes for lactic acid biosynthesis and have achieved GRAS status as well [86]. Engineering microbial species of C. utilis [95] and L. lactis LM0230 [96] also increased lactic acid production. Citric acid is broadly used in the cosmetics, food, and detergent industries owing to its buffering characteristics, water solubility, and reduced toxicity. Presently, microbial fermentation using Y. lipolytica and A. niger is being carried out for citric acid production on an industrial scale [97]. A. succinogens also produce succinic acid naturally [98]; however, several microbial strains can also be metabolically engineered for enhanced production, such as engineering L. plantarum sp. NCIMB 8826 [99], C. glutamicum, [100], and C. synechococcus elongates sp. PCC 7942 [101], which provide higher succinic acid yields. Succinic acid serves as a precursor for preparing polyester, a surfactant, and several other valuable derivatives. Notably, succinic acid is among the few examples of biosynthesizing carboxylic acids by industrial manufacturers, for instance, Reverdia and BioAmber successfully commercialized succinic acid, accounting for half of the global annual production [102].
5.2. Vitamins
Vitamins are essential nutrients that play a crucial role in maintaining optimal health and metabolic activities. Vitamins show wide-ranging applications in the food, feed, cosmetics, and pharmaceutical industries. The requirement of pressurized reactors and high temperatures, along with the safety concerns regarding increased pollution from hazardous waste, have shifted the focus toward utilizing microbial sources instead of chemical synthesis methods for the production of valuable vitamins [46]. Several microbes serve as cell factories for this purpose, for instance, E. coli has emerged as a preferred approach for the production of vitamin B1 (also known as thiamine), which has vast applications in preventing skin inflammation and eczema [103]. Furthermore, the emergence of synthetic biology and metabolic engineering approaches has further strengthened the concept of utilizing microbial cell factories for the production of vitamins, such as the increased yields of vitamin B1 and vitamin A (β-carotene), which can be attained from A. oryzae and Y. lipolytica by overexpressing genes and heterologous enzymes [43,44], respectively. L-ascorbic acid (LAA), also known as vitamin C, can also be produced from several microbial species. Vitamin C is an antioxidant and also acts as a significant co-factor in multiple reactions that occur in the body. LAA is presently manufactured commercially by utilizing B. megaterium and G. oxydans [46]. P. shermanii, and P. denitrificans produce cobalamin (vitamin B12), while vitamin K can be attained from Sinorhizobium meliloti, B. subtilis [104,105,106,107]. Recently, MK-7, which is an effective subtype of vitamin K produced from B. subtilis natto, became an FDA certified-safe food [107]. From the perspectives of the absorption rate, biological activity, and safety concerns, the biological synthesis of vitamins by using microbial strains is a promising approach for achieving a significant yield improvement of vitamins.
5.3. Amino Acids
Amino acids serve as attractive metabolites in the food industry and pharmaceutical fields. They also play a role in enhancing flavor formation in several fermented foods. At present, C. glutamicum, P. ananatis, and E. coli are extensively utilized host bacterial strains for the industrial production of amino acids. Moreover, a glutamate secreting bacterial isolate, C. glutamicum, is a preferred bacterium to be exploited in engineering approaches for the increased production of lysine, glutamate, and flavor-active amino acids at vast scales [108,109]. Lysine and valine are two vital amino acids with varied applications. Lysine is an essential amino acid and is used as a feed additive, with a market size reaching around 1 million tons per annum [109], while valine, a branched amino acid, is an essential nutrient for animals. Valine is also utilized as a raw material for the production of various herbicides and drugs, with an annual fermentative production of 500 tons [110]. E. coli and C. glutamicum are considered as the most powerful industrial microbes for both lysine and valine production [111,112,113,114]. Considering the fact that industrial processes often operate at high temperatures, thermotolerant microbes have also been developed. For instance, thermotolerant bacteria, C. efficiens and B. methanolicus, are being developed as lysine producers [115]. Another semi-essential amino acid, L-Arginine (L-Arg), has widespread applications in the pharmaceutical industry as it promotes the secretion of insulin, growth hormones, and prolactin [108,116]. Several engineered microbes, including B. subtilis, E. coli, C. glutamicum, S. cerevisiae, and C. crenatum, have been used as model organisms for achieving L-Arg overproduction [117]. Moreover, L-ornithine has also been synthesized by a high-glutamate-producing strain, C. glutamicum S9114, through pathway engineering [118]. L-Ornithine is a non-protein amino acid and is universally used for the treatment of trauma and liver protection, in addition to strengthening the heart. High-level L-ornithine production is also achieved by carrying out modular pathway rewiring [119]. Constructing microbial workhorses on a large scale aims at satisfying the demand for amino acids as bulk biochemicals. Moreover, amino acid secretion from microbial sources is economically and industrially relevant to biotechnological fermentation processes, as it involves the simplified extraction and purification of metabolites.
5.4. Plant Natural Products
Plant natural products exhibit significant pharmacological activities and have widespread utilization in healthcare products, food additives, and cosmetics. Most PNPs are extracted from cultivated plants; however, the yield is restricted due to complex processing steps, a long growth cycle, climate change, and seasonal availability, making the process quite unsustainable. Moreover, a complex PNP structure also affects its chemical synthesis efficiency [120]. With the development of modern approaches, the biosynthesis of PNPs from microbial alternatives has gained considerable attention. Several microbial species have been utilized for PNP production, including artemisinin, resveratrol, and many carotenoids as well. Artemisinin is referred to as an effective pharmaceutical compound for treating malaria. It is naturally synthesized by plant A. annua; however, the concentration obtained from the plant source is very low, i.e., 0.01–1.1%, and improving the yield through total organic synthesis or plant breeding remains a contest, shifting the focus toward production via microbial sources [121]. Several microbial strains have been genetically engineered for its production. For meeting large-scale demands, the de novo reconstitution of artemisinin biosynthesis in heterologous microbes, including S. cerevisiae and E. coli, has noteworthy achievements, for instance, engineering the carbon flux and genotype of S. cerevisiae yields high quantities of artemisinic acid. Moreover, S. cerevisiae has also demonstrated itself as a heterologous host for producing artemisinin precursors, such as artemisinic acid, dihydro-artemisinic acid, and amorphadiene [122].
Resveratrol is a polyphenolic compound and is also found in various plant species, such as peanuts, grapes, cranberries, and bush berries. It also exhibits wide applications in the cosmetic, health, and medicine industries, and is reported to reduce the risks of cancer, heart diseases, and diabetes [123]. It is among the fastest-growing nutritional supplements in the flavonoid market [124]. Like artemisinin, the resveratrol concentration in plants is limited and is commercially unsustainable as well. Engineered strains of E. coli and budding yeast S. cerevisiae result in increased resveratrol titers by the introduction or modification of STS enzymes (stilbene synthase) [125,126]. Evolva has effectively developed the production of resveratrol on an industrial scale by utilizing a yeast cell factory [121]. The two strains serve as platform organisms and excellent hosts for the production of a wide array of PNPs, such as isoprenoids (l-histidine, l-phenylalanine, and l-tyrosine), phenylpropanoids (flavonoids, coumarins, and lignans), and alkaloids (carotenoids and sterols) [127]. Although the eco-friendly and resource-conserving synthesis of PNPs has gained much attention, several challenges are associated with this approach as well, regarding their large-scale application. Moreover, a low enzyme catalytic activity, poor precursor supply, and unknown PNP biosynthesis pathways have limited the heterologous production of microorganisms due to the high fermentative costs [121].
5.5. Carotenoids
Carotenoids are lipid-soluble, naturally occurring pigments and are widely used in industries owing to their antioxidant activities and capacity as natural colorants [128]. By the year 2025, the carotenoids global market is anticipated to reach USD 1.68 billion [129]. Photosynthetic cyanobacteria serve as natural microbial factories for the synthesis of carotenoid pigments as cellular antioxidants. Many successful demonstrations have also been reported for genetically engineering cyanobacteria to achieve an improved carotenoid content [130]. At the industrial scale, the filamentous fungi Phycomyces blakesleeanus and Blakeslea trispora serve as potential candidates for carotenoid production [131]. Moreover, other carotenogenic microbes, including Haematococcus pluvialis and X. dendrorhous, have also been widely exploited in large-scale processes. Moreover, transforming carotenoid genes in non-carotenogenic microbes, such as C. utilis, E. coli, S. cerevisiae, and Zymomonas mobilis, can elevate carotenoid production levels [132]. Several recombinant or engineered microbes also demonstrated the successful production of carotenoids, including recombinant S. elongatus sp. PCC 7942 [133], Planococcus faecalis [134], Halobacillus halophilus [135], B. indicus, and B. firmus [136].
Cyanobacteria produce a diverse range of metabolites in the carotenoid biosynthetic pathway, and zeaxanthin is among these principal carotenoids. Zeaxanthin is a naturally occurring xanthophyll carotenoid and is widely produced by algae, plants, and microorganisms. It is utilized in the food, nutraceutical, and pharmaceutical industries due to its antioxidant and anti-cancer properties. Moreover, it also plays a critical role in preventing cataracts and macular degeneration. Many bacterial isolates, predominantly belonging to the Paracoccus and Flavobacterium genera, demonstrate the active accumulation of zeaxanthin. The Flavobacteriaceae family primarily comprises zeaxanthin-producing bacteria, including Kordia aquimaris sp. CC-AMZ-301T 150 [137], Aquibacter zeaxanthinifaciens sp. CC-AMZ-304T 151 [138], G. oceani sp. CC-AMSZ-TT 152 [139], and G. planctonica sp. CC-AMWZ-3T 153 [50]. Also, non-photosynthetic Flavobacterium sp. produces an abundant amount of zeaxanthin, accounting for 95% of total carotenoid production [140].
Another carotenoid, astaxanthin, can also be produced from microbial workhorses. Astaxanthin is an ideal source of pigmentation in the food and aquaculture industries. It also exhibits several health benefits owing to its anti-inflammatory, anti-cancer, antioxidant, and neuroprotective activities [141]. Several microbial strains serve as cell factories for astaxanthin biosynthesis, such as Brevundimonas sp. [142], Paracoccus sp. [143], and Sphingomonas sp. [144]. Among yeast species, X. dendrorhous is a major astaxanthin producer [145]. Metabolic engineering has developed potential commercial interests in enhancing carotenoids’ yield and titer to attain sustainable production. As an instance, E. coli and S. cerevisiae have also been exploited as cell factories for astaxanthin production by carrying out modular engineering and the membrane-fused expression of β-carotene hydroxylase [77,146,147]. Another xanthophyll pigment, i.e., lutein, can also be biosynthesized in S. cerevisiae. The lutein pigment is widely used in aquaculture, healthcare, food processing, and poultry farming industries [52]. The production of carotenoids by microbes provides an insight into the economically viable, environmentally friendly, and renewable production of natural products, depicting the industrial viability of MCFs for carotenoid biosynthesis.
5.6. Flavors and Fragrances
In addition to several valuable chemicals, flavor and aroma compounds are also synthesized by MCFs, which include indole, terpenoids, β-Ionone, geraniol, 3-phenylpropanol, vanillin, and patchoulol. The global flavor and fragrance market was valued at USD 28,193.1 million in 2019 and is anticipated to achieve USD 35,914.3 million by 2027 with a CAGR of 4.7% [148,149]. Moreover, the cosmetic industry has presented an even higher market value and is expected to reach USD 363.80 billion by 2030 [150]. Thus, the inclination toward utilizing natural and sustainable products drives the focus of ingredient and fragrance firms toward finding alternative sources to create natural products, i.e., microbial biosynthesis [151].
Indole is a nitrogen-containing aromatic compound and is famed for its jasmine-like floral odor [152]. Corynebacterium strains display the sustainable production of indole from tryptophan to achieve industrially pertinent indole titers [153]. Terpenoids are vital aroma components for perfumery and flavor products. β-Ionone, a fragrant terpenoid, possesses a pleasing floral scent. Microbial fermentation using GRAS species, such as Y. lipolytica, is an effective approach for β-Ionone production as it is oleaginous, economically viable, and a sustainable natural aroma compound [154]. Several monoterpenoids can be generated by using recombinant microbial strains, for instance, the metabolic engineering of P. putida to produce geranic acid. Geraniol is an acyclic monoterpene alcohol and it is a standard constituent of many fragrance and flavor products. E. coli engineering for expressing the heterologous mevalonate pathway can result in an increased titer of geraniol [155]. E. coli cells have also been engineered to produce 3-Phenylpropanol and vanillin. Both flavor compounds are used in the beverages, food, fragrance, and cosmetic industries [156,157]. Moreover, patchoulol is a naturally occurring sesquiterpene and is found naturally in Pogostemon cablin. It has a wide range of applications in cosmetics and daily use products, including shampoo, hair spray, perfumes, and essential oils. Its limited production from natural resources shifted the focus toward engineered microbes, such as mitochondria-engineered yeast cells (S. cerevisise), which reported the elevated production of patchoulol [158]. Thus, a consequential upsurge in aroma chemicals requires efficient renewable production strategies. For this purpose, biotechnological production from microbial sources is regarded as the best alternative to generate flavor and fragrant molecules [159].
5.7. Bioenergy
Increasing energy and food demands, the deterioration of the environment, and climate change are the major challenges of this era. Among several other natural sources, biofuels also have immense potential to harmonize the trilemma of food, energy, and environment. Using MCFs for the production of bioenergy is not a new concept, yet it has received great attention for creating novel bioproducts with better productivity rates [160]. Several microbes serve as bioenergy sources, such as the production of 1-butanol, which is a potential fuel and chemical feedstock, by both native and engineered Clostridium species [161], and bioethanol production from Pichia stipites sp., C. albicans, and S. cerevisiae sp. KL17 [162,163,164]. Apart from fuels, microbes also serve as factories for biogas and biohydrogen production. Methanogens are integral to biogas production and high-performance methanogens include Methanocaldococcoccus jannaschii and the methanococci Methanotorris igneus [165]. Methanogens can also be employed as autobiocatalysts for converting molecular hydrogen (H2) and carbon dioxide (CO2) to biological CO2-based methane, also referred to as the CO2-BMP process. This process has varied applications, comprising the decentralized production of energy, power-to-gas applications, and biogas upgrading [166]. The most studied microbe for this purpose is Methanothermobacter marburgensis. It has several advantages, including flexibility toward substrate gas impurities and elevated CH4 productivity rates [167]. Biohydrogen is also an environmentally benign fuel, having no CO2 emissions during combustion. It can be produced by the biological conversion processes of photofermentation and dark fermentation. Several microbes can be utilized for biohydrogen production, including Caldicellulosiruptor saccharolyticus [168], Desulfurococcus amylolyticus DSM 16532, and Rhodobacter sphaeroides [169]. Moreover, biodiesel, a derivative of fatty acids, such as methyl, ethyl, or propyl esters (fatty acid methyl esters (FAMEs), fatty acid ethyl esters (FAEEs), and fatty acid propyl esters (FAPEs)) has an annual consumption rate of 2 billion gallons. Several studies have reported the engineered E. coli strain as an effective microbial source for producing structurally tailored fatty esters (i.e., biodiesel) [170,171,172]
The metabolic engineering of microbes has led to the production of next-generation biofuels. The biofuel molecules with petroleum replica structures exhibit greater advantages than others as they possess elevated energy densities and can also be utilized as drop-in fuels because of no modification requirements in the internal combustion engines, destined for petro-diesel and gasoline [170]. These biofuels are referred to as fourth-generation (4G) or advanced biofuels. Few microorganisms naturally produce such biofuels, for example, the production of butanol from B. subtilis, P. putida, and Clostridium species. [171]. However, at times, these native producers face limitations in terms of their slow growth rates, low production titers, and the overall yield of the product. Moreover, it is also a challenge to engineer native microbes because of ineffective genetic elements, the unidentified endogenous regulation of synthetic pathways, redox potential, cellular toxicity, and the presence of inhibitors. Metabolic pathway engineering impels the need to reconstruct biofuel biosynthesis in genetically tractable microbes to carry out the heterologous synthesis of 4G biofuels [172]. The point mutation of the cydC gene in E. coli has the capacity to completely restore the production of D-limonene and biogasoline, along with the platform chemical isopentenol [173].
MCFs are attracting considerable attention as next-generation energy sources because of their intended use for recovering energy in the form of electricity. Microbial fuel cells convert solar or chemical energy to electrical energy by utilizing microbial cell factories as catalysts [174]. Several electricigens are used for MFCs’ construction as pure cultures, including Natrialba magadii, Haloferax volcanii, R. sphaeroides [175], Rhodospirillum rubrum [176], and Acidiphilium cryptum [177]. However, pure cultures mandate the requirement of strict operating settings and selective substrates. Therefore, using miscellaneous consortiums as anodic biocatalysts is a preferred approach, as mixed communities are highly suitable for complex substrates as well. Several co-cultured electricigens reported high peak densities, including co-cultures of Geobacter sulfurreducens + C. cellulolyticum [178], Klebsiella pneumonia + Lipomyces starkeyi [179], and P. aeruginosa + K. variicola [180], demonstrating the potential of mixed cultures for effective electricity generation.
6. Industrial Progress, Commercial Limitations, and Future Prospects
The commercialization and industrial applicability of microbial products are very crucial issues. A number of microbe-based bioproducts are available on the market. For instance, as mentioned above, Novo Nordisk successfully synthesized recombinant insulin (Insulin Human AF) from S. cerevisiae [181]. Moreover, Quorn also developed Quorn mycoprotein by utilizing the strain of Fusarium venenatum [182]. Another company, Sunsonzymes, synthesized different enzymatic products named heat-stable acid pullulanase sunson® PUL, acid protease sunson® PRA100, and glucose amylase Sunson® GA. The former product was synthesized by B. subtilis, while the latter two were produced by A. niger [183], depicting the potential of MCFs for the manufacturing of industrially vital products. A range of microbial-based bioproducts that are available on the market are provided in Table 3.

Table 3.
Bio-compounds attained via microbial cell factories: commercially available products and major manufacturers.
Despite all the advancements in the field of metabolic engineering, still only a few microbial products and biotechnological processes have reached the industrial scale. While industrial biotechnology is currently seen as a key technology for the pharmaceutical and chemical sectors, microbial production must be economically viable to compete with the traditional chemical methods. Thus, to boost the production of microbial cell factories at the industrial level, long-term approaches and the deployment of integration strategies are needed. Firstly, the microbial strain should be developed appropriately in a way that it can efficiently utilize massive substrates for the production of desired products. Secondly, there should be a careful selection of microorganisms for creating a particular product [15]. The market development for microbial products is the foremost concern. There exists a dire need to establish a new market for commercializing bioproducts and replacing petroleum-based industries. All the advancements should be accompanied by market establishments as, even if efficient microbial products are developed, they will be of no use if there is no willingness regarding the use of bioproducts due to higher prices or any other reason. For example, 2,3-butanediol (2,3-BDO) has several applications in the agriculture, cosmetics, food, and polymer industries. The product development was challenging on an industrial scale due to the difficult and costly chemical processes required to derive pure 2,3-BDO from fossil resources. However, optically pure 2,3-BDO is currently being produced by utilizing engineered microbes, creating firsthand applications in the cosmetics and agriculture markets. The example clearly validates the necessity of the market establishment of bioproducts for potentially replacing petroleum-based materials [196].
Author Contributions
Conceptualization: R.C., A.N. and I.u.H.; formal analysis: R.C., L.D. and M.F.; investigation: R.C. and A.N.; methodology: R.C. and A.N.; resources: R.C., H.M. and A.N.; supervision: L.D. and M.F.; validation: L.D. and M.F.; writing—original draft preparation: R.C. and A.N.; writing—review and editing: L.D., M.F. and R.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wilbanks, T.J.; Fernandez, S. Climate Change and Infrastructure, Urban Systems, and Vulnerabilities: Technical Report for the US Department of Energy in Support of the National Climate Assessment; Island Press: Washington, DC, USA, 2014. [Google Scholar]
- Shi, T.Q.; Darvishi, F.; Cao, M.; Ji, B.; Ji, X.J. Design and construction of microbial cell factories for the production of fuels and chemicals. Front. Bioeng. Biotechnol. 2023, 11, 1198317. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Wittmann, C. Advanced biotechnology: Metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products. Angew. Chem. Int. Ed. 2015, 54, 3328–3350. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.L.; Zhou, J.J.; Quan, C.S.; Xiu, Z.L. Advances in industrial microbiome based on microbial consortium for biorefinery. Bioresour. Biproc. 2017, 4, 11. [Google Scholar] [CrossRef]
- Zuroff, T.R.; Curtis, W.R. Developing symbiotic consortia for lignocellulosic biofuel production. Appl. Microbiol. Biotechnol. 2012, 93, 1423–1435. [Google Scholar] [CrossRef]
- He, Y.; Xie, K.; Xu, P.; Huang, X.; Gu, W.; Zhang, F.; Tang, S. Evolution of microbial community diversity and enzymatic activity during composting. Res. Microbiol. 2013, 164, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 2014, 41, 185–201. [Google Scholar] [CrossRef]
- Abedi, E.; Hashemi, S.M.B. Lactic acid production–producing microorganisms and substrates sources-state of art. Heliyon 2020, 6, 04974. [Google Scholar] [CrossRef]
- Piwowarek, K.; Lipińska, E.; Hać-Szymańczuk, E.; Kieliszek, M.; Ścibisz, I. Propionibacterium spp.—Source of propionic acid, vitamin B12, and other metabolites important for the industry. Appl. Microbiol. Biotechnol. 2018, 102, 515–538. [Google Scholar] [CrossRef]
- Chubukov, V.; Mukhopadhyay, A.; Petzold, C.J.; Keasling, J.D.; Martín, H.G. Synthetic and systems biology for microbial production of commodity chemicals. NPJ Syst. Biol. Appl. 2016, 2, 16009. [Google Scholar] [CrossRef]
- Fisher, A.K.; Freedman, B.G.; Bevan, D.R.; Senger, R.S. A review of metabolic and enzymatic engineering strategies for designing and optimizing performance of microbial cell factories. Comput. Struct. Biotechnol. J. 2014, 11, 91–99. [Google Scholar] [CrossRef]
- Novo Nordisk Global. Available online: https://www.novonordisk.com/ (accessed on 4 January 2024).
- DSM. Available online: https://www.dsm.com/corporate/home.html (accessed on 13 December 2023).
- Cargill. Available online: https://www.cargill.com/home (accessed on 11 January 2024).
- Navarrete, C.; Jacobsen, I.H.; Martínez, J.L.; Procentese, A. Cell factories for industrial production processes: Current issues and emerging solutions. Processes 2020, 8, 768. [Google Scholar] [CrossRef]
- Locey, K.J.; Lennon, J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 5970–5975. [Google Scholar] [CrossRef]
- Petrova, P.; Petrov, K. Lactic acid fermentation of cereals and pseudocereals: Ancient nutritional biotechnologies with modern applications. Nutrients 2020, 12, 1118. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- Meruvu, H.; Harsa, S.T. Lactic acid bacteria: Isolation–characterization approaches and industrial applications. Crit. Rev. Food Sci. Nutr. 2022, 63, 8337–8356. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Kunze, G. Yeast Biotechnology: Diversity and Applications; Springer Science: Berlin/Heidelberg, Germany, 2009; Volume 78. [Google Scholar]
- Abdel-Azeem, A.M.; Abdel-Azeem, M.A.; Abdul-Hadi, S.Y.; Darwish, A.G. Aspergillus: Biodiversity, ecological significances, and industrial applications. In Recent Advancement in White Biotechnology through Fungi; Springer: Cham, Switzerland, 2019; pp. 121–179. [Google Scholar]
- Baeshen, N.A.; Baeshen, M.N.; Sheikh, A.; Bora, R.S.; Ahmed, M.M.M.; Ramadan, H.A.; Saini, K.S.; Redwan, E.M. Cell factories for insulin production. Microb. Cell Factories 2014, 13, 141. [Google Scholar] [CrossRef]
- Bomgardner, M. Food Ingredients Cargill, DSM start up stevia factory. Chem. Eng. News. 2019, 97, 14–15. [Google Scholar]
- Tolieng, V.; Tanaka, N.; Shiwa, Y.; Thitiprasert, S.; Kanchanasin, P.; Phongsopitanun, W.; Booncharoen, A.; Thongchul, N.; Tanasupawat, S. Weizmannia acidilactici sp. nov., a lactic acid producing bacterium isolated from soils. Syst. Appl. Microbiol. 2023, 46, 126389. [Google Scholar] [CrossRef]
- Romero-Mota, D.I.; Estrada-García, J.; Sales-Pérez, R.E.; Méndez-Contreras, J.M. Valorization of Bovine Manure and Molasses by the Production of Lactic Acid and Biomass through Probiotic Anaerobic Cofermentation with Lactobacillus acidophilus, Lactobacillus fermentum, and Bacillus subtilis. J. Environ. Eng. 2024, 150, 04023100. [Google Scholar] [CrossRef]
- Sansatchanon, K.; Sudying, P.; Promdonkoy, P.; Kingcha, Y.; Visessanguan, W.; Tanapongpipat, S.; Runguphan, W.; Kocharin, K. Development of a Novel D-Lactic Acid Production Platform Based on Lactobacillus saerimneri TBRC 5746. J. Microbiol. 2023, 61, 853–863. [Google Scholar] [CrossRef]
- Tong, Z.; Tong, Y.; Wang, D.; Shi, Y.C. Whole maize flour and isolated maize starch for production of citric acid by Aspergillus niger: A review. Starch-Stärke 2023, 75, 2000014. [Google Scholar] [CrossRef]
- Diamantopoulou, P.; Sarris, D.; Tchakouteu, S.S.; Xenopoulos, E.; Papanikolaou, S. Growth Response of Non-Conventional Yeasts on Sugar-Rich Media: Part 1: High Production of Lipid by Lipomyces starkeyi and Citric Acid by Yarrowia lipolytica. Microorganisms 2023, 11, 1863. [Google Scholar] [CrossRef] [PubMed]
- Behera, B.C.; Mishra, R.; Mohapatra, S. Microbial citric acid: Production, properties, application, and future perspectives. Food Front. 2021, 2, 62–76. [Google Scholar] [CrossRef]
- Saichana, N.; Matsushita, K.; Adachi, O.; Frébort, I.; Frebortova, J. Acetic acid bacteria: A group of bacteria with versatile biotechnological applications. Biotechnol. Adv. 2015, 33, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Chang, Y.; Xie, S.; Song, J.; Wang, M. Impacts of bioprocess engineering on product formation by Acetobacter pasteurianus. Appl. Microbiol. Biotechnol. 2018, 102, 2535–2541. [Google Scholar] [CrossRef]
- Kuenz, A.; Krull, S. Biotechnological production of itaconic acid—Things you have to know. Appl. Microbiol. Biotechnol. 2018, 102, 3901–3914. [Google Scholar] [CrossRef]
- Salgado, J.M.; Abrunhosa, L.; Venâncio, A.; Domínguez, J.M.; Belo, I. Integrated use of residues from olive mill and winery for lipase production by solid state fermentation with Aspergillus sp. Appl. Biochem. Biotechnol. 2014, 172, 1832–1845. [Google Scholar] [CrossRef]
- Tanyildizi, M.S.; Özer, D.; Elibol, M. Production of bacterial α-amylase by B. amyloliquefaciens under solid substrate fermentation. Biochem. Eng. J. 2007, 37, 294–297. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Lazarus, S.; Vincent, S.G.P. De-hairing protease production by an isolated Bacillus cereus strain AT under solid-state fermentation using cow dung: Biosynthesis and properties. Saudi J. Biol. Sci. 2014, 21, 27–34. [Google Scholar] [CrossRef]
- Rajkumar, R.; Jayappriyan, K.R.; Rengasamy, R. Purification and characterization of a protease produced by Bacillus megaterium RRM2: Application in detergent and dehairing industries. J. Basic Microbiol. 2011, 51, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.A.; Yusof, N.Z.; Nordin, N.; Zakaria, Z.A.; Rezali, M.F. Production and characterization of violacein by locally isolated Chromobacterium violaceum grown in agricultural wastes. Appl. Biochem. Biotechnol. 2012, 167, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiang, P.; Lu, Y.; Ruan, Z.; Jiang, R.; Xing, X.H.; Wei, D. Optimization of culture conditions for violacein production by a new strain of Duganella sp. B2. Biochem. Eng. J. 2009, 44, 119–124. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, L.; Xu, J.; Yang, R.; He, S.; Yan, X. Determination of oxidized scytonemin in Nostoc commune Vauch cultured on different conditions by high performance liquid chromatography coupled with triple quadrupole mass spectrometry. J. Appl. Phycol. 2013, 25, 1001–1007. [Google Scholar] [CrossRef]
- Kurbanoglu, E.B.; Ozdal, M.; Ozdal, O.G.; Algur, O.F. Enhanced production of prodigiosin by Serratia marcescens MO-1 using ram horn peptone. Braz. J. Microbiol. 2015, 46, 631–637. [Google Scholar] [CrossRef]
- Borić, M.; Danevčič, T.; Stopar, D. Prodigiosin from Vibrio sp. DSM 14379; a new UV-protective pigment. Microb. Ecol. 2011, 62, 528–536. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Q.; Sun, T.; Zhu, X.; Xu, H.; Tang, J.; Zhang, X.; Ma, Y. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metabol Eng. 2013, 17, 42–50. [Google Scholar] [CrossRef]
- Larroude, M.; Celinska, E.; Back, A.; Thomas, S.; Nicaud, J.M.; Ledesma-Amaro, R. A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol. Bioeng. 2018, 115, 464–472. [Google Scholar] [CrossRef]
- Tokui, M.; Kubodera, T.; Gomi, K.; Yamashita, N.; Nishimura, A. Construction of a thiamine pyrophosphate high-producing strain of Aspergillus oryzae by overexpression of three genes involved in thiamine biosynthesis. J. Biosci. Bioeng. 2011, 111, 388–390. [Google Scholar] [CrossRef]
- Schwechheimer, S.K.; Park, E.Y.; Revuelta, J.L.; Becker, J.; Wittmann, C. Biotechnology of riboflavin. Appl. Microbiol. Biotechnol. 2016, 100, 2107–2119. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Jin, Z.; Zhang, D. Microbial Cell Factories for Green Production of Vitamins. Front. Bioeng. Biotechnol. 2021, 9, 473. [Google Scholar] [CrossRef] [PubMed]
- Dmytruk, K.V.; Ruchala, J.; Fayura, L.R.; Chrzanowski, G.; Dmytruk, O.V.; Tsyrulnyk, A.O.; Andreieva, Y.A.; Fedorovych, D.V.; Motyka, O.I.; Mattanovich, D.; et al. Efficient production of bacterial antibiotics aminoriboflavin and roseoflavin in eukaryotic microorganisms, yeasts. Microb. Cell Factories 2023, 22, 132. [Google Scholar] [CrossRef] [PubMed]
- Chandra, N.; Kumar, S. Antibiotics Producing Soil Microorganisms. In Antibiotics and Antibiotics Resistance Genes in Soils; Hashim, M.Z., Strezov, V., Varma, A., Eds.; Springer International Publishing AG: Berlin/Heidelberg, Germany, 2017; Volume 51, pp. 1–18. [Google Scholar]
- Bhosale, P.; Larson, A.J.; Bernstein, P.S. (Factorial analysis of tricarboxylic acid cycle intermediates for optimization of zeaxanthin production from Flavobacterium multivorum. J. Appl. Microbiol. 2004, 96, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Shahina, M.; Hameed, A.; Lin, S.Y.; Lee, R.J.; Lee, M.R.; Young, C.C. Gramella planctonica sp. nov., a zeaxanthin-producing bacterium isolated from surface seawater, and emended descriptions of Gramella aestuarii and Gramella echinicola. Anton. Van Leeu. 2014, 105, 771–779. [Google Scholar] [CrossRef]
- Sowmya, R.; Sachindra, N.M. Carotenoid production by Formosa sp. KMW, a marine bacteria of Flavobacteriaceae family: Influence of culture conditions and nutrient composition. Biocatal. Agri Biotechnol. 2015, 4, 559–567. [Google Scholar] [CrossRef]
- Joshi, C.; Singhal, R.S. Modelling and optimization of zeaxanthin production by Paracoccus zeaxanthinifaciens ATCC 21588 using hybrid genetic algorithm techniques. Biocatal. Agri Biotechnol. 2016, 8, 228–235. [Google Scholar] [CrossRef]
- Barredo, J.L.; García-Estrada, C.; Kosalkova, K.; Barreiro, C. Biosynthesis of astaxanthin as a main carotenoid in the heterobasidiomycetous yeast Xanthophyllomyces dendrorhous. J. Fungi 2017, 3, 44. [Google Scholar] [CrossRef]
- Krupa, D.; Nakkeeran, E.; Kumaresan, N.; Vijayalakshmi, G.; Subramanian, R. Extraction, purification and concentration of partially saturated canthaxanthin from Aspergillus carbonarius. Bioresour. Technol. 2010, 101, 7598–7604. [Google Scholar] [CrossRef] [PubMed]
- Hojjati, M.; Razavi, S.H.; Rezaei, K.; Gilani, K. Stabilization of canthaxanthin produced by Dietzia natronolimnaea HS-1 with spray drying microencapsulation. J. Food Sci. Technol. 2014, 51, 2134–2140. [Google Scholar] [CrossRef]
- Market Research Future. Available online: https://www.marketresearchfuture.com/reports/bio-based-chemicals-market-5706 (accessed on 17 January 2024).
- Ko, Y.S.; Kim, J.W.; Lee, J.A.; Han, T.; Kim, G.B.; Park, J.E.; Lee, S.Y. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem. Soc. Rev. 2020, 49, 4615–4636. [Google Scholar] [CrossRef]
- Runguphan, W.; Keasling, J.D. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014, 21, 103–113. [Google Scholar] [CrossRef]
- Patra, P.; Das, M.; Kundu, P.; Ghosh, A. Recent advances in systems and synthetic biology approaches for developing novel cell-factories in non-conventional yeasts. Biotechnol. Adv. 2021, 47, 107695. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Wortel, M.T.; Molenaar, D.; Teusink, B. Metabolic shifts: A fitness perspective for microbial cell factories. Biotechnol. Lett. 2012, 34, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Archer, D.B. Filamentous fungi as microbial cell factories for food use. Curr. Opin. Biotechnol. 2000, 11, 478–483. [Google Scholar] [CrossRef]
- Woolston, B.M.; Edgar, S.; Stephanopoulos, G. Metabolic engineering: Past and future. Annu. Rev. Chem. Biomol. Eng. 2013, 4, 259–288. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Teo, W.; Chen, B.; Leong, S.S.J.; Chang, M.W. Microbial tolerance engineering toward biochemical production: From lignocellulose to products. Curr. Opin. Biotechnol. 2014, 29, 99–106. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, H.U.; Chae, T.U.; Cho, J.S.; Kim, J.W.; Shin, J.H.; Kim, D.I.; Ko, Y.S.; Jang, W.D.; Jang, Y.S. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2019, 2, 18–33. [Google Scholar] [CrossRef]
- Rangel, A.E.; Gomez Ramirez, J.M.; Gonzalez Barrios, A.F. From industrial by-products to value-added compounds: The design of efficient microbial cell factories by coupling systems metabolic engineering and bioprocesses. Biofuels. Bioprod. Biorefin. 2020, 14, 1228–1238. [Google Scholar] [CrossRef]
- Li, Y.; Lin, Z.; Huang, C.; Zhang, Y.; Wang, Z.; Tang, Y.J.; Chen, T.; Zhao, X. Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metab. Eng. 2015, 31, 13–21. [Google Scholar] [CrossRef]
- Tong, Y.; Charusanti, P.; Zhang, L.; Weber, T.; Lee, S.Y. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol. 2015, 4, 1020–1029. [Google Scholar] [CrossRef]
- Blin, K.; Pedersen, L.E.; Weber, T.; Lee, S.Y. CRISPy-web: An online resource to design sgRNAs for CRISPR applications. Synth. Syst. Biotechnol. 2016, 1, 118–121. [Google Scholar] [CrossRef]
- Choi, S.; Song, H.; Lim, S.W.; Kim, T.Y.; Ahn, J.H.; Lee, J.W.; Lee, M.H.; Lee, S.Y. Highly selective production of succinic acid by metabolically engineered Mannheimia succiniciproducens and its efficient purification. Biotechnol. Bioeng. 2016, 113, 2168–2177. [Google Scholar] [CrossRef]
- Zhang, X.; Tervo, C.J.; Reed, J.L. Metabolic assessment of E. coli as a biofactory for commercial products. Metab. Eng. 2016, 35, 64–74. [Google Scholar] [CrossRef]
- Otero, J.M.; Cimini, D.; Patil, K.R.; Poulsen, S.G.; Olsson, L.; Nielsen, J. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS ONE 2013, 8, e54144. [Google Scholar] [CrossRef]
- Huang, H.; Liu, H.; Gan, Y.R. Genetic modification of critical enzymes and involved genes in butanol biosynthesis from biomass. Biotechnol. Adv. 2010, 28, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, V.; Nampoothiri, K.M. Corynebacterium glutamicum. In Encyclopedia of Food Microbiology, 1st ed.; Batt, C.A., Tortorello, M.L., Eds.; Elsevier Ltd, Academic Press: Amsterdam, Netherlands, 2007; Volume 1, pp. 504–517. [Google Scholar]
- Kim, H.T.; Baritugo, K.A.; Oh, Y.H.; Hyun, S.M.; Khang, T.U.; Kang, K.H.; Jung, S.H.; Song, B.K.; Park, K.; Kim, K.; et al. Metabolic engineering of Corynebacterium glutamicum for the high-level production of cadaverine that can be used for the synthesis of biopolyamide 510. ACS Sustain. Chem. Eng. 2018, 6, 5296–5305. [Google Scholar] [CrossRef]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Choi, O.; Wu, C.Z.; Kang, S.Y.; Ahn, J.S.; Uhm, T.B.; Hong, Y.S. Biosynthesis of plant-specific phenylpropanoids by construction of an artificial biosynthetic pathway in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2011, 38, 1657–1665. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Binkley, R.M.; Kim, W.J.; Lee, M.H.; Lee, S.Y. Metabolic engineering of Escherichia coli for high-level astaxanthin production with high productivity. Metabol Eng. 2018, 49, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Chae, T.U.; Ko, Y.S.; Hwang, K.S.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of four-, five-and six-carbon lactams. Metabol Eng. 2017, 41, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kao, E.; Wang, G.; Baidoo, E.E.; Chen, M.; Keasling, J.D. Metabolic engineering of Escherichia coli for the biosynthesis of 2-pyrrolidone. Metabol Eng. Commun. 2016, 3, 1–7. [Google Scholar] [CrossRef]
- Choi, O.; Lee, J.K.; Kang, S.Y.; Pandey, R.P.; Sohng, J.K.; Ahn, J.S.; Hong, Y.S. Construction of artificial biosynthetic pathways for resveratrol glucoside derivatives. J. Microbiol. Biotechnol. 2014, 24, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Yim, H.; Haselbeck, R.; Niu, W.; Pujol-Baxley, C.; Burgard, A.; Boldt, J.; Khandurina, J.; Trawick, J.D.; Osterhout, R.E.; Stephen, R.; et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011, 7, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Nielsen, J.; Zhou, Y.J. Engineering robustness of microbial cell factories. Biotechnol. J. 2017, 12, 1700014. [Google Scholar] [CrossRef]
- Cho, J.S.; Kim, G.B.; Eun, H.; Moon, C.W.; Lee, S.Y. Designing microbial cell factories for the production of chemicals. JACS Au 2022, 2, 1781–1799. [Google Scholar] [CrossRef]
- Choi, K.R.; Jang, W.D.; Yang, D.; Cho, J.S.; Park, D.; Lee, S.Y. Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019, 37, 817–837. [Google Scholar] [CrossRef]
- Thomford, N.E.; Senthebane, D.A.; Rowe, A.; Munro, D.; Seele, P.; Maroyi, A.; Dzobo, K. Natural products for drug discovery in the 21st century: Innovations for novel drug discovery. Int. J. Mol. Sci. 2018, 19, 1578. [Google Scholar] [CrossRef]
- Li, J.; Rong, L.; Zhao, Y.; Li, S.; Zhang, C.; Xiao, D.; Foo, J.L.; Yu, A. Next-generation metabolic engineering of non-conventional microbial cell factories for carboxylic acid platform chemicals. Biotechnol. Adv. 2020, 43, 107605. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.J.; de Fatima Borges, M.; de Freitas Rosa, M.; Castro-Gómez, R.J.H.; Spinosa, W.A. Acetic acid bacteria in the food industry: Systematics, characteristics and applications. Food Technol. Biotechnol. 2018, 56, 139. [Google Scholar] [CrossRef]
- Song, H.; Lee, S.Y. Production of succinic acid by bacterial fermentation. Enzym. Microb. Technol. 2006, 39, 352–361. [Google Scholar] [CrossRef]
- Dwidar, M.; Park, J.Y.; Mitchell, R.J.; Sang, B.I. The future of butyric acid in industry. Sci. World. J. 2012, 2012, 471417. [Google Scholar] [CrossRef]
- Jiang, L.; Cui, H.; Zhu, L.; Hu, Y.; Xu, X.; Li, S.; Huang, H. Enhanced propionic acid production from whey lactose with immobilized Propionibacterium acidipropionici and the role of trehalose synthesis in acid tolerance. Green Chem. 2015, 17, 250–259. [Google Scholar] [CrossRef]
- Wang, Z.; Ammar, E.M.; Zhang, A.; Wang, L.; Lin, M.; Yang, S.T. Engineering Propionibacterium freudenreichii subsp. Shermanii for enhanced propionic acid fermentation: Effects of overexpressing propionyl-CoA: Succinate CoA transferase. Metabol Eng. 2015, 27, 46–56. [Google Scholar] [CrossRef]
- Guan, N.; Li, J.; Shin, H.D.; Du, G.; Chen, J.; Liu, L. Metabolic engineering of acid resistance elements to improve acid resistance and propionic acid production of Propionibacterium jensenii. Biotechnol. Bioeng. 2016, 113, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Rabah, H.; Carmo, F.L.R.D.; Jan, G. Dairy propionibacteria: Versatile probiotics. Microorganisms. 2017, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, B.P.; DeVeaux, L.C.; Christopher, L.P. Metabolic engineering as a tool for enhanced lactic acid production. Trends Biotechnol. 2014, 32, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Tamakawa, H.; Ikushima, S.; Yoshida, S. Efficient production of L-lactic acid from xylose by a recombinant Candida utilis strain. J. Biosci. Bioeng. 2012, 113, 73–75. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M.; Avramidis, N. Lactococcus lactis as a cell factory: A twofold increase in phosphofructokinase activity results in a proportional increase in specific rates of glucose uptake and lactate formation. Enzym. Microbiol. Technol. 2011, 49, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, G.S.; Brar, S.K.; Kaur, S.; Verma, M. Bioproduction and extraction optimization of citric acid from Aspergillus niger by rotating drum type solid-state bioreactor. Ind. Crop. Prod. 2013, 41, 78–84. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Laivenieks, M.; Schindler, B.D.; McKinlay, A.A.; Siddaramappa, S.; Challacombe, J.F.; Lowry, S.R.; Clum, A.; Lapidus, A.L.; Burkhart, K.B.; et al. genomic perspective on the potential of Actinobacillus succinogenes for industrial succinate production. BMC Genom. 2010, 11, 680. [Google Scholar] [CrossRef]
- Tsuji, A.; Okada, S.; Hols, P.; Satoh, E. Metabolic engineering of Lactobacillus plantarum for succinic acid production through activation of the reductive branch of the tricarboxylic acid cycle. Enzym. Microb. Technol. 2013, 53, 97–103. [Google Scholar] [CrossRef]
- Uchikura, H.; Ninomiya, K.; Takahashi, K.; Tsuge, Y. Requirement of de novo synthesis of pyruvate carboxylase in long-term succinic acid production in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2020, 104, 4313–4320. [Google Scholar] [CrossRef]
- Lan, E.I.; Wei, C.T. Metabolic engineering of cyanobacteria for the photosynthetic production of succinate. Metabol Eng. 2016, 38, 483–493. [Google Scholar] [CrossRef]
- Cok, B.; Tsiropoulos, I.; Roes, A.L.; Patel, M.K. Succinic acid production derived from carbohydrates: An energy and greenhouse gas assessment of a platform chemical toward a bio-based economy. Biofuel Bioprod. Biorefin. 2016, 8, 16–29. [Google Scholar] [CrossRef]
- Cardinale, S.; Tueros, F.G.; Sommer, M.O.A. Genetic-metabolic coupling for targeted metabolic engineering. Cell Rep. 2017, 20, 1029–1037. [Google Scholar] [CrossRef]
- Sych, J.M.; Lacroix, C.; Stevens, M.J.A. Vitamin B12 : Physiology, production and application. In Industrial Biotechnology of Vitamins, Biopigments, and Antioxidants; Vandamme, E.J., Revuelta, J.L., Eds.; Wiley-VCH: Hoboken, NJ, USA, 2016; pp. 129–159. [Google Scholar]
- Xia, W.; Chen, W.; Peng, W.F.; Li, K.T. Industrial vitamin B12 production by Pseudomonas denitrificans using maltose syrup and corn steep liquor as the cost-effective fermentation substrates. Bioprocess Biosyst. Eng. 2015, 38, 1065–1073. [Google Scholar] [CrossRef]
- Cai, Y.; Xia, M.; Dong, H.; Qian, Y.; Zhang, T.; Zhu, B.; Wu, J.; Zhang, D. Engineering a vitamin B12 high-throughput screening system by riboswitch sensor in Sinorhizobium meliloti. BMC Biotechnol. 2018, 18, 27. [Google Scholar] [CrossRef]
- Wang, H.; Sun, X.; Wang, L.; Wu, H.; Zhao, G.; Liu, H.; Wang, P.; Zheng, Z. Coproduction of menaquinone-7 and nattokinase by Bacillus subtilis using soybean curd residue as a renewable substrate combined with a dissolved oxygen control strategy. Ann. Microbiol. 2018, 68, 655–665. [Google Scholar] [CrossRef]
- Georgi, T.; Rittmann, D.; Wendisch, V.F. Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: Roles of malic enzyme and fructose-1, 6-bisphosphatase. Metabol Eng. 2005, 7, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Bommareddy, R.R.; Chen, Z.; Rappert, S.; Zeng, A.P. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase. Metabol Eng. 2014, 25, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, T.; Shimizu, H. Recent advances in amino acid production by microbial cells. Curr. Opin. Biotechnol. 2016, 42, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Hajnal, I. Meeting Report: Cold Spring Harbor Asia Synthetic Biology Meeting. Biotechnol. J. 2016, 11, 197–198. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Wendisch, V.F. Engineering microbial cell factories: Metabolic engineering of Corynebacterium glutamicum with a focus on non-natural products. Biotechnol. J. 2015, 10, 1170–1184. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Suda, M.; Uematsu, K.; Natsuma, Y.; Hiraga, K.; Jojima, T.; Inui, M.; Yukawa, H. Engineering of Corynebacterium glutamicum for high-yield L-valine production under oxygen deprivation conditions. Appl. Environ. Microbiol. 2013, 79, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jang, Y.S.; Lee, J.W.; Lee, S.Y. Escherichia coli W as a new platform strain for the enhanced production of L-Valine by systems metabolic engineering. Biotechnol. Bioeng. 2011, 108, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Ito, H.; Yasueda, H. Amino acid production: L-Lysine. In Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology, 1st ed.; Flickinger, M.C., Ed.; Wiley Online Library: Hoboken, NJ, USA, 2009; Volume 7, pp. 1–10. [Google Scholar]
- Xu, J.M.; Li, J.Q.; Zhang, B.; Liu, Z.Q.; Zheng, Y.G. Fermentative production of the unnatural amino acid L-2-aminobutyric acid based on metabolic engineering. Microb. Cell Factories 2019, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chen, X.; Wan, F.; Zhang, B.; Chen, J.; Xiong, Y. Effect of Tween 40 and DtsR1 on l-arginine overproduction in Corynebacterium crenatum. Microb. Cell Factories 2015, 14, 119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, B.; Yu, M.; Zhou, Y.; Li, Y.; Ye, B.C. Systematic pathway engineering of Corynebacterium glutamicum S9114 for L-ornithine production. Microb. Cell Factories 2017, 16, 158. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zhou, Y.J.; Krivoruchko, A.; Huang, M.; Liu, L.; Khoomrung, S.; Nielsen, J. Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine. Nat. Commun. 2015, 6, 8224. [Google Scholar] [CrossRef]
- Yang, B.; Feng, X.; Li, C. Microbial Cell Factory for Efficiently Synthesizing Plant Natural Products via Optimizing the Location and Adaptation of Pathway on Genome Scale. Front. Bioeng. Biotechnol. 2020, 8, 969. [Google Scholar] [CrossRef]
- Liu, X.; Ding, W.; Jiang, H. Engineering microbial cell factories for the production of plant natural products: From design principles to industrial-scale production. Microb. Cell Factories 2017, 16, 125. [Google Scholar] [CrossRef]
- Beyraghdar Kashkooli, A.; Farmanpour-Kalalagh, K.; Babaei, A. Yeast Synthetic Biology for Production of Artemisinin as an Antimalarial Drug. In Synthetic Biology of Yeasts: Tools and Applications; Springer International Publishing: Cham, Switzerland, 2022; pp. 157–180. [Google Scholar]
- Wu, C.-F.; Yang, J.-Y.; Wang, F.; Wang, X.-X. Resveratrol: Botanical origin, pharmacological activity and applications. Chin. J. Nat. Med. 2013, 11, 1–15. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.; Yu, O. Metabolic engineering of flavonoids in plants and microorganisms. Appl. Microbiol. Biotechnol. 2011, 91, 949–956. [Google Scholar] [CrossRef]
- Beekwilder, J.; Wolswinkel, R.; Jonker, H.; Hall, R.; de Vos, C.R.; Bovy, A. Production of resveratrol in recombinant microorganisms. Appl. Environ. Microbiol. 2006, 72, 5670–5672. [Google Scholar] [CrossRef]
- Lim, C.G.; Fowler, Z.L.; Hueller, T.; Schaffer, S.; Koffas, M.A. High-yield resveratrol production in engineered Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 3451–3460. [Google Scholar] [CrossRef]
- Marienhagen, J.; Bott, M. Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol. 2013, 163, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Mata-Gómez, L.C.; Montañez, J.C.; Méndez-Zavala, A.; Aguilar, C.N. Biotechnological production of carotenoids by yeasts: An overview. Microb. Cell Factories 2014, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Global Carotenoids Market. Available online: https://www.bccresearch.com/partners/verified-market-research/global-carotenoids-market.html (accessed on 17 January 2024).
- Sarnaik, A.; Sawant, K.; Khadilkar, J.; Pillai, G.; Pandit, R.; Lali, A. Cyanobacterial Cell Factories for Improved Carotenoid Biosynthesis through a Synthetic Biology Approach. In Next Generation Biomanufacturing Technologies; American Chemical Society: Washington, DC, USA, 2019; pp. 23–39. [Google Scholar]
- Kuzina, V.; Cerdá-Olmedo, E. Ubiquinone and carotene production in the Mucorales Blakeslea and Phycomyces. Appl. Microbiol. Biotechnol. 2007, 76, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Yoon, S.H.; Lee, S.H.; Kim, J.Y.; Oh, D.K.; Kim, S.W. An update on microbial carotenoid production: Application of recent metabolic engineering tools. Appl. Microbiol. Biotechnol. 2007, 77, 505–512. [Google Scholar] [CrossRef]
- Sarnaik, A.; Nambissan, V.; Pandit, R.; Lali, A. Recombinant Synechococcus elongatus PCC 7942 for improved zeaxanthin production under natural light conditions. Algal Res. 2018, 36, 139–151. [Google Scholar] [CrossRef]
- Kim, J.H.; Kang, H.J.; Yu, B.J.; Kim, S.C.; Lee, P.C. Planococcus faecalis sp. nov., a carotenoid-producing species isolated from stools of Antarctic penguins. Int. J. Syst. Evol. Microbiol. 2015, 65, 3373–3378. [Google Scholar] [CrossRef] [PubMed]
- Köcher, S.; Breitenbach, J.; Müller, V.; Sandmann, G. Structure, function and biosynthesis of carotenoids in the moderately halophilic bacterium Halobacillus halophilus. Arch. Microbiol. 2009, 191, 95–104. [Google Scholar] [CrossRef]
- Steiger, S.; Perez-Fons, L.; Cutting, S.M.; Fraser, P.D.; Sandmann, G. Annotation and functional assignment of the genes for the C30 carotenoid pathways from the genomes of two bacteria: Bacillus indicus and Bacillus firmus. Microbiology 2015, 161, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Shahina, M.; Lin, S.Y.; Cho, J.C.; Lai, W.A.; Young, C.C. Kordiaaquimaris sp. nov., a zeaxanthin-producing member of the family Flavobacteriaceae isolated from surface seawater, and emended description of the genus Kordia. Int. J. Syst. Evol. Microbiol. 2013, 63, 4790–4796. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Shahina, M.; Lin, S.Y.; Lai, W.A.; Hsu, Y.H.; Liu, Y.C.; Young, C.C. Aquibacter zeaxanthinifaciens gen. nov., sp. nov., a zeaxanthin-producing bacterium of the family Flavobacteriaceae isolated from surface seawater, and emended descriptions of the genera Aestuariibaculum and Gaetbulibacter. Int. J. Syst. Evol. Microbiol. 2014, 64, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.; Shahina, M.; Lin, S.Y.; Liu, Y.C.; Lai, W.A.; Young, C.C. Gramella oceani sp. nov., a zeaxanthin-producing bacterium of the family Flavobacteriaceae isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2014, 64, 2675–2681. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, R.; Thiagarajan, R.; Goutham, G.; Arumugam, M.; Beulaja, M.; Rastrelli, L.; Woźniak, K.S.; Habtemariam, S.; Orhan, I.E.; Nabavi, S.F.; et al. Zeaxanthin and ocular health, from bench to bedside. Fitoterapia 2016, 109, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Ambati, R.R.; Phang, S.M.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Mar. Drugs 2014, 12, 128–152. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Rouvière, P.E.; Cheng, Q. A carotenoid synthesis gene cluster from a non-marine Brevundimonas that synthesizes hydroxylated astaxanthin. Gene 2006, 379, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.S.; Choi, T.J.; Lee, W.J.; Kim, Y.T. Paracoccus haeundaensis sp. nov., a Gram-negative, halophilic, astaxanthin-producing bacterium. Int. J. Syst. Evol. Microbiol 2004, 54, 1699–1702. [Google Scholar] [CrossRef]
- Asker, D.; Beppu, T.; Ueda, K. Sphingomonas astaxanthinifaciens sp. nov., a novel astaxanthin-producing bacterium of the family Sphingomonadaceae isolated from Misasa, Tottori, Japan. FEMS Microbiol. Lett. 2007, 273, 140–148. [Google Scholar] [CrossRef]
- Li, C.; Swofford, C.A.; Sinskey, A.J. Modular engineering for microbial production of carotenoids. Metab. Eng. Commun. 2020, 10, e00118. [Google Scholar] [CrossRef]
- Jin, J.; Wang, Y.; Yao, M.; Gu, X.; Li, B.; Liu, H.; Ding, M.; Xiao, W.; Yuan, Y. Astaxanthin overproduction in yeast by strain engineering and new gene target uncovering. Biotechnol. Biofuels 2018, 11, 230. [Google Scholar] [CrossRef] [PubMed]
- Basiony, M.; Ouyang, L.; Wang, D.; Yu, J.; Zhou, L.; Zhu, M.; Wang, X.; Feng, J.; Dai, J.; Shen, Y.; et al. Optimization of microbial cell factories for astaxanthin production: Biosynthesis and regulations, engineering strategies and fermentation optimization strategies. Synth. Syst. Biotechnol. 2022, 7, 689–704. [Google Scholar] [CrossRef] [PubMed]
- Global Premium Fragrances Market to Reach $31.4 Billion by 2027. Report Linker. Available online: https://www.reportlinker.com/p06032650/?utm_source=GNW (accessed on 11 January 2024).
- Flavors and Fragrance Market by Type (Flavors and Fragrance), Nature (Natural and Synthetic), and Application (Food & Beverages, Cosmetics & Personal Care, Home Care and Fabric Care): Global Opportunity Analysis and Industry Forecast, 2021–2027. Available online: https://www.alliedmarketresearch.com/flavors-and-fragrances-market (accessed on 14 January 2024).
- Cosmetics Market Size, Share & Trends Analysis Report By Product (Skin Care, Hair Care, Makeup, Fragrance), by End-user (Men, Women), by Distribution Channel, by Region, and Segment Forecasts, 2023–2030. Research and Markets. Available online: https://www.researchandmarkets.com/reports/5648046/cosmetics-market-size-share-and-trends-analysis (accessed on 7 January 2024).
- Van Wyk, N.; Kroukamp, H.; Pretorius, I.S. The smell of synthetic biology: Engineering strategies for aroma compound production in yeast. Fermentation 2018, 4, 54. [Google Scholar] [CrossRef]
- Barden, T.C. Indoles: Industrial, agricultural and over-the-counter uses. Heterocyclic Scaffolds II. ChemInform 2014, 42, 31–46. [Google Scholar]
- Mindt, M.; Beyraghdar Kashkooli, A.; Suarez-Diez, M.; Ferrer, L.; Jilg, T.; Bosch, D.; Santos, V.M.; Wendisch, V.F.; Cankar, K. Production of indole by Corynebacterium glutamicum microbial cell factories for flavor and fragrance applications. Microb. Cell Factories 2022, 21, 45. [Google Scholar] [CrossRef] [PubMed]
- Czajka, J.J.; Nathenson, J.A.; Benites, V.T.; Baidoo, E.E.; Cheng, Q.; Wang, Y.; Tang, Y.J. Engineering the oleaginous yeast Yarrowia lipolytica to produce the aroma compound β-ionone. Microb. Cell Factories 2018, 17, 136. [Google Scholar] [CrossRef]
- Mi, J.; Becher, D.; Lubuta, P.; Dany, S.; Tusch, K.; Schewe, H.; Buchhaupt, M.; Schrader, J. De novo production of the monoterpenoid geranic acid by metabolically engineered Pseudomonas putida. Microb. Cell Factories 2014, 13, 170. [Google Scholar] [CrossRef][Green Version]
- Barghini, P.; Di Gioia, D.; Fava, F.; Ruzzi, M. Vanillin production using metabolically engineered Escherichia coli under non-growing conditions. Microb. Cell Factories 2007, 6, 13. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, X.; Lei, D.; Qiao, B.; Zhao, G.R. Metabolic engineering of Escherichia coli for de novo production of 3-phenylpropanol via retrobiosynthesis approach. Microb. Cell Factories 2021, 20, 121. [Google Scholar] [CrossRef]
- Tao, X.Y.; Lin, Y.C.; Wang, F.Q.; Liu, Q.H.; Ma, Y.S.; Liu, M.; Wei, D.Z. Production of sesquiterpene patchoulol in mitochondrion-engineered Saccharomyces cerevisiae. Biotechnol. Lett. 2022, 44, 571–580. [Google Scholar] [CrossRef]
- Kampranis, S.C.; Makris, A.M. Developing a yeast cell factory for the production of terpenoids. Comput. Struct. Biotechnol. J. 2012, 3, e201210006. [Google Scholar] [CrossRef]
- Tilman, D.; Socolow, R.; Foley, J.A.; Hill, J.; Larson, E.; Lynd, L.; Pacala, S.; Reilly, J.; Searchinger, T.; Somerville, C.; et al. Beneficial biofuels—The food, energy, and environment trilemma. Science 2009, 325, 270–271. [Google Scholar] [CrossRef]
- Lan, E.I.; Liao, J.C. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 2011, 13, 353–363. [Google Scholar] [CrossRef]
- Silva, J.P.A.; Mussatto, S.I.; Roberto, I.C.; Teixeira, J.A. Ethanol production from xylose by Pichia stipitis NRRL Y-7124 in a stirred tank bioreactor. Braz. J. Chem. Eng. 2011, 28, 151–156. [Google Scholar]
- Aruna, A.; Nagavalli, M.; Girijashankar, V.; Ponamgi, S.P.D.; Swathisree, V.; Venkateswar, R.L. Direct bioethanol production by amylolytic yeast C andida albicans. Lett. Appl. Microbiol. 2015, 60, 229–236. [Google Scholar] [CrossRef]
- Kim, J.H.; Ryu, J.; Huh, I.Y.; Hong, S.K.; Kang, H.A.; Chang, Y.K. Ethanol production from galactose by a newly isolated Saccharomyces cerevisiae KL17. Bioprocess Biosyst. Eng. 2014, 37, 1871–1878. [Google Scholar] [CrossRef]
- Mauerhofer, L.M.; Zwirtmayr, S.; Pappenreiter, P.; Bernacchi, S.; Seifert, A.H.; Reischl, B.; Schmider, T.; Taubner, R.S.; Paulik, C.; Rittmann, S.K.M. Hyperthermophilic methanogenic archaea act as high-pressure CH4 cell factories. Commun. Biol. 2021, 4, 289. [Google Scholar] [CrossRef] [PubMed]
- Seifert, A.H.; Rittmann, S.; Bernacchi, S.; Herwig, C. Method for assessing the impact of emission gasses on physiology and productivity in biological methanogenesis. Bioresour. Technol. 2013, 136, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Simon, K.M.R.; Seifert, A.H.; Bernacchi, S. Kinetics, multivariate statistical modelling, and physiology of CO2-based biological methane production. Appl. Energy 2018, 216, 751–760. [Google Scholar]
- Willquist, K.; Zeidan, A.A.; van Niel, E.W. Physiological characteristics of the extreme thermophile Caldicellulosiruptor saccharolyticus: An efficient hydrogen cell factory. Microb. Cell Factories 2010, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Gabrielyan, L.; Sargsyan, H.; Trchounian, A. Novel properties of photofermentative biohydrogen production by purple bacteria Rhodobacter sphaeroides: Effects of protonophores and inhibitors of responsible enzymes. Microb. Cell Factories 2015, 14, 131. [Google Scholar] [CrossRef] [PubMed]
- Callegari, A.; Bolognesi, S.; Cecconet, D.; Capodaglio, A.G. Production technologies, current role, and future prospects of biofuels feedstocks: A state-of-the-art review. Crit. Rev. Environ. Sci. Technol. 2020, 50, 384–436. [Google Scholar] [CrossRef]
- Singh, V.; Mani, I.; Chaudhary, D.K.; Dhar, P.K. Metabolic engineering of biosynthetic pathway for production of renewable biofuels. Appl. Biochem. Biotechnol. Appl. 2014, 172, 1158–1171. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.R.; Jiao, S.; Lee, S.Y. Metabolic engineering strategies toward production of biofuels. Curr. Opin. Chem. Biol. 2020, 59, 1–14. [Google Scholar] [CrossRef]
- Eng, T.; Demling, P.; Herbert, R.A.; Chen, Y.; Benites, V.; Martin, J.; Lipzen, A.; Baidoo, E.E.; Blank, L.M.; Petzold, C.J.; et al. Restoration of biofuel production levels and increased tolerance under ionic liquid stress is enabled by a mutation in the essential Escherichia coli gene cydC. Microb. Cell Factories 2018, 17, 159. [Google Scholar] [CrossRef]
- Cao, Y.; Mu, H.; Liu, W.; Zhang, R.; Guo, J.; Xian, M.; Liu, H. Electricigens in the anode of microbial fuel cells: Pure cultures versus mixed communities. Microb. Cell Factories 2019, 18, 39. [Google Scholar] [CrossRef]
- Cho, Y.K.; Donohue, T.J.; Tejedor, I.; Anderson, M.A.; McMahon, K.D.; Noguera, D.R. Development of a solar-powered microbial fuel cell. J. Appl. Microbiol. 2008, 104, 640–650. [Google Scholar] [CrossRef]
- Gomez, M.V.; Mai, G.; Greenwood, T.; Mullins, J.P. The development and maximization of a novel photosynthetic microbial fuel cell using Rhodospirillum rubrum. J. Emerg. Invest. 2014, 3, 1–7. [Google Scholar] [CrossRef]
- Borole, A.P.; O’Neill, H.; Tsouris, C.; Cesar, S. A microbial fuel cell operating at low pH using the acidophile Acidiphilium cryptum. Biotechnol. Lett. 2008, 30, 1367–1372. [Google Scholar] [CrossRef]
- Ren, Z.; Ward, T.E.; Regan, J.M. Electricity production from cellulose in a microbial fuel cell using a defined binary culture. Environ. Sci. Tech. 2007, 41, 4781–4786. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Ethiraj, B.; Cheng, C.K.; Yousuf, A.; Thiruvenkadam, S.; Prasad, R.; Rahman Khan, M.M. Enhanced current generation using mutualistic interaction of yeast-bacterial coculture in dual chamber microbial fuel cell. Ind. Eng. Chem. Res. 2018, 57, 813–821. [Google Scholar] [CrossRef]
- Islam, M.A.; Ong, H.R.; Ethiraj, B.; Cheng, C.K.; Khan, M.M.R. Optimization of co-culture inoculated microbial fuel cell performance using response surface methodology. J. Environ. Manag. 2018, 225, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Recombinant Insulin. Novo Nordisk Pharmatech A/S. Available online: https://novonordiskpharmatech.com/products/recombinant-insulin/ (accessed on 23 November 2023).
- Quorn Mycoprotein. Available online: https://www.quorn.co.uk/mycoprotein (accessed on 4 January 2024).
- Sunsonzymes. Available online: https://www.sunsonzyme.com/ (accessed on 4 January 2024).
- Biosuccinium™ Reverdia. Available online: https://www.reverdia.com/biosuccinium/ (accessed on 7 January 2024).
- BioAstin® Cyanotech. Available online: https://www.cyanotech.com/astaxanthin/bioastin-water-dispersible-astaxanthin/ (accessed on 11 January 2024).
- Astapure® Algatech. Available online: https://www.algatech.com/algatech-product/astapure-natural-astaxanthin/ (accessed on 7 December 2023).
- Fucovital™ AlgaTech. Available online: https://www.algatech.com/algatech-product/fucovital/ (accessed on 13 January 2024).
- Purecane™ Amyris. Available online: https://amyris.com/2427/purecane-launches-in-kroger-stores-in-time-for-healthy-holiday-baking (accessed on 15 January 2024).
- Aji-No-Moto®. Available online: https://www.ajinomoto.com/msg/what-is-aji-no-moto-and-how-is-it-made#cf_block_1_cf_heading_txt (accessed on 31 December 2023).
- Nootkatone by Evolva™. Available online: https://evolva.com/nootkatone/ (accessed on 21 January 2024).
- Geno BDO™. Genomatica. Available online: https://www.genomatica.com/products/ (accessed on 21 January 2024).
- AvelaTM. Genomatica. Available online: https://www.genomatica.com/products/ (accessed on 18 December 2023).
- CaroCare® DSM. Available online: https://www.dsm.com/food-beverage/en_US/ingredients/beverages-and-brewing/beverage/carocare.html# (accessed on 7 January 2024).
- Biolys® Evonik. Available online: https://animal-nutrition.evonik.com/en/products-and-solutions/amino-acids/l-lysine/biolys-131967.html (accessed on 17 January 2024).
- Threamino® Evonik. Available online: https://animal-nutrition.evonik.com/en/products-and-solutions/amino-acids/l-threonine/threamino-131965.html (accessed on 17 January 2024).
- Song, C.W.; Park, J.M.; Chung, S.C.; Lee, S.Y.; Song, H. Microbial production of 2, 3-butanediol for industrial applications. J. Ind. Microbiol. 2019, 46, 1583–1601. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).