Abstract
During the COVID-19 pandemic, microbiological controls neglected the spread of viruses through the air. Techniques to identify this threat required additional research to enable control measures to be introduced to protect against the spread of disease through this route. Due to the very high level of risk occurring during research on the COVID-19 and SARS-CoV-2 viruses, it seems necessary to use analogous microorganisms that will allow, through an experiment, to validate or challenge a method that stops the spread of infectious microorganisms, without unnecessary risk to research staff. The presented work was carried out to assess the possibility of using airborne microorganisms that are safe for humans for this type of research. The work presents the selection process of bacteria and viruses (bacteriophages) that have the greatest potential for use in experimental studies on airborne-droplet transmission indoors, especially in hospital facilities. In the study, it was assumed that determining the survival rates of groups of organisms would allow them to be used as a proxy for studying more dangerous bacteria and viruses. Survival studies of selected microorganisms were carried out, and the paper selected microorganisms with the highest survival rate in a given environment.
1. Introduction
Since the outbreak of the COVID-19 pandemic in December 2019, the world has continued to face the acute upper respiratory syndrome caused by SARS-CoV-2, which is currently spreading. To date, research worldwide in the field of evaluating the transmission of SARS-CoV-2 microorganisms, conducted through numerous scientific experiments, has allowed for a deeper understanding of this subject. Studies in the area of computational fluid dynamics (CFD) simulations have allowed the analysis of the risk of releasing airborne pathogens in enclosed spaces [1,2,3,4,5,6,7]. Research has also been conducted on the use of aerosol particle generators and counters to assess the quantity and shape of particles [8,9,10,11] or has been based on statistical data related to infection rates correlated with the level of protection and isolation [12,13]. The COVID-19 pandemic has revealed numerous knowledge gaps in our understanding and the need to update the traditional perspective on viral transmission methods.
Previous definitions of droplet and airborne transmission do not account for the mechanisms by which contaminated droplets and aerosols from the respiratory tract move through the air and lead to infections. Aerosols are so small that they can remain suspended in the air, accumulate in poorly ventilated spaces, and be transmitted over short and long distances [14,15]. Defining the problem in this way creates an urgent need to include aerosol precautions in current respiratory infectious disease control protocols. During the COVID-19 pandemic, controls focused mainly on preventing droplet and germ-borne transmission [16,17]; airborne transmission required further research before control measures could be implemented to prevent disease spread through this route. Due to the high risk associated with the study of COVID-19 and SARS-CoV-2 viruses, it is necessary to employ similar microorganisms that, through experimentation, allow us to confirm or refute a method that stops the spread of infectious microorganisms without unnecessarily exposing research personnel. The role and mechanisms of airborne transmission in spreading infectious viral diseases are not well understood, and the level of scientific information available about their airborne transmission differs substantially for each specific virus. In laboratories working with aerosol-transmitted microorganisms, it is usual to apply control measures at the appropriate biosafety level (BSL), depending on the pathogens you are working with. It is important to follow strict safety procedures and adhere to the guidelines specified for the BSL level. A critical review of what is known about the airborne transmission of dangerous infectious viruses from an engineer’s perspective will help researchers understand the similarities and general identification gaps in knowledge about airborne virus transmission, as well as gaps specific to individual viruses. But to make this possible, a repeatable research method is needed that allows for research in conditions that are safe for research personnel. This method will enable the development of safety systems for patients and medical staff in various rooms individually adapted to the conditions. This creates the basis for the development of unconventional but effective methods of preventing the spread of viruses. In the work, microorganisms that do not threaten human life were used and all safety measures were observed.
The selection of microorganisms for the study was preceded by an analysis that concluded that bacteriophage phi6 and SARS-CoV-2 virus are two different categories of viruses, but there are some similarities in their structure and function, although they are two completely different classes of pathogens. Both bacteriophage phi6 and SARS-CoV-2 are viruses. This means that they are microscopic organisms that do not have their own metabolism or ability to multiply on their own. Instead, they must infect host cells to multiply. Both phi6 and SARS-CoV-2 have a capsid, which is an outer protective layer. In the case of phi6, it is the capsid that surrounds its RNA genome, while in the case of SARS-CoV-2, it is the protein envelope that surrounds its RNA genome. Both phi6 and SARS-CoV2 contain an RNA nucleic acid-based genome. The phi6 bacteriophage has a three-segmented RNA genome, while SARS-CoV-2 is a single-stranded RNA virus. Both phi6 and SARS-CoV-2 need to infect host cells to multiply. Phi6 infects bacteria, while SARS-CoV-2 infects human cells. Both viruses bind to receptors on the surface of host cells, allowing them to enter the cell and begin the multiplication process. Phi6 is a bacteriophage that infects bacteria, while SARS-CoV-2 is a virus that infects humans. They have completely different replication mechanisms and effects on their hosts. In addition, SARS-CoV-2 is responsible for causing COVID-19 disease in humans, making it a pathogen of great health importance, while phi6 is relatively less harmful to humans because it infects bacteria [18,19,20].
The work presented here was carried out to assess the feasibility of using air-sprayed, human-safe microorganisms for this type of research. The process of selecting bacteria and a virus (bacteriophage) with the highest potential for use in experimental studies of airborne transmission within indoor environments is presented.
2. Materials and Methods
For the study, three bacterial strains were selected: Escherichia coli ATCC 26922 (PCM 2057), Micrococcus luteus ATCC 7468 (PCM 1143), and Staphylococcus aureus ATCC 6538 P (PCM 1932), as well as three bacteriophages (phage phi6, Escherichia phage phiX174, and phage T4). The bacterial strains were obtained from the Polish Microorganisms Collection of the Polish Academy of Sciences and stored in a biobank, while the phages were obtained from the German Collection of Microorganisms and Cell Cultures DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen). The bacterial strains (including those that act as hosts for the bacteriophages) were stored in a freezer at −20 °C, on agar slants and agar plates. Prior to freezing the strains, reduction inoculation was performed to verify their purity and surface inoculation was performed on the appropriate media (AO, TSB, LB). After incubation at the appropriate temperature (25 °C, 30 °C, 37 °C), the biomass was washed with 20% glycerol until an OD600 optical density of 1.5 was achieved; 1 mL of the suspension was poured into cryotubes. For ongoing studies, strains were screened on slants with AO and plates with TSB (Pseduomonas sp.). The DSMZ biobank phage suspension (1 mL) was stored in a refrigerator at 8 °C, according to the manufacturer’s requirements. Bacteriophages for ongoing research were stored in 10 mL aliquots in a refrigerator at 8 °C. Before actual phage propagation, host surface inoculation was performed on two TSB/LB plates (biomass) and incubated for 24 h at 25/37 °C. The biomass was collected in a bottle with sterile TSB broth and incubated for 2–3 h in a water bath with shaking at 25/37 °C. After the initial incubation, 50–100 µL of the phage suspension was added, followed by another 2–3 h of incubation with shaking in a water bath. Subsequently, the host plate biomass was added again and incubated in a water bath with shaking for 12 h at 25/37 °C. After incubation, the phage suspension was centrifuged (10,000–12,000 rpm, 30 min), and the supernatant was filtered through a sterile syringe filter (0.22 µm). The finished phage suspension was stored in the refrigerator. To determine the phage abundance in the suspension, phage nomination was performed. Sterile semi-solid agar was prepared in glass test tubes. Before nomination, the agars were dissolved in a Koch apparatus or drier and then stored in a water bath at 60 °C. The host culture was renewed (by collecting biomass into LB/TSB broth) and incubated for 2–3 h at 25/37 °C. A number of dilutions of the phage suspension in sterile saline solution (0.85% NaCl) were performed. Proper phage nomination was carried out as follows: after cooling the agar to 40–45 °C, 100µL of rejuvenated host culture and 100 µL of diluted phage were added, mixed on a shaker, and then poured onto an LB/TSB agar plate. After 24 h of incubation, plaques (clear zones) were counted and converted to pfu/ml (plaque formation unit) (Table 1).

Table 1.
Cultivation Conditions for Test Bacterial Strains and Bacteriophages.
The tests were carried out on a test stand, a diagram of which is shown in Figure 1. The following test elements were used in the test room: fans with a remote control and an oscillation of 90 degrees (placed on tripods with a height of 35 cm from the floor); an ozone generator with a capacity of 5–10 g/h ozone (with the possibility of switching on by remote control from outside the test room); an ozone meter with the possibility of remote start-up, reading and generation of a report (time of reaching the maximum concentration and its concentration); a probe for measuring low concentrations in the range of 0–20 ppm ozone (electrochemical sensor, accuracy ±15%) and an optionally (interchangeable) probe for measuring high concentrations of 10–1000 ppm (semiconductor sensor, accuracy ±15%); a device for actuating the electrovalve for compressed air cylinders (programmed times: 30 s, 90 s, 180 s); a temperature and humidity sensor with remote reading capability; wall-mounted stands for microbiological samplers (impactors); impactors (microbiological samplers) SAS Duo taking 3 dm3 per minute (180 L/min) simultaneously for two heads, placed on a wall-mounted stand. The apparatus allowed for the maintenance of repeatable experimental conditions. The test stand was supplemented with a survival analysis station, which allowed for the collection of bioaerosol samples at specified time intervals while avoiding the influence of the person conducting the test on the homogenization of the bioaerosol in the air. The bioaerosol survival analysis station included a table and sleeves that allowed the operator to change plates in the samplers outside the infectious room (bioaerosol station), as indicated in Figure 1.
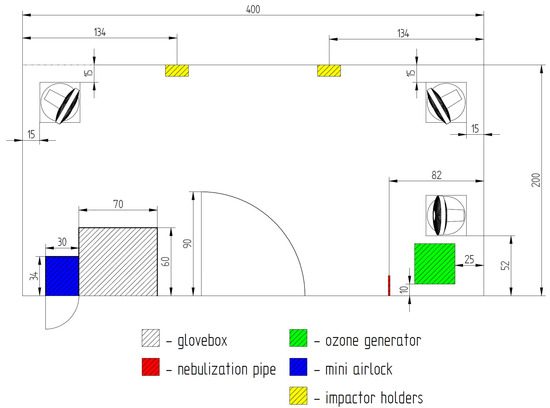
Figure 1.
Diagram of the construction of the bioaerosol test stand.
2.1. Survival Studies of Bacterial Strains
Survival studies of bacterial strains in physiological fluid were performed as a surface culture on the appropriate medium incubated at the appropriate temperature (30/37 °C) and then a suspension was prepared with an OD600 optical density of 1.5. At time “0” and every hour, a series of 10-fold dilutions were performed in two replicates on AO plates, after incubation at the appropriate temperature (30/37 °C), colonies were counted and converted to cfu/mL. The analysis was carried out at a temperature of 22 ° C. To determine the survival rate of bacteria in the air, a test suspension was prepared with the appropriate optical density on the McFarland scale. The suspension was nebulized at a pressure of 20 psi for 30 s (other parameters were chosen so that quantifiable results could be obtained). After initial homogenization (1 min), samples were taken at time “0” and at 5, 10, 15, 20, 25, 30, and 35 min. The analysis was carried out using a survival analysis station, fans were turned on during the analysis to obtain a homogeneous bioaerosol. The McFarland scale, which is used to standardize microbiological methods by preparing standardized solutions (suspensions) was used to study bacterial strains. To determine the abundance of bacteria in the suspension according to the scale, a surface culture was performed on the appropriate medium, and incubated at the appropriate temperature (30/37 °C). After the specified incubation time (24/28 h), suspensions were prepared with OD550 optical densities of: 0.125; 0.25; 0.5; 0.75; 1; 1.25; 1.5; 1.75. From each prepared suspension, a series of 10-fold dilutions were made in two replicates on AO plates; after incubation at the appropriate temperature (30/37 °C), colonies were counted and converted to cfu/mL.
2.2. Survival Studies of Bacteriophages
To determine the amount of phage in suspension, phage nomination was performed. For this purpose, sterilized semiliquid agars were prepared in glass tubes. Before nomination, the agars were dissolved in a Koch apparatus and then stored in a water bath at 60 °C. The host culture was renewed (collecting the biomass into LB/TSB broth) and incubated at 25/37 °C. Several dilutions of the phage suspension were performed in sterile saline (0.85% NaCl). The actual nomination was performed as follows: after the agar was cooled to 40–45 °C, 100 µL of the rejuvenated host culture and 100 µL of the phage dilution were added to the agar, mixed well on a shaker and the suspension was transferred to an LB/TSB media plate. It was incubated for 24 h and the bald spots (translucencies) were counted and converted to pfu/mL. Preliminary analyses performed for different nebulization parameters in the infectious room (bioaerosol test stand; Figure 1) for phages included:
- the phage used, the titer of the phage in the initial suspension;
- the titer of the phage in the test suspension to be nebulized (dependent on the prepared starting suspension, which was obtained after the phage was cultured from the biobank);
- the suspension of the dilution factor of the phage test (factor 1; 2.5; 5);
- nebulization time in the test room (30/90/180 s);
- homogenization (distribution) time of bioaerosol in the infectious room (bioaerosol test bed);
- the volume of air taken by the microbiological sampler.
Based on the phage characteristics, the nebulization parameters were adjusted for verification on the bioaerosol analysis station. The aim of the preliminary studies was to verify the appropriate nebulization parameters for a given phage. If the phage count in the air sample was too low or too high, the test was repeated with adjusted parameters.
To assess the survival of the bioaerosol, a host strain test suspension and a phage suspension were prepared. The suspension was nebulized at a pressure of 20 psi for a time determined as optimal for the given bacteriophage (allowing for countable results). After an initial homogenization (1 min), samples were collected at “time 0” and at 5, 10, 15, 20, 25, 30, and 35 min. The analysis was carried out using the survival analysis station, with fans running during the analysis to ensure a uniform distribution of the bioaerosol. According to ISO 16000-36:2018, the result was presented as the number of colonies per cubic meter of air (cfu/m3) or the number of plaques per cubic meter of air (cfu/m3), taking into account correction tables (available in the standard) and using Formula (1). The colony count/plaque count after incubation (r), according to the impactor manufacturer’s recommendation, should be corrected to the most probable number (Pr) according to the table provided in the impactor manual [19].
where C is the number of colonies per cubic meter; V is the volume of the collected sample; Pr is the most probable number of colonies.
The research carried out aimed to identify groups of microorganisms and their testing possibilities in various settings, with a focus on result reproducibility, personnel safety, and testing conditions similar to real-world scenarios.
3. Results
3.1. Results of the Survival Analysis of Bacterial Strains in Physiological Saline and Air
The analysis of the results of the experiments with bacterial strains allowed for the development of a procedure demonstrating the survivability of each strain under test conditions. The following pictures show a reduction culture (Figure 2A,D,G), the purpose of which is to check the purity of the strain and describe its morphology; a plate after nebulization with normal results (Figure 2B,E,H) and a plate after nebulization with abnormal results (Figure 2C,F,I). The correct plates display individual bacterial colonies after air sampling from the bioaerosol using a microbiological probe. A proper reading indicates a countable number of colonies ranging from 20 to 219 (216 is based on the number of holes in the probe head and the probable number of colonies table (as cited)). Based on the results analysis, an acceptable number of colonies between 50 and 216 and an optimal range between 100 and 150 were adopted. The analysis of the abundance of bacterial strains in the saline fluid allowed us to determine the curve of the relationship between the number of colonies (cfu/mL) and the McFarland scale (Table 2). In subsequent studies, suspensions were prepared specifying the degree on the McFarland scale. Based on the survival results of the bacterial strains in saline, all strains were found to be stable in saline for up to 4 h. Based on the results of the survival of bacterial strains in the air, it was found that although all strains have a high survival rate in the fluid (reduction in abundance by no more than 20% in 2 h), the strain with the highest survival rate in the air is M. luteus TCC 7468. Strains E. coli ATCC 26922 and S. aureus ATCC6538P had a reduction in abundance after nebulization in the test room of more than 98%; M. luteus ATCC 7468 had a reduction of about 38% within 30 min (Table 3).
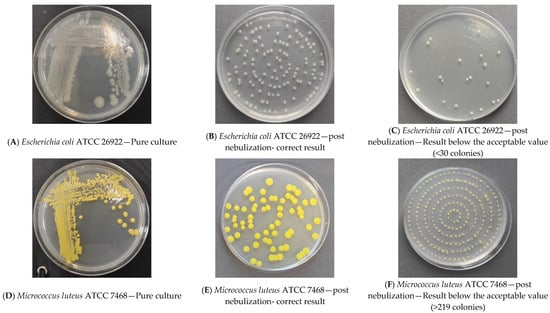
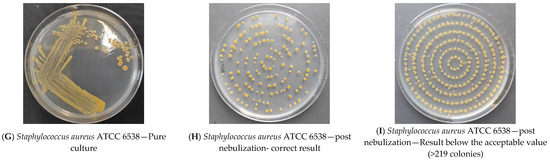
Figure 2.
Sample bacterial strain streak plates.

Table 2.
Survival of bacterial strains in physiological saline.

Table 3.
Survival of bacterial strains in the air.
3.2. Results of Bacteriophage Survival Tests in Physiological Fluid and Air
During the preliminary analyses, effective nebulization (yielding countable plaques on the nutrient agar with the host from the air sample) was achieved only when the phage was undiluted and the maximum nebulization time was applied without homogenization (Table 4). This indicates insufficient nebulization of this phage and a very short survival rate, not enough to perform the test. Despite a very good survival rate in physiological fluid for the phage phiX174, no survival analysis was performed in the air and saliva (synthetic saliva reflects the spread of bioaerosol between the patients in the room). During the preliminary analyses performed, effective nebulization (obtaining countable bald spots on host nutrient agar with the host) was not achieved even in the absence of phage dilution and at the maximum nebulization time. The T4 phage exhibits low survivability in physiological fluid (reduction in bacteriophage quantity by more than 99% in 2 h) (Table 5). Due to unsatisfactory survival in physiological fluid and a lack of effective nebulization even at maximum nebulization parameters, a survival analysis was not performed in air (Table 6).

Table 4.
Bacteriophage Survivability at Various Bioaerosol Nebulisation Parameters.

Table 5.
Bacteriophage Survivability in Physiological Solution.

Table 6.
Bacteriophage Survivability in Air.
Based on the results of the survival tests of bacteriophages in physiological fluid, the phages phi6 and phiX174 were found to be stable in physiological fluid for up to 4 h (reduction in the abundance of bacteriophages by no more than 30% during 2 h). In the case of phage T4, a significant decrease in phage abundance was observed in the suspension as early as the first hour of storage in physiological fluid (Table 7). Based on the results of bacteriophage survival in air, only bacteriophage phi6 showed survival in air after spraying in the bioaerosol form. Bacteriophage phi6 is able to remain stable in air for no longer than 10 min, with a noticeable decrease in abundance after 5 min.

Table 7.
Bacteriophage Phi6 Survivability in air within the 0–10 min Range.
4. Discussion
The analysis of the survivability of bacterial strains in physiological fluid and air showed that all bacterial strains remain stable in physiological fluid; however, in the air, only the M. luteus ATCC 7468 strain exhibits a slight decrease in quantity over 30 min. The initial population of strains is due to differences in suspension counts at the same turbidity on the McFarland scale. In the case of the M. luteus strain, the difference in the population in physiological fluid (the lowest quantity in physiological fluid but the best survivability in the air) may be due to variations in cell size compared to other strains, the formation of tetrads by cells, and the production of pigments that protect cells from UV light in the atmosphere [21]. This confirms that M. luteus is the best strain for testing in bioaerosol studies (Figure 3). In the case of phages, their survivability in the air is much shorter due to the capsid structure and higher sensitivity to environmental conditions (Figure 4). Research has shown that phage phi6 can also be used for bioaerosol studies, provided that the test is carried out in 5 min due to the decline in phage population over time and maintaining the appropriate parameters at the bioaerosol test station (such as humidity and temperature) [20].
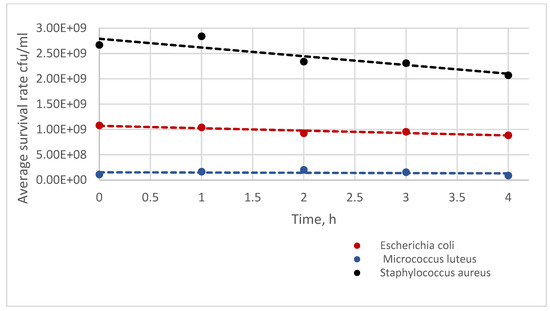
Figure 3.
Comparison of the Survival of Bacterial Strains in Physiological Fluid.
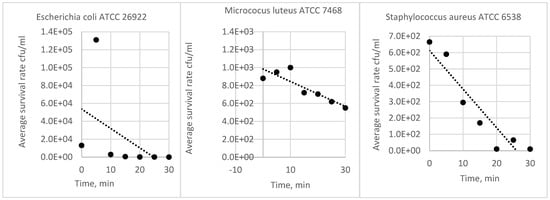
Figure 4.
Comparison of survival rates of bacterial strains in air.
In order to select test strains for further studies using a test bioaerosol, the following characteristics of bacterial strains were compared: the ability to obtain a quantifiable bioaerosol from a suspension with turbidity in the McFarland scale range, ease of colony description (color, shape, colony size), survival in physiological fluid, and survival in air. All assumptions were best met by Micrococus luteus strain ATCC 7468 (Table 8). For bacteriophages, the analysis included the ability to obtain a quantifiable bioaerosol, the number of phages in suspension (phage titer), ease of reading baldness (amount of transparency), survival in physiological fluid, and survival in air. Of this group, the bacteriophage phage phi6 scored the highest (Table 9).

Table 8.
Comparison of characteristics of bacterial strains (none; + poor; ++ good; +++ very good).

Table 9.
Comparison of bacteriophage characteristics (- none; + poor; ++ good; +++ very good).
5. Conclusions
Analyzing the results obtained for the tested bacterial strains, it was determined that the Micrococus luteus strain exhibits the highest survivability and, consequently, the greatest research potential. Taking into account the assessed characteristics of the bacteriophages, it was concluded that the phage phi6 strain has the best diagnostic characteristics because its survival in physiological fluids is comparable to the other examined phages (phiX174, T4), while the possibility of obtaining a ‘countable’ bioaerosol is significantly higher. It was also noted that due to the short survival time of phage phi6 in the air, the test should be performed within 5 min, as after this time there is a decrease in the phage by half.
Based on the conducted research, it was determined that the most effective strains to assess the effectiveness of the method to prevent the spread of microorganisms through nebulization are the bacteria strain Micrococus luteus ATCC 7468 and the bacteriophage phi6. The use of these microorganisms also ensures the safety of research personnel while maintaining the highest standards of repeatability of the experiment. The use of non-hazardous bacteriophages, with an accepted time limitation, will much better reflect the way pathogenic viruses propagate through the air (similar size). Having at one’s disposal the characteristics presented in the publication, it is possible to correct the concentration values for the effect of survival, isolating the influence of the tested protection agents. The results of the study will be used in the future to develop a universal procedure and testbed dedicated to devices for reducing the concentration of microorganisms in the air when exiting a room. Then, the effectiveness of the devices will be tested in different variants using selected bacteriophages and bacteria.
Author Contributions
Conceptualization, G.S.; Methodology, J.M. (Justyna Molska), A.S. and R.W. (Radosław Włostowski); Software, J.M. (Jędrzej Matla); Investigation, A.W.; Writing—review & editing, M.R.-G.; Visualization, M.A.-Z.; Supervision, R.W. (Radosław Wróbel). All authors have read and agreed to the published version of the manuscript.
Funding
The project is co-financed by the European Union under he action of the National Center for Research and Development and the Intelligent Development 2014–2020 program, competition 5/1.1.1/2020 Fast Track 5_2020 with the number POIR.01.01.01-00-2513/20 (Mobile and adaptive antroom with automatic disinfection system increasing patient isolation efficacy).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
All authors have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Motamedi, H.; Shirzadi, M.; Tominaga, Y.; Mirzaei, P.A. CFD modeling of airborne pathogen transmission of COVID-19 in confined spaces under different ventilation strategies. Sustain. Cities Soc. 2022, 76, 103397. [Google Scholar] [CrossRef] [PubMed]
- Basu, S. Computational characterization of inhaled droplet 1 transport in the upper airway leading to SARS-CoV-2 2 infection 3. medRxiv 2020. [Google Scholar] [CrossRef]
- Mirzaei, P.A.; Moshfeghi, M.; Motamedi, H.; Sheikhnejad, Y.; Bordbar, H. A simplified tempo-spatial model to predict airborne pathogen release risk in enclosed spaces: An Eulerian-Lagrangian CFD approach. Build. Environ. 2022, 207, 108428. [Google Scholar] [CrossRef] [PubMed]
- Sheikhnejad, Y.; Aghamolaei, R.; Fallahpour, M.; Motamedi, H.; Moshfeghi, M.; Mirzaei, P.A.; Bordbar, H. Airborne and aerosol pathogen transmission modeling of respiratory events in buildings: An overview of computational fluid dynamics. Sustain. Cities Soc. 2022, 79, 103704. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, S.; Yin, Y.; Zhang, T.; Chen, Q. Airborne transmission of COVID-19 virus in enclosed spaces: An overview of research methods. Indoor Air 2022, 32, e13056. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. J. Am. Med. Assoc. 2020, 323, 1406–1407. [Google Scholar] [CrossRef]
- Tang, S.; Mao, Y.; Jones, R.M.; Tan, Q.; Ji, J.S.; Li, N.; Shen, J.; Lv, Y.; Pan, L.; Ding, P.; et al. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ. Int. 2020, 144, 106039. [Google Scholar] [CrossRef]
- Sang-Nourpour, N.; Olfert, J.S. Calibration of optical particle counters with an aerodynamic aerosol classifier. J. Aerosol. Sci. 2019, 138, 105452. [Google Scholar] [CrossRef]
- Zhang, R.; Li, Y.; Zhang, A.L.; Wang, Y.; Molina, M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. USA 2020, 117, 14857–14863. [Google Scholar] [CrossRef]
- Alghamdi, W.; Neamatallah, A.A.; Alshamrani, M.M.; Al Mehmadi, F.; El-Saed, A. Distribution and the trend of airborne particles and bio-aerosol concentration in pediatric intensive care units with different ventilation setting at two hospitals in Riyadh, Saudi Arabia. J. Infect. Public Health 2023, 16, 588–595. [Google Scholar] [CrossRef]
- Bolookat, F.; Hassanvand, M.S.; Faridi, S.; Hadei, M.; Rahmatinia, M.; Alimohammadi, M. Assessment of bioaerosol particle characteristics at different hospital wards and operating theaters: A case study in Tehran. MethodsX 2018, 5, 1588–1596. [Google Scholar] [CrossRef]
- Ali, S.; McDermott, S. Impact of the COVID-19 pandemic on the incidence of healthcare-associated Clostridioides difficile infection in a tertiary healthcare facility in the Republic of Ireland. Infect. Prev. Pract. 2023, 5, 100300. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Q.; Yang, F.; Zhang, J.; Fu, Y.; Zhu, Z.; Guo, J.; Li, X.; Yang, L. Associations between COVID-19 infection experiences and mental health problems among Chinese adults: A large cross-section study. J. Affect. Disord. 2023, 340, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ji, L.; Xiong, G.; Ning, K. The distinct microbial community patterns and pathogen transmission routes in intensive care units. J. Hazard. Mater. 2023, 441, 129964. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. J. Am. Med. Assoc. 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed]
- Cook, T.M. Personal protective equipment during the coronavirus disease (COVID) 2019 pandemic–a narrative review. Anaesthesia 2020, 75, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Cowling, B.J.; Leung, G.M. Epidemiological research priorities for public health control of the ongoing global novel coronavirus (2019-nCoV) outbreak. Eurosurveillance 2020, 25, 2000110. [Google Scholar] [CrossRef] [PubMed]
- Macher, J.M. Positive-Hole Correction of Multiple-Jet Impactors for Collecting Viable Microorganisms. Am. Ind. Hyg. Assoc. J. 1989, 50, 561–568. [Google Scholar] [CrossRef]
- Sanmark, E.; Kuula, J.; Laitinen, S.; Oksanen, L.M.A.H.; Bamford, D.H.; Atanasova, N.S. Safe use of PHI6 IN the experimental studies. Heliyon 2023, 9, e13565. [Google Scholar] [CrossRef]
- Bailey, R.; Fielding, L.; Young, A.; Griffith, C. Effect of Ozone and Open Air Factor against Aerosolized Micrococcus luteus. J. Food Prot. 2007, 70, 2769–2773. [Google Scholar] [CrossRef]
- Borkar, S.B.; Negi, M.; Jaiswal, A.; Raj Acharya, T.; Kaushik, N.; Choi, E.H.; Kaushik, N.K. Plasma-generated nitric oxide water: A promising strategy to combat bacterial dormancy (VBNC state) in environmental contaminant Micrococcus luteus. J. Hazard. Mater. 2024, 461, 132634. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).