1. Introduction
Candida auris (
C. auris) emerged as an opportunistic budding yeast in 2009 in Japan. However, retrospective studies on a culture collection proved the earlier existence of the strain in the mid-1990s [
1,
2]. Unlike other fungal pathogens,
C. auris has been able to progressively spread over three continents with mortality rates as high as 60% [
3]. It is believed that
C. auris has a similar ability to colonize the skin and gastrointestinal tract (GIT); however, there is compelling evidence that
C. auris preferentially colonizes the skin rather than the GIT, hence its tendency to cause hospital-acquired infections (HAIs) [
4]. Additionally, its acquired antifungal resistance compounded by biofilm formation enables the pathogen to persist on hospital surfaces and medical devices for up to 14 days, challenging hospital cleaning protocols and causing infections [
3,
5].
Multiple strains of
C. auris have been identified across 41 countries, including several countries in the Middle East. These strains have been classified based on whole genome sequencing into five clades: South Asian (Clade I), East Asian (Clade II), African (Clade III), South American (Clade IV), and Iranian (Clade V) clades [
6]. This growing concern worldwide was recognized by the US Centers for Disease Control and Prevention (CDC), which designated
C. auris as an urgent threat [
7], and by the WHO’s Global Antimicrobial Resistance Surveillance System (GLASS) that developed the GLASS-FUNGI module for GLASS-focused surveillance to encourage countries “to strengthen or build their national fungal AMR surveillance [
8]. Given the intricacies of the identification and antifungal susceptibility testing of
C. auris compounded by the inadequacy of national expertise in mycology and the lack of functional surveillance systems, disproportionate reporting among countries is expected [
3].
To the best of our knowledge, this is the first case of C. auris reported in Jordan. Reporting of this case was made possible through a large network of hospitals and affiliated laboratories constituting the network of the national surveillance program of hospital-acquired infections (HAIs) and antimicrobial resistance (AMR) in Jordan. The program was established in 2011 as a joint collaboration between the Ministry of Health (MOH) and the US Naval Medical Research Unit EURAFCENT (formerly known as NAMRU-3), with the aim to identify the burden of HAIs in participating hospitals and to institute evidence-based interventions in Jordan.
To date, the program has been implemented across five governorates with the participation of nine hospitals: five MOH hospitals, three hospitals affiliated with the private sector, and one university teaching hospital.
2. External Quality Assessment (EQA) Program at the Central Public Health Laboratory (CPHL) Amman
Each participating hospital is served by a well-equipped laboratory. Hospital laboratories detect pathogens for the sources of four HAIs (bloodstream infection, urinary tract infection, pneumonia, and surgical site infection). Once the pathogen is isolated, identification (ID) and antimicrobial susceptibility testing (AST) are carried out. The colonies of the causative pathogen(s) are preserved as pure colonies in trypticase soya broth (TSB) vials at −20 °C to be later dispatched to the CPHL Amman for confirmation and antifungal susceptibility testing.
The CPHL Amman was officially designated as the national reference laboratory for AMR in 2018 and is currently equipped with a VITEK-2 automated identification system (BioMérieux, Craponne, France) and a PCR unit for genomic analysis.
Laboratory data (ID and AST) generated by each surveillance site laboratory are uploaded onto a secured web-based surveillance database hosted on the MOH server to be correlated with confirmatory data from the CPHL. Following the automated matching process, agreement reports are instantly generated for each laboratory noting erroneous results, if any, and advising on the corrective actions to be taken.
3. Case Presentation
On 17 June 2021 (day one), a 48-year-old female was admitted to the emergency room at the Specialty Hospital (SH) Amman complaining of fever and bilateral knee pain that had lasted for two days. The patient had a history of diabetes mellitus (DM), hypertension (HTN), hypothyroidism, and SARS-CoV-2 infection that required hospitalization in another hospital between 2 and 23 May 2021.
Upon physical examination, the patient was feverish (temperature 39 °C). Multiple purulent pressure ulcers were observed, and a urinary catheter was in place for an unknown duration. The total leucocyte count was 17,600/mm3 with 90% neutrophils, a creatinine level of 0.76 mg/dL, a sodium level of 139 mEq/L, a potassium level of 4.39 mEq/L, and a CRP level of 378 mg/dL. Urine analysis revealed a specific gravity of 1.037, a pH of 5, protein +2, glucose +4, urobilinogen +2, negative ketone nitrate and bilirubin, a WBC count of 60/HPF, an RBC count of 8/HPF, and no casts.
The patient was diagnosed with sepsis, and culture samples were obtained from pressure ulcers, blood, and urine. Empirical antibacterial therapy was initiated immediately after the collection of laboratory samples and consisted of 2 g of IV ceftriaxone once daily, 1 g of IV vancomycin twice daily, and 4.5 g of IV piperacillin–tazobactam every eight hours.
On day two of admission (18 June 2021), the patient complained of dyspnea, and her oxygen saturation level dropped. Pulmonary computed tomography angiography (PCTA) revealed a pulmonary embolism, and the patient was transferred to the intensive care unit (ICU). On day four (20 June 2021), culture and sensitivity results reported the isolation of carbapenem-resistant Klebsiella pneumoniae (KPC) from the urine, pressure ulcers, and blood samples. Candida auris was first identified at the genus level from a urine sample cultured on Sabouraud dextrose agar medium incubated at 37 °C for up to two weeks (count 10,000/HPF). Species identification was possible using the Vitek-2 yeast identification system (version 8.01) (BioMérieux, France). Pure colonies were isolated and preserved onto (TSB) for confirmatory testing at the CPHL.
On day five (21 June 2021) and in concordance with the AST results of KPC, antimicrobial therapy was modified to 4.5 million units of colistin twice daily. However, the patient’s condition rapidly deteriorated, and she succumbed to the disease due to overwhelming sepsis. Therefore, the hospital laboratory discontinued antifungal susceptibility testing.
4. Confirmatory Testing at the CPHL
At the CPHL, retesting of
C. auris isolates was carried out in October 2022 using Chromogenic Candida agar (Condalab, Madrid, Spain), blood, and Sabouraud dextrose agar media. Colonies were identified by the Vitek2 yeast identification system (version 8.01) (BioMérieux, France) and further confirmed by real-time PCR. The extraction and PCR settings were used according to the manufacturer’s instructions. DNA was extracted from a single pure colony of
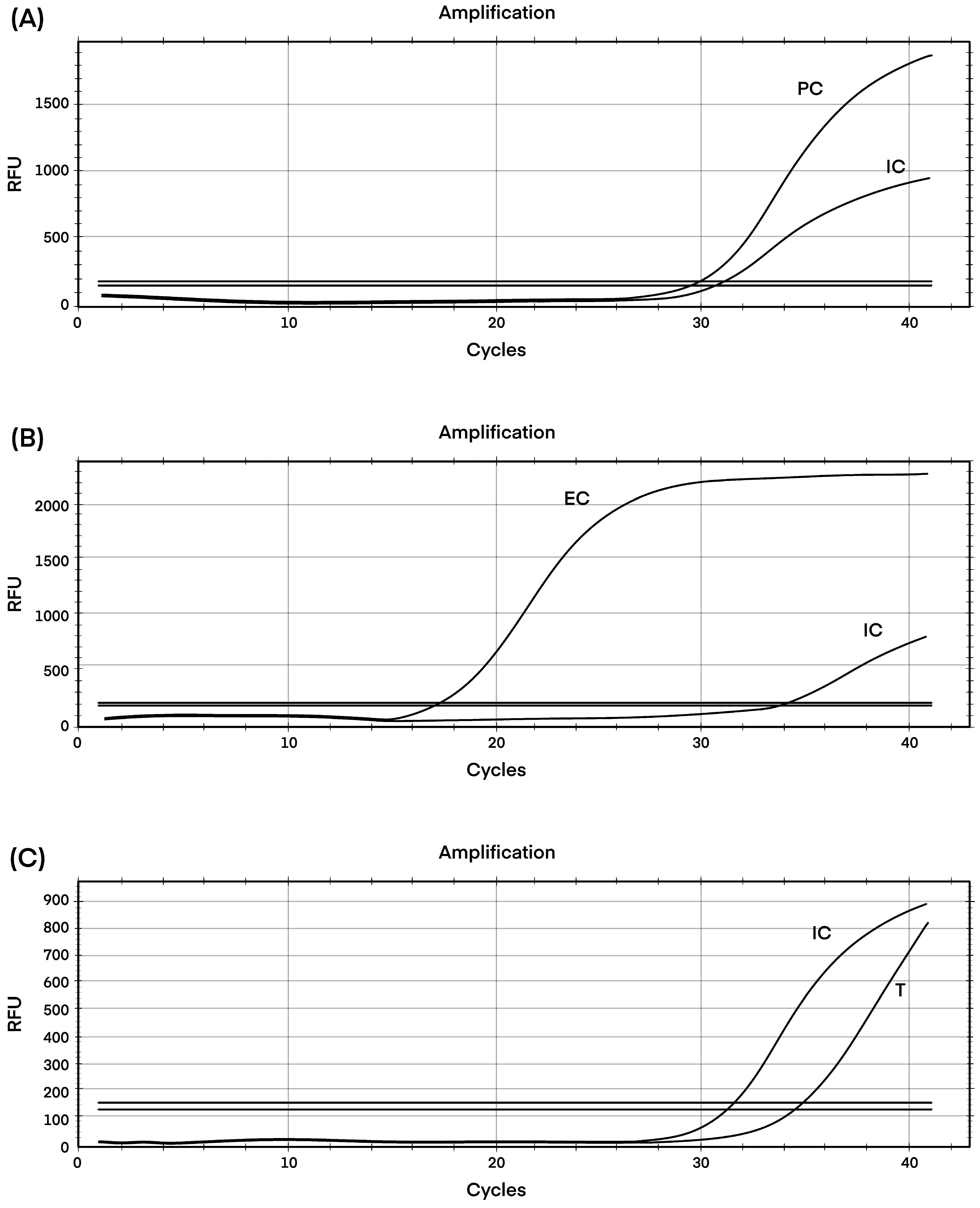
C. auris using the YeaStar Genomic DNA kit (ZYMO RESEARCH, Irvine, CA, USA). In brief, 1.5 mL of cells was centrifuged at 500 g for 2 min, and the supernatant was removed completely. Approximately 120 μL of YD Digestion Buffer and 5 μL of R-Zymolyase™1(RNase A + Zymolyase™) were added followed by incubation at 37 °C for 40–60 min. Approximately 120 μL of YD Lysis Buffer2 was added and mixed well by gently vertexing, and centrifuged >10,000 rpm for 2 min. The supernatant was loaded onto the Zymo-Spin™ III Column and centrifuged at >10,000 rpm for 1 min. Approximately 300 μL of DNA Wash Buffer was added and centrifuged for 1 min at ≥10,000 rpm to wash. An additional 300 μL of DNA Wash Buffer was added to repeat the wash and centrifuged for 1 min. The Zymo-Spin™ III Column was transferred to a new 1.5 mL centrifuge tube and 60 μL of water was added. The final concentration of DNA was 3.3 ng/μL. The extraction step was followed by quantitative PCR (qPCR) with the GPS™ CanAur MONODOSE dtec-qPCR Test (Alicante, Spain) using 5 μL of DNA extract. This system employs ready-to-use tubes with dehydrated components for which only the extracted DNA needs to be added before thermocycling (
Figure 1). A standard curve dilution series was prepared from the kit’s standard template where a cycle threshold (Ct) value of ≤35 was reported as positive and >35 was reported as negative for the isolates. The fluorescent signal was collected by using the FAM channel for the target, and the HEX channel for the internal control. The cycling conditions of the assay were initiated by 2 min activation at 95 °C followed by 40 cycles of 95 °C for 5 s and 60 °C for 20 s.
5. Antifungal Susceptibility Testing
In vitro antifungal susceptibility testing was carried out in March 2023 using broth microdilution (
Figure 2) according to the Clinical and Laboratory Standards Institute (CLSI) (M27-A3 CLSI) [
9].
The minimum inhibitory concentrations were 1 µg/mL for amphotericin B, 2 µg/mL for fluconazole, 0.12 µg/mL for itraconazole, 0.12 µg/mL for anidulafungin, 0.06 µg/mL for caspofungin, 0.12 µg/mL for micafungin, 0.015 µg/mL for voriconazole, and 0.03 µg/mL for posaconazole, denoting susceptibility to all tested antifungal drugs. To ensure the reliability and the reproducibility of the technique, the microbroth dilution test was repeated and the same results were obtained.
6. Discussion
Candid auris is inarguably one of the most urgent threats that humankind is currently facing owing to its unconventional character of multidrug resistance [
10]. Although it was first identified from the external canal of a Japanese patient in 2019, the literature has reported its involvement in a vast array of deep-seated infections, including UTIs, with high mortalities and co-morbidities [
11].
Herein, we report the first case of
C. auris identified in Jordan through the national HAI and AMR surveillance program in Jordan. This adds Jordan to the eight other countries in the Middle East Region that have reported
C. auris cases (Oman, Kuwait, Saudi Arabia, the United Arab Emirates, Iran, Sudan, Lebanon and Qatar) [
6,
12,
13,
14].
The patient in this case had a history of risk factors including diabetes mellitus, hypertension, and hypothyroidism. Furthermore, her previous hospitalization for COVID-19 increased the risk of acquiring
C. auris. Her weakened immune status and the intake of steroids and at least one antibiotic (notably azithromycin) created the perfect environment for increasing the colonization of Candida species including
C. auris [
15,
16]. Furthermore, several studies have shown that the association of SARS-CoV-2 infection with
C. auris is attributed to the persistence of
C. auris on hospital surfaces, biofilm formation, and its resistance to disinfectants [
17]. ICU admission has contributed notably to acquiring infections with
C. auris. The use of invasive devices including urinary catheters and prolonged hospital stays in addition to compromising infection control measures due to the overcrowding of hospitals have augmented the risks for the colonization of
C. auris [
6,
18].
Similar to other
Candida species, the isolation of
C. auris from the urinary tract is not solid proof of a UTI. In fact, it may indicate colonization rather than infection, particularly since the distinction between colonization and infection has not yet been agreed upon and is reliant on the clinical decision of the physician at each facility [
19]. Once identified and following CDC guidelines, isolation is recommended, and decolonization using a suitable disinfectant like chlorhexidine or others is the recommended practice to prevent horizontal transmission among patients [
20].
Reliable identification of
C. auris is considered a real challenge for laboratories mostly because
C. auris is misidentified as
Candida haemulonii by common commercial systems such as the VITEK-2, BD Phoenix, and the API-20 [
21]. However, the reliability of
C. auris detection by the Vitek-2 yeast identification system (version 8.01) was reported by Ambaraghassi et al., 2019 [
22], and this finding was also consistent with the CDC’s algorithm which stated that “no further testing is needed if a
C. auris identification is obtained by the Vitek 2 8.01 system” [
23]. Currently, MALDI-TOF-MS (Bruker Inc., MA, USA) is the most reliable method for the identification of
C. auris [
21], which is unfortunately not feasible in our surveillance system. Nevertheless, confirmatory testing by real-time PCR was carried out at the CPHL. The isolate demonstrated a Cycle threshold (Ct) value of 35. A similar result was obtained by Martínez-Murcia et al., 2018, who detected one
C. auris isolate (Ct = 40) with the same PCR kit. This high Ct value was explained as a “weak concentration of extracted DNA resulting in a weak signal” [
24].
The availability of sophisticated techniques such as MALDI-TOF MS and molecular techniques is still limited to well-resourced laboratories. Many laboratories in developing countries rely on conventional methods for fungal identification and reporting. Therefore, identification many times stops at the genus level without reaching the species level [
25]. To further complicate the situation, the breakpoints for antifungal susceptibility are not clearly defined by the CLSI or European committee on antimicrobial susceptibility testing (EUCAST). Nevertheless, the CDC and food and drug administration (FDA) have proposed guidance for interpretation most laboratorians rely on when it comes to reporting to clinicians [
26]. Therefore, reference laboratories play a key role in providing not only confirmatory testing but also up-to-date guidance on antifungal susceptibility protocols that should be followed by laboratories.
In the present case and in accordance with CDC breakpoints, the isolate was surprisingly sensitive to common antifungal drugs used in the battery for assessing the sensitivity of
C. auris, including azoles, which are known to be inactive against strains of
C. auris in 90% of cases. This uncommon pattern of susceptibility has also been reported in India [
27], Iran [
28], and Colombia [
29], where low minimum inhibitory concentrations (MICs) against fluconazole and other antifungal drugs have been reported. Notably, the diversity observed in the patterns of antifungal susceptibility in different countries suggests “localized resistance development” [
28].
To date, there is no consensus on treatment protocols. Echinocandin and liposomal amphotericin B are recommended as first-line agents for patients with UTIs. Combination therapies such as flucytosine and amphotericin B bladder irrigations are considered potential alternatives in the case of the recurrence or persistence of infection [
3].
Crude mortality in
C. auris-associated infections has been reported to vary from 33.33% to 100% worldwide [
10]. However, the cause of death in our case was probably not due to
C. auris infection because of a concomitant bloodstream infection with carbapenem-resistant
K. pneumoniae (KPC). Unfortunately, the patient passed away before colistin could be administered as recommended by her physician and before receiving the antifungal susceptibility results from the CPHL.
7. Limitations
We were not able to obtain the patient’s medical records from her previous hospitalization to glean information about her previous diagnosis, laboratory results, and the treatment protocol she received.
Considering that this isolate was confirmed a year later, in addition to the unknown history of her previous hospitalization, it was almost impossible to identify the possible source of colonization. Further environmental studies are recommended to identify the prevalence of C. auris in hospitals in Jordan and guide prevention efforts.
Additionally, it should be noted that clade information for the C. auris isolate was not provided in this study as this requires the use of whole-genome sequencing and phylogenetic analysis, which are not currently part of the standard methodology for the surveillance of HAI and AMR in Jordan. However, there are plans in place to enhance the next-generation sequencing capacities at the CPHL in Jordan to conduct whole genome sequencing for all surveillance isolates of public health importance including this isolate.
8. Conclusions
The findings of this report indicate the potential threat of C. auris in Jordan. This case was reported through the national surveillance network and confirmed by the national reference laboratory.
We highlight the fact that the inaccurate diagnosis and misidentification of C. auris in laboratories are probably reasons for underreporting, in addition to a lack of awareness of the threat posed by C. auris in hospitals.
It is of the utmost importance to ensure that functional surveillance systems are in place to identify and report emerging pathogens and alert public health hospitals to halt their spread and contain any potential outbreaks.
The susceptibility pattern of C. auris isolated in this case represents an uncommon pattern, and more strains need to be tested and aligned with previously identified clades through genome sequencing to better understand the characteristics of C. auris strains in Jordan. Moreover, studies with larger sample sizes can help guide treatment protocols and infection control and prevention measures.
Author Contributions
Conceptualization, J.W.A.-R. and R.A.G.; methodology, O.H.S., D.A.-J., and M.E.-S.; validation, M.M.S., O.H.S., and T.S.O.; resources, S.N. and O.H.S.; original draft preparation and writing; M.E.-S., review and editing, T.S.O.; project administration, M.G. and T.S.O.; visualization and supervision, T.S.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Armed Forces Health Surveillance Division, Global Emerging Infections Surveillance (GEIS) Branch, ProMIS ID P0166_22_N3 and P0126_23_N3.
Institutional Review Board Statement
The study protocol was approved by the Naval Medical Research Unit 3 Institutional Review Board in compliance with all applicable federal regulations governing the protection of human subjects. The study protocol number “NAMRU3.PJT.2011.0014” was approved as an activity not involving human research.
Informed Consent Statement
Not applicable. The study was declared as public health activity that does not involve human research activity and it does not require written informed consent.
Data Availability Statement
This is a case report. All available data was reported in this article. Data sharing is not applicable to this article.
Acknowledgments
The authors would like to acknowledge the effort of the bacteriology and the molecular staff at the CPHL of Jordan, and the effort of the staff of the Infection Control department at the MOH of Jordan.
Conflicts of Interest
The authors declare no conflict of interest. Author Said M is an employee of the U.S. Government. This work was prepared as part of her official duties. Title 17, U.S.C. §105 provides that copyright protection under this title is not available for any work of the U.S. Government. Title 17, U.S.C. §101 defines a U.S. Government work as work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties. The views expressed in this article reflect the results of research conducted by the author and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the United States Government.
References
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog 2020, 16, e1008921. [Google Scholar] [CrossRef]
- Ahmad, S.; Alfouzan, W. Candida auris: Epidemiology, Diagnosis, Pathogenesis, Antifungal Susceptibility, and Infection Control Measures to Combat the Spread of Infections in Healthcare Facilities. Microorganisms 2021, 9, 807. [Google Scholar] [CrossRef] [PubMed]
- Griffith, N.; Danziger, L. Candida auris Urinary Tract Infections and Possible Treatment. Antibiotics 2020, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Voss, A.; Meis, J.F. Multidrug-resistant Candida auris: ‘New kid on the block’ in hospital-associated infections? J. Hosp. Infect. 2016, 94, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Kumar, V.A.; Sharma, C.; Prakash, A.; Agarwal, K.; Babu, R.; Dinesh, K.R.; Karim, S.; Singh, S.K.; Hagen, F.; et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 919–926. [Google Scholar] [CrossRef]
- Alfouzan, W.B.; Dhara, R.; Albarrag, A.; Al-Abdelyda, H. The emerging pathogen Candida auris: A focus on the Middle-Eastern countries. Microbiol. J. Infect. Public Health 2019, 12, 451–459. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States. 2019. Available online: https://stacks.cdc.gov/view/cdc/82532 (accessed on 22 July 2023).
- WHO’s Global Antimicrobial Resistance Surveillance System (GLASS)-Fungi; GLASS Focused Surveillance. Available online: https://www.who.int/initiatives/glass/glass-modules-5 (accessed on 22 July 2023).
- Clinical and Laboratory Standards Institute: Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard. CLSI Doc M27-A3 Suppl S. 2008, 3, 6–12.
- Osei Sekyere, J. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen 2018, 7, e00578. [Google Scholar]
- Gajdács, M.; Dóczi, I.; Ábrók, M.; Lázár, A.; Burián, K. Epidemiology of candiduria and candida urinary tract infections in inpatients and outpatients: Results from a 10-year retrospective survey. Cent. Eur. J. Urol. 2019, 72, 209–214. [Google Scholar]
- Badri, A.M.; Sherfi, S.A. First Detection of Emergent Fungal Pathogen Candida auris in Khartoum State, Sudan. Am. J. Biomed. Sci. Res. 2019, 6, 4–7. [Google Scholar] [CrossRef]
- Allaw, F.; Kara Zahreddine, N.; Ibrahim, A.; Tannous, J.; Taleb, H.; Bizri, A.R.; Dbaibo, G.; Kanj, S.S. First Candida auris Outbreak during a COVID-19 Pandemic in a Tertiary-Care Center in Lebanon. Pathogens 2021, 10, 157. [Google Scholar] [CrossRef] [PubMed]
- Koleri, J.; Petkar, H.M.; Al Soub, H.A.R.S.; AlMaslamani, M.A.R.S. Candida auris Blood stream infection—A descriptive study from Qatar. BMC Infect Dis. 2023, 23, 513. [Google Scholar] [CrossRef] [PubMed]
- Janniger, E.J.; Kapila, R. Public health issues with Candida auris in COVID-19 patients. World Med. Health Policy 2021, 13, 766–772. [Google Scholar] [CrossRef]
- Machado, M.; Estévez, A.; Sánchez-Carrillo, C.; Guinea, J.; Escribano, P.; Alonso, R.; Valerio, M.; Padilla, B.; Bouza, E.; Muñoz, P. Incidence of Candidemia Is Higher in COVID-19 versus Non-COVID-19 Patients, but Not Driven by Intrahospital Transmission. J. Fungi 2022, 8, 305. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Carvalho, A.; Nguyen, M.H.; Hedayati, M.T.; Netea, M.G.; Perlin, D.S.; Hoenigl, M. Covid-19-associated candidiasis (CAC): An underestimated complication in the absence of immunological predispositions? J. Fungi 2020, 6, 211. [Google Scholar] [CrossRef] [PubMed]
- Thoma, R.; Seneghini, M.; Seiffert, S.N.; Vuichard Gysin, D.; Scanferla, G.; Haller, S.; Flury, D.; Boggian, K.; Kleger, G.R.; Filipovic, M.; et al. The challenge of preventing and containing outbreaks of multidrug-resistant organisms and Candida auris during the coronavirus disease 2019 pandemic: Report of a carbapenem-resistant Acinetobacter baumannii outbreak and a systematic review of the literature. Antimicrob. Resist. Infect. Control 2022, 11, 12. [Google Scholar] [PubMed]
- Fisher, J.F.; Sobel, J.D.; Kauffman, C.A.; Newman, C.A. Candida urinary tract Infections Treatment. Clin. Infect. Dis. 2011, 52, S457–S466. [Google Scholar] [CrossRef]
- Das, S.; Tigga, R.; Rai, G.; Singh, P.K.; Datt, S.; Tyagi, A.; Singh, N.P. Candida auris colonization in an immunocompetent patient: A new threat in medical ICU. Med. Mycol. Case Rep. 2018, 21, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Kathuria, S.; Singh, P.K.; Sharma, C.; Prakash, A.; Masih, A.; Kumar, A.; Meis, J.F.; Chowdhary, A. Multidrug-resistant Candida auris misidentified as Candida haemulonii: Characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J. Clin. Microbiol. 2015, 53, 1823–1830. [Google Scholar]
- Ambaraghassi, G.; Dufresne, P.J.; Dufresne, S.F.; Vallières, É.; Muñoz, J.F.; Cuomo, C.A.; Berkow, E.L.; Lockhart, S.R.; Luong, M.L. Identification of Candida auris by Use of the Updated Vitek 2 Yeast Identification System, Version 8.01: A Multilaboratory Evaluation Study. J. Clin. Microbiol. 2019, 57, e00884-19. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Algorithm to Identify Candida Auris Based on Phenotypic Laboratory Method and Initial Species Identification. Centers for Disease Control and Prevention, Atlanta, GA. 2018. Available online: https://www.cdc.gov/fungal/diseases/candidiasis/pdf/Testing-algorithm-by-Method-temp.pdf (accessed on 5 July 2023).
- Martínez-Murcia, A.; Navarro, A.; Bru, G.; Chowdhary, A.; Hagen, F.; Meis, J.F. Internal validation of GPS™ MONODOSE CanAur dtec-qPCR kit following the UNE/EN ISO/IEC 17025:2005 for detection of the emerging yeast Candida auris. Mycoses 2018, 61, 877–884. [Google Scholar] [CrossRef]
- Garcia-Bustos, V.; Cabanero-Navalon, M.D.; Ruiz-Saurí, A.; Ruiz-Gaitán, A.C.; Salavert, M.; Tormo, M.Á.; Pemán, J. What Do We Know about Candida auris? State of the Art, Knowledge Gaps, and Future Directions. Mcroorganisms 2021, 9, 2177. [Google Scholar] [CrossRef]
- Chastain, D.B.; King, S.T.; Stover, K.R. Rethinking urinary antibiotic breakpoints: Analysis of urinary antibiotic concentrations to treat multidrug resistant organisms. BMC Res. Notes 2018, 11, 497. [Google Scholar] [CrossRef]
- Mathur, P.; Hasan, F.; Singh, P.K.; Malhotra, R.; Walia, K.; Chowdhary, A. Five-year profile of candidemia at an Indian trauma centre: High rates of Candida auris blood stream infections. Mycoses 2018, 61, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Abastabar, M.; Haghani, I.; Ahangarkani, F.; Rezai, M.S.; Taghizadeh Armaki, M.; Roodgari, S.; Kiakojuri, K.; Al-Hatmi, A.M.S.; Meis, J.F.; Badali, H. Candida auris otomycosis in Iran and review of recent literature. Mycoses 2019, 62, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Escandón, P.; Chow, N.A.; Caceres, D.H.; Gade, L.; Berkow, E.L.; Armstrong, P.; Rivera, S.; Misas, E.; Duarte, C.; Moulton-Meissner, H.; et al. Molecular epidemiology of Candida auris in Colombia reveals a highly related, country-wide colonization with regional patterns in amphotericin B resistance. Clin. Infect. Dis. 2019, 68, 15–21. [Google Scholar] [PubMed]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).