Natural Products from Red Algal Genus Laurencia as Potential Inhibitors of RdRp and nsp15 Enzymes of SARS-CoV-2: An In Silico Perspective
Abstract
1. Introduction
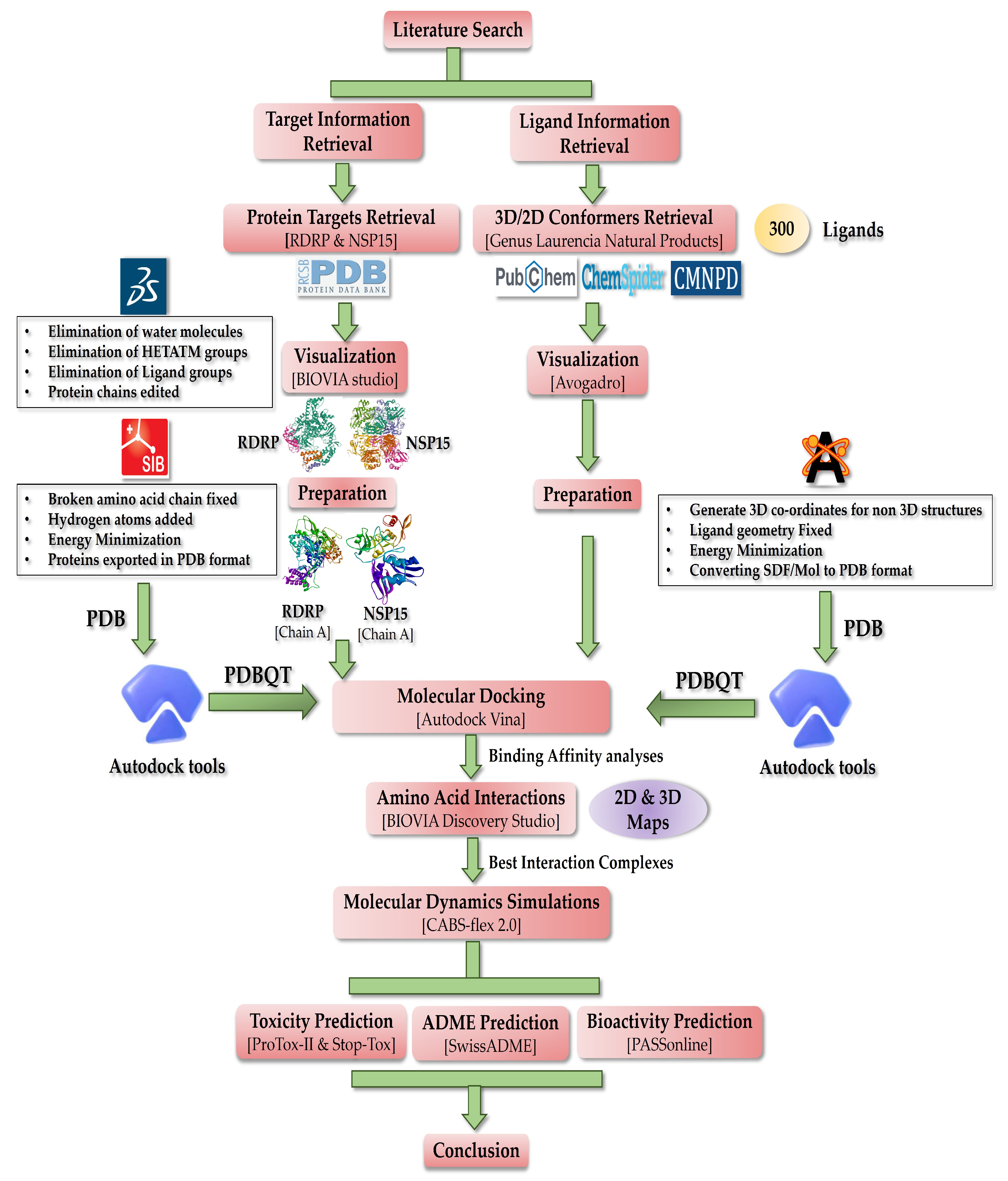
2. Materials and Methods
2.1. Retrieval of Proteins
2.2. Retrieval of Ligands
2.3. Visualization Tools
2.4. Pre-Docking Preparations
2.5. Docking Protocol Validation
2.6. Molecular Docking
2.7. MD Simulation
2.8. Toxicity Profile Assessment
2.9. Pharmacokinetics Analysis
3. Results and Discussion
3.1. Binding Affinity Studies
3.1.1. Docking β-Hairpin Motif Region (RdRp): N-Terminal
| RdRp Protein | ||||
|---|---|---|---|---|
| Low | Moderate | High | ||
| −4.0 to −4.9 kcal/mol | −5 to −5.9 kcal/mol | −6 to −6.9 kcal/mol | −7 to −7.9 kcal/mol | −8.0 kcal/mol and Above |
| 1-methyl-2,3,5-tribromoindole | (−)-3-(E)-bromomethylidene-10beta-bromo-beta-chamigrene | 1,2-Dehydro-3,4-epoxypalisadin B | 5-alpha-cholestane-3,6-dione | Bromophycoic acid B |
| 6,8-cycloeudesmane | (+)-3-(Z)-bromomethylidene-10beta-bromo-beta-chamigrene | 2-Hydroxyluzofuranone A, B | 6-hydroxycholest-4-en-3-one | Bromophycoic acid C |
| 9-octadecanoic acid | (5S)-5-Acetoxy-beta-bisabolene | 3 alpha-hydroperoxy-3-epiaplysin | 10-acetoxyangasiol | Bromophycolide E |
| 14-methylpentadecanoic acid | (6R,9R,10S)-10-bromo-9-hydroxychamigra-2,7(14)-diene | 3-Bromo-4,5-dihydroxybenzaldehyde | 10-epi-Dehydrothyrsiferol | Bromophycolide H |
| beta-Synderol | (10R)-10-Bromo-alpha-chamigrene | 3-Bromobarekoxide | 13-Hydroxyprethyrsenol A | Bromophycolide K |
| Halomon | 2-bromospironippol | 3-epi-Perforenone A | 15,16-Dehydrovenustatriol | Bromophycolide L |
| Luzonensin | (3Z)-laurenyne | 3R,4S-luzonolone | 15-DehydroxythyrsenolA | Bromophycolide P |
| Hordenine | 3,4-epoxypalisadin B | 3S, 4R-luzonolone | 16-Hydroxydehydrothyrsiferol | Bromophycolide R |
| Laurencenyne | 3,7-dihydroxydihydrolaurene | 5-acetoxypalisadin B | Beta cryptoxanthin | Bromophycolide S |
| Trans-Laurencenyne | 3-alpha-Hydroxydebromoaplysin | 5-alpha-Hydroxyaplysistatin | Brassicasterol | Callicladol |
| 3-beta-Hydroperoxyaplysin | 9-hydroxy-3-epi-perforenone A | Bromophycoic acid A, D, E | Dehydrothyrsiferol | |
| 4-Hydroxy-1,8-epi-isotenerone | 11,14-dihydroaplysia-5,11,14,15-Tetrol | Bromophycolide A–D, F, J, M–O, Q, T, U | Isodehydrothyrsiferol | |
| 4-hydroxypalisidin C | 15-hydroxypalisadin A | Callophycoic acid C–E | Laurebiphenyl | |
| 5-acetoxyoxachamigrene | Acetylmajapolene A, B | Campesterol | Lithothamin A | |
| 5-epi-maneolactone | Aldingenin D | Cholest-4-en-3,6-dione | Mammeisin | |
| 7-acetyl-aplysiol | Aplysistatin | Dehydrovenustatriol | Thyrsenol A | |
| 7-hydroxylaurene | Aristolan-10-ol-9-one | Lactodehydrothyrsiferol | Thyrsenol B | |
| 8,10-dibromoisoaplysin | Aplysiodiol | Neurymenolide A, B | Thyrsiferol | |
| 9-Deoxyelatol | Barekoxide | Neoirietetraol | ||
| 10-Bromo-beta-chamigrene | beta-Sitosterol | Predehydrovenustatriolacetate | ||
| 10-Bromosoaplyin | Bromophycolide G, I | Prethyrsenol A | ||
| 10-hydroxyisolaurene | Caespitol | Pseudodehydrothyrsiferol | ||
| 10-Hydroxyaplysin | Callophycoic acid A, B, G, H, I, J | Stigmasterol | ||
| 12-hydroxy Isolaurene | Callophycol A, B | |||
| 12-Hydroxypalisadin B | Chamigrene Lactone | |||
| 15-Hydroxylaurene | Compositacin B, E, F, I, N | |||
| 15-Oxolaurene | Cholest-4-en-3-one | |||
| Almadioxide | Cholest-5-en-3alpha-ol | |||
| Aromadendrene | Cholest-5-en-3beta-ol | |||
| Aldingenin A, B, C | Debromoisocalenzanol | |||
| Allolaurinterol | Japonenyne A | |||
| Allolaurinterolacetate | Johnstonol | |||
| Aplysinol | Laurecomin A, D | |||
| Aristolan-8-en-1-one | Laurenokomarin | |||
| Aristolane | Laurepoxyene | |||
| Axinysone B | Laureacetal C | |||
| Aplysiol-7-one | Laurefurenyne B | |||
| Aplysiolic acid | Laurinterolacetate | |||
| (−)-BisezakyneA | Luzondiol | |||
| Brasilenol | Majapolene A | |||
| Bromocuparene | Oryzalexin S | |||
| Bromocyclococanol | Pacifenol | |||
| Bosseopentanoic acid | Palisadin D | |||
| Bromlaurenidificin | Perforenone A | |||
| Caespitane | Saringosterol | |||
| Caesspitenone | Tiomanene | |||
| Cycloeudesmol | ||||
| Cyclolaurenol | ||||
| Callophycoic acid F | ||||
| Callenzanol | ||||
| Chamigrene epoxide | ||||
| Chinzallene | ||||
| Compositacin A, C, D, G, H, J, K, L, M | ||||
| Cycloelatanene A, B | ||||
| Cycloisoallolaurinterol | ||||
| Chlorofucin | ||||
| Cupalaurenol | ||||
| Dactylyne | ||||
| Debromoaplysin | ||||
| Debromoepiaplysinol | ||||
| Debromolaurinterol | ||||
| Debromolaurinterolacetate | ||||
| Dendroidiol | ||||
| Dendroidone | ||||
| Deoxyprepacifenol | ||||
| Deschloroelatol | ||||
| Elatenyne | ||||
| Elatol | ||||
| Epiaplysinol | ||||
| Epibrasilenol | ||||
| Floridoside | ||||
| Filiformin | ||||
| Filiforminol | ||||
| Guimarediol | ||||
| Heterocladol | ||||
| Intricenyne | ||||
| Isocaespitol | ||||
| Isodihydrolaurene | ||||
| Isolaurallene | ||||
| Isolaureatin | ||||
| Isolaurene | ||||
| Itomanindole A | ||||
| Isolaurenidificin | ||||
| Isoafricanol | ||||
| Isoallolaurinterol | ||||
| Isoaplysin | ||||
| Isodactyloxene A | ||||
| Isodebromolaurinterol | ||||
| Isolaurenisol | ||||
| Isoobtusol | ||||
| Isopalisol | ||||
| Isorigidol | ||||
| Kumausallene | ||||
| Laurallene | ||||
| Laureacetal A, B | ||||
| Laurefurenyne A, C, D, E, F | ||||
| Laurencial | ||||
| Laurendecumallene A, B | ||||
| Laurendecumenyne A, B | ||||
| Laurenenyne | ||||
| Laurenisol | ||||
| Laurenone A | ||||
| Laureoxanyne | ||||
| Laurecomin B, C | ||||
| Laurencomposidiene | ||||
| Laurene | ||||
| Laurentristich-4-ol | ||||
| Laureperoxide | ||||
| Laurokamurene A-D | ||||
| Luzofuran | ||||
| Luzonenone | ||||
| Luzonensol | ||||
| Luzonensol acetate | ||||
| Laurepinnacin | ||||
| Laurinterol | ||||
| Ma’iliohydrin | ||||
| Mailione | ||||
| Majapolene B | ||||
| Microcladallene A-C | ||||
| Neoisoprelaurefucin | ||||
| Neolaurallene | ||||
| Nidificene | ||||
| Notoryne | ||||
| Octadecanedioic acid | ||||
| Obtusane | ||||
| Okamurene A-E | ||||
| Omaezallene | ||||
| Oxachamigrene | ||||
| Palisadin A-C | ||||
| Pannosane | ||||
| Pannosanol | ||||
| Perforatone | ||||
| Perforenol | ||||
| Prepacifenol | ||||
| Prelaureatin | ||||
| Rhodophytin | ||||
| Scopariol | ||||
| Seco-Laurokamurone | ||||
| Spirolaurenone | ||||
| Trans-Deacetylkumausyne | ||||
| Trans-Kumausyne | ||||
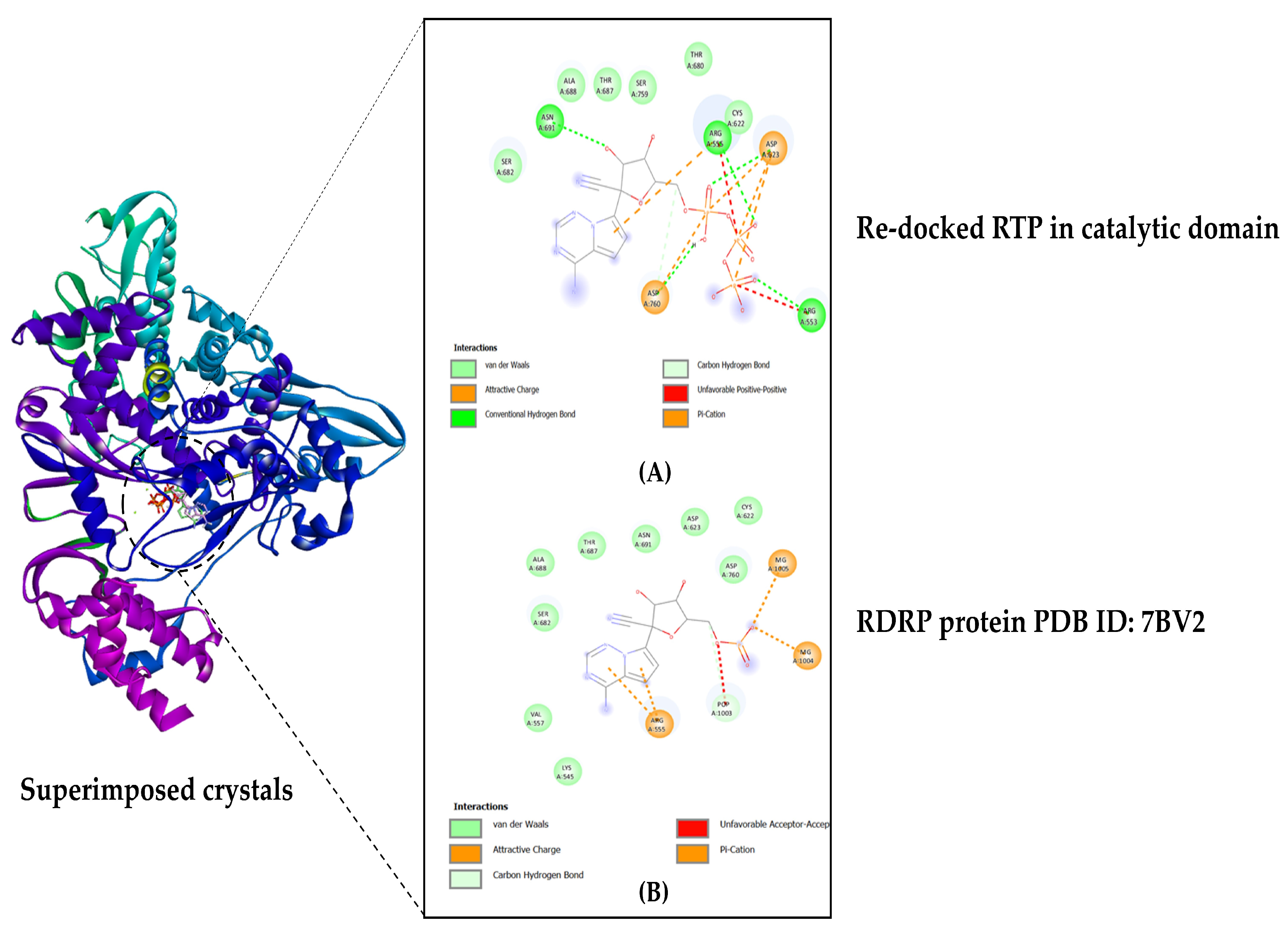
3.1.2. Validation of RdRp Catalytic Domain Docking Protocol
3.1.3. Docking Catalytic Domain (RdRp): C-Terminal
3.1.4. Docking nsp15
| Categorization of Ligands—nsp15 | ||||
|---|---|---|---|---|
| Low | Moderate | High | ||
| −4.0 to −4.9 kcal/mol | −5 to −5.9 kcal/mol | −6 to −6.9 kcal/mol | −7 to −7.9 kcal/mol | −8.0 kcal/mol and Above |
| 9-Octadecanoic acid | 1-Methyl-2,3,5-tribromoindole | (−)-3-(E)-Bromomethylidene-10-beta-bromo-beta-chamigrene | 2-Hydroxyluzofuranone A | 5-alpha-Cholestane-3,6-dione |
| 14-Methylpentadecanoic acid | 2-Bromospironippol | (+)-3-(Z)-Bromomethylidene-10-beta-bromo-beta-chamigrene | 3-alpha-Hydroperoxy-3-epiaplysin | 6-hydroxycholest-4-en-3-one |
| Bosseopentanoic acid | 3,4-epoxypalisadin B | (5S)-5-Acetoxy-beta-bisabolene | 5-alpha-Hydroxyaplysistatin | 15-Dehydroxythyrsenol A |
| Halomon | 3-alpha-Hydroxydebromoaplysin | (6R,9R,10S)-10-Bromo-9-hydroxychamigra-2,7(14)-diene | 10-epi-Dehydrothyrsiferol | 16Hydroxydehydrothyrsiferol |
| Hordenine | 4-Hydroxy-1,8-epi-isotenerone | (10R)-10-Bromo-alpha-chamigrene | 11,14-Dihydroaplysia-5,11,14,15-tetrol | Brassicasterol |
| Octadecanedioic acid | 5-Acetoxyoxachamigrene | 1,2-Dehydro-3,4-epoxypalisadin B | 13-Hydroxyprethyrsenol A | beta-sitosterol |
| Tiomanene | 5-Acetoxypalisadin B | 2-Hydroxyluzofuranone B | 15,16-Dehydrovenustatriol | Bromophycoic acid B |
| 6,8-Cycloeudesmane | 3,7-dihydroxydihydrolaurene | Aplysistatin | Bromophycoic acid C | |
| 7-Acetyl-aplysiol | 3-beta-Hydroperoxyaplysin | Beta cryptoxanthin | Bromophycolide G | |
| 9-Deoxyelatol | 3-Bromo-4,5-dihydroxybenzaldehyde | Bromophycoic acid A, D, E | Bromophycolide K | |
| 10-Bromo-beta-chamigrene | 3-epi-PerforenoneA | Bromophycolide A–F, H, I, J, M, N, R | Bromophycolide L | |
| Aristolan-8-en-1-one | 3R,4S-Luzonolone | Barekoxide | Bromophycolide O | |
| Aristolan-10-ol-9-one | 3S, 4R-Luzonolone | Bromophycolide S | Bromophycolide P | |
| Aristolane | (3Z)-Laurenyne | Callophycoic acid A, C, D–F, H, I | Bromophycolide Q | |
| Almadioxide | 3-Bromobarekoxide | Cholest-5-en-3-alpha-ol | Bromophycolide T | |
| Aromadendrene | 4-Hydroxypalisidin C | Dehydrovenustatriol | Bromophycolide U | |
| Aplysiol-7-one | 5-epi-Maneolactone | Isodehydrothyrsiferol | Callicladol | |
| Aplysiolic acid | 7-Hydroxylaurene | Lactodehydrothyrsiferol | Callophycoic acid B | |
| beta-Synderol | 8,10-Dibromoisoaplysin | Neurymenolide B | Campesterol | |
| Brasilenol | 9-Hydroxy-3-epi-perforenone A | Predehydrovenustatriolacetate | Cholest-4-en-3,6-dione | |
| (−)-BisezakyneA | 10-Acetoxyangasiol | Prethyrsenol A | Cholest-4-en-3-one | |
| Callenzanol | 10-Bromosoaplyin | Pseudodehydrothyrsiferol | Cholest-5-en-3-beta-ol | |
| Chamigrene epoxide | 10-Hydroxyaplysin | Saringosterol | Dehydrothyrsiferol | |
| Chinzallene | 10-hydroxyisolaurene | Laurebiphenyl | ||
| Compositacin A, D-H, L, M, N | 12-hydroxy Isolaurene | Lithothamin A | ||
| Cycloelatanene A, B | 12-Hydroxypalisadin B | Neurymenolide A | ||
| Cycloeudesmol | 15-hydroxypalisadin A | Stigmasterol | ||
| Cyclolaurenol | 15-Hydroxylaurene | Thyrsenol A | ||
| Dactylyne | 15-Oxolaurene | Thyrsenol B | ||
| Debromoepiaplysinol | Acetylmajapolene A | Thyrsiferol | ||
| Dendroidiol | Acetylmajapolene B | |||
| Dendroidone | Aldingenin A-D | |||
| Deoxyprepacifenol | Allolaurinterol | |||
| Deschloroelatol | Allolaurinterolacetate | |||
| Elatol | Aplysinol | |||
| Epibrasilenol | Axinysone B | |||
| Elatenyne | Aplysiodiol | |||
| Floridoside | Bromocyclococanol | |||
| Guimarediol | Bromlaurenidificin | |||
| Heterocladol | Bromocuparene | |||
| Isolaurenidificin | Callophycoic acid G, J | |||
| Isoafricanol | Callophycol A, B | |||
| Isoaplysin | Chamigrene Lactone | |||
| Isodactyloxene A | Compositacin B, C, I, J, K | |||
| Isoobtusol | Cycloisoallolaurinterol | |||
| Isopalisol | Chlorofucin | |||
| Isorigidol | Cupalaurenol | |||
| Intricenyne | Caespitane | |||
| Isolaurallene | Caespitol | |||
| Itomanindole A | Caesspitenone | |||
| Kumausallene | Debromoisocalenzanol | |||
| Laurecomin B, C | Debromolaurinterol | |||
| Laurenokomarin | Debromolaurinterolacetate | |||
| Luzondiol | Debromoaplysin | |||
| Luzonensin | Epiaplysinol | |||
| Luzonensol | Filiformin | |||
| Luzonensolacetate | Filiforminol | |||
| Laurallene | Isoallolaurinterol | |||
| Laureacetal B, C | Isodebromolaurinterol | |||
| Laurefurenyne A, B, D, E, F | Isolaurenisol | |||
| Laurencenyne | Isocaespitol | |||
| Laurencial | Isodihydrolaurene | |||
| Laurendecumallene A, B | Isolaureatin | |||
| Laurendecumenyne B | Isolaurene | |||
| Laurenenyne | Japonenyne A | |||
| Laurenisol | Johnstonol | |||
| Laureoxanyne | Laurecomin A, D | |||
| Laurepinnacin | Laurencomposidiene | |||
| Ma’iliohydrin | Laurene | |||
| Mailione | Laurentristich-4-ol | |||
| Microcladallene A-C | Laureperoxide | |||
| Neoisoprelaurefucin | Laurepoxyene | |||
| Neolaurallene | Laurinterolacetate | |||
| Nidificene | Laurokamurene A-D | |||
| Obtusane | Luzofuran | |||
| Okamurene C, E | Luzonenone | |||
| Omaezallene | Laurinterol | |||
| Pacifenol | Laureacetal A | |||
| Palisadin B, C, D | Laurefurenyne C | |||
| Pannosane | Laurendecumenyne A | |||
| Perforatone | Laurenone A | |||
| Perforenol | Majapolene A, B | |||
| Prelaureatin | Mammeisin | |||
| Rhodophytin | Neoirietetraol | |||
| Scopariol | Notoryne | |||
| trans-Deacetylkumausyne | Okamurene A, B, D | |||
| trans-Kumausyne | Oxachamigrene | |||
| trans-Laurencenyne | Oryzalexin S | |||
| Palisadin A | ||||
| Pannosanol | ||||
| Perforenone A | ||||
| Prepacifenol | ||||
| Seco-Laurokamurone | ||||
| Spirolaurenone | ||||
3.2. Selection of Best Interaction Complexes—RdRp and nsp15
3.2.1. Selection Criteria
- It must interact with the majority of catalytic amino acids;
- It must form many hydrogen bonds with catalytic amino acids (RMSD ≤ 3.00);
- For electrostatic connections, the pre-specified threshold RMSD cutoff distance (Å ≤ 5.00) must not be exceeded.
3.2.2. Rejection Criteria
- No interactions with key amino acids;
- No hydrogen bonds are connected to important residues or exceeded RMSD ≥ 3.00;
- Mostly Van der Waals forces are involved in the interactions;
- For electrostatic bonding, the maximum pre-defined RMSD cutoff distance (Å ≤ 5.00) was surpassed.
3.3. Best Interaction Complexes—RdRp and nsp15
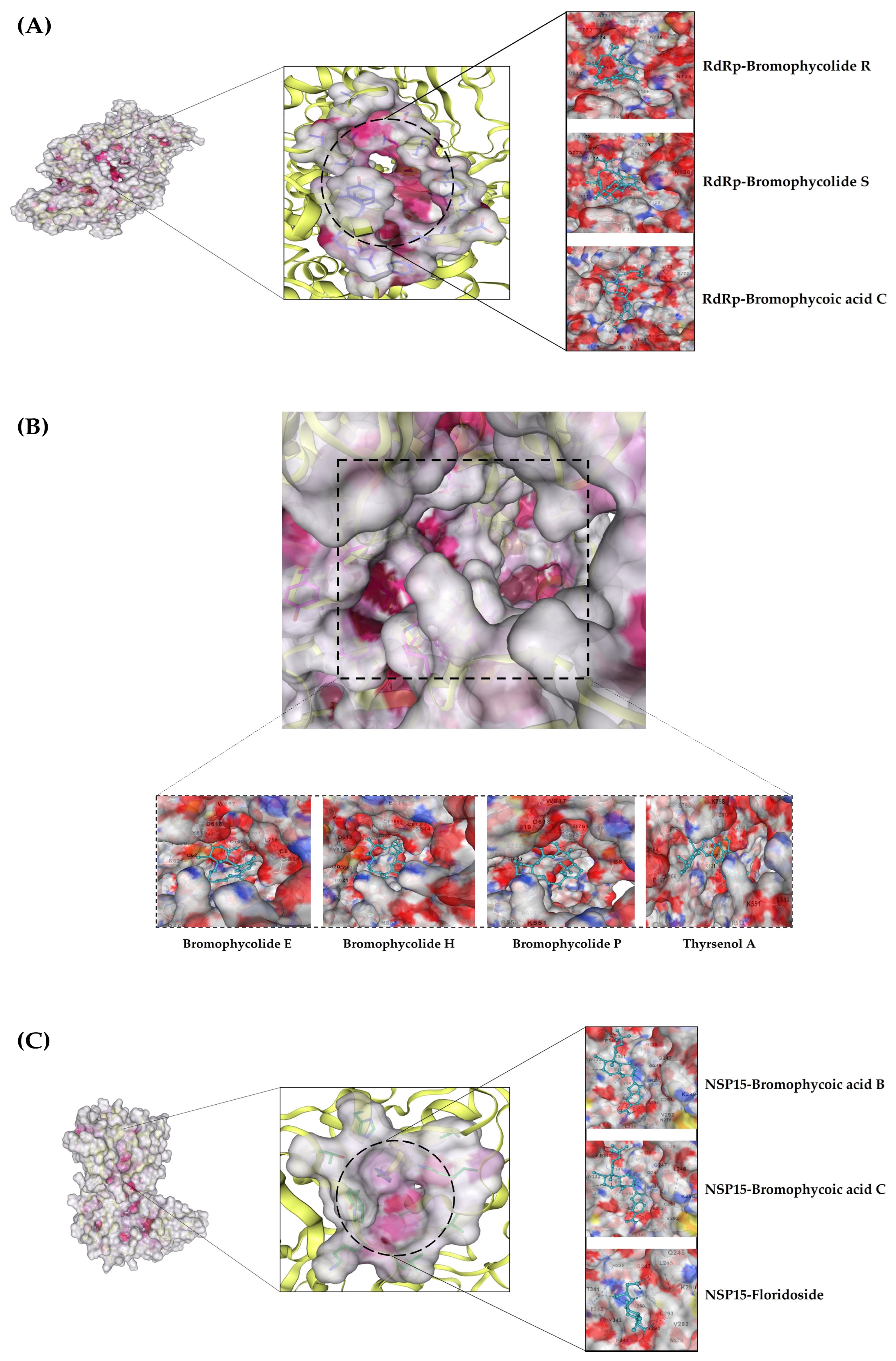
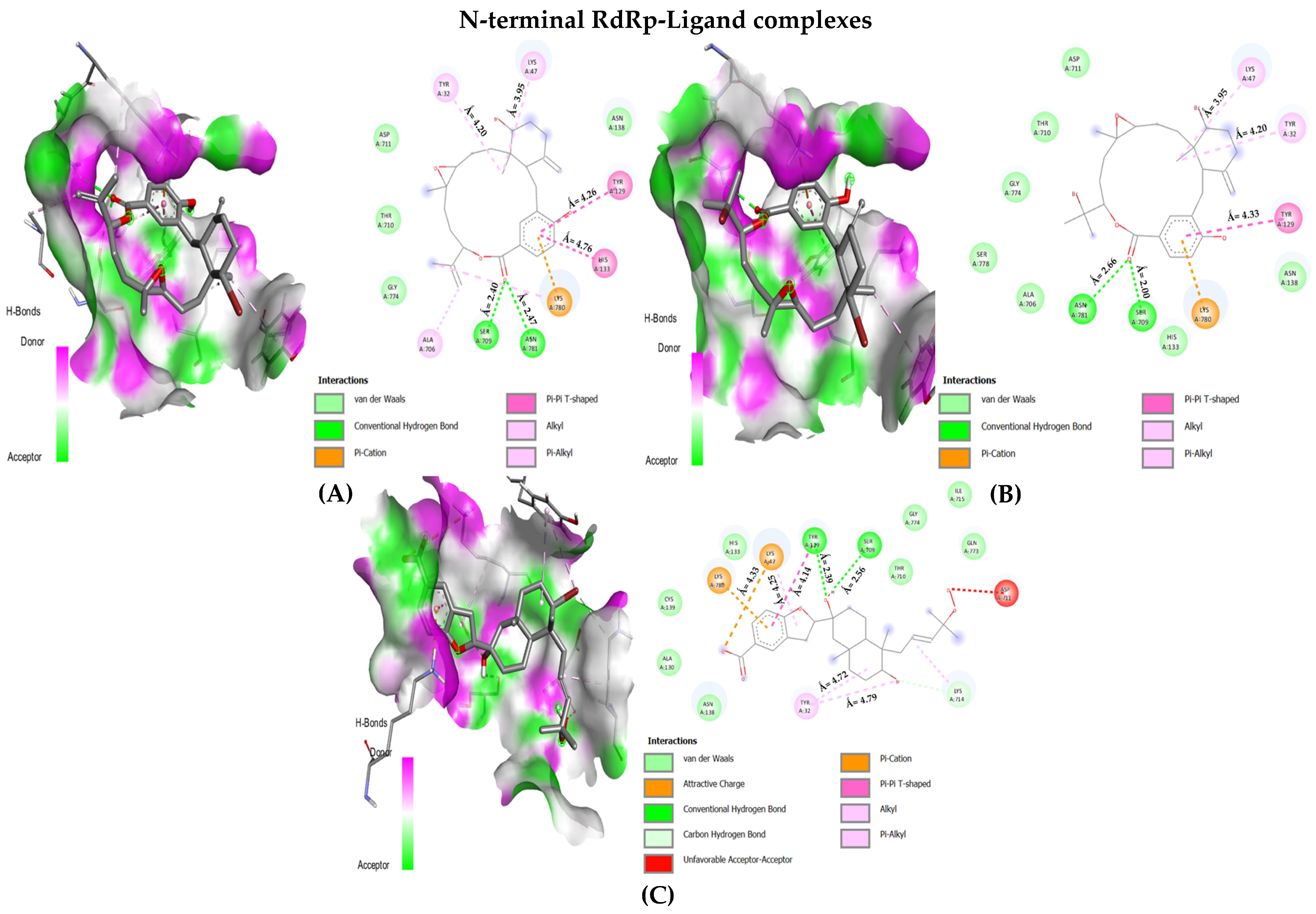
| Favourable Residue Interactions | |
|---|---|
| Filtered Ligands | Involved Amino Acids |
| RdRp (N-Terminal β-Hairpin Motif) | |
| Bromophycolide R | TYR32 LYS47 TYR129 HIS133 ASN138 ALA706 SER709 THR710 ASP711 GLY774 LYS780 ASN781 |
| Bromophycolide S | TYR32 LYS47 TYR129 HIS133 ASN138 ALA706 SER709 THR710 ASP711 GLY774 SER778 LYS780 ASN781 |
| Bromophycoic acid C | TYR32 LYS47 TYR129 ALA130 HIS133 ASN138 CYS139 SER709 THR710 ASP711 LYS714 ILE715 GLN773 GLY774 LYS780 |
| RdRp (C-terminal catalytic core) | |
| Bromophycolide E | ARG553 ARG555 ASP618 TYR619 PRO620 CYS622 ASP623 THR680 SER682 THR687 ALA688 ASN691 LEU758 SER759 ASP760 ASP761 CYS813 |
| Bromophycolide H | ARG553 ARG555 ASP618 LYS621 CYS622 ASP623 LEU758 SER759 ASP760 ASP761 CYS813 |
| Bromophycolide P | ARG553 ARG555 ASP618 TYR619 PRO620 LYS621 CYS622 ASP623 ASP760 ASP761 SER814 |
| Thyrsenol A | ARG555 ASP618 TYR619 PRO620 LYS621 CYS622 ASP623 THR687 ALA688 ASN691 LEU758 SER759 ASP760 ASP761 CYS813 |
| nsp15 enzyme | |
| Bromophycoic acid B | HIS235 GLY247 GLY248 HIS250 LYS290 VAL292 CYS293 SER294 TRP333 GLU340 THR341 TYR343 PRO344 LYS345 LEU346 |
| Bromophycoic acid C | HIS235 GLY247 GLY248 HIS250 LYS290 VAL292 CYS293 SER294 TRP333 GLU340 THR341 TYR343 PRO344 LYS345 LEU346 |
| Floridoside | HIS235 GLN245 LEU246 GLY247 GLY248 HIS250 LYS290 VAL292 CYS293 SER294 THR341 PHE342 TYR343 |


3.4. MD Simulations
MD Simulations for Selected RdRp and nsp15 Complexes
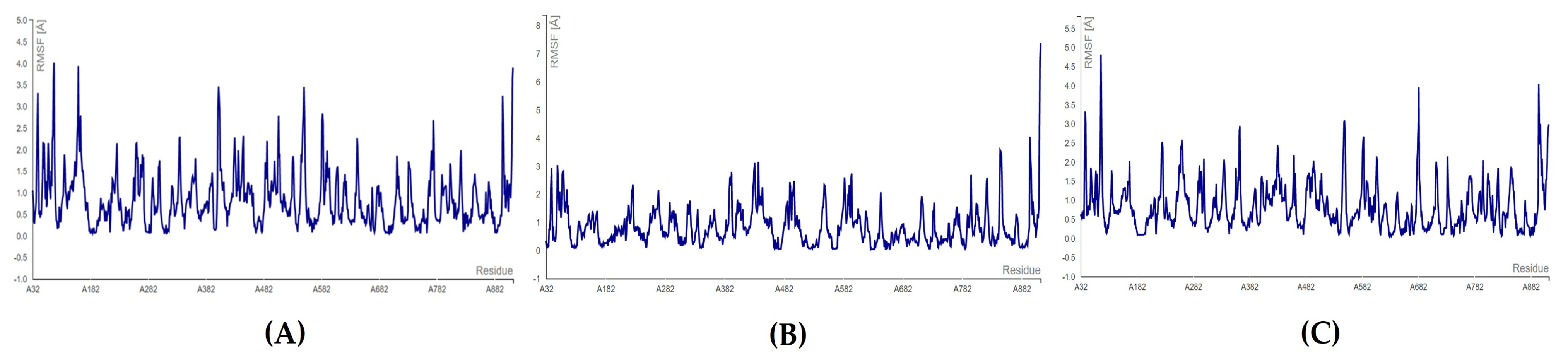
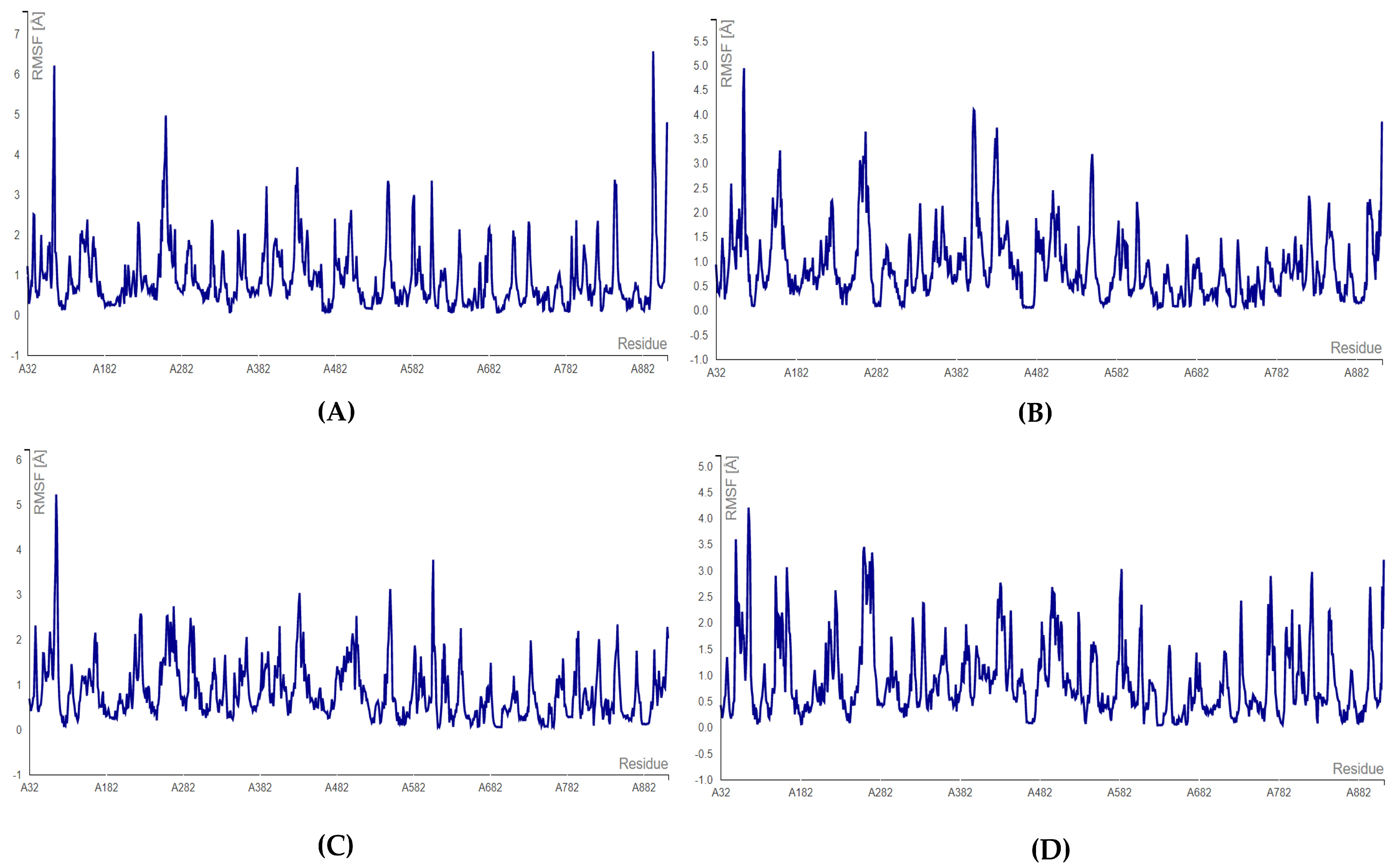
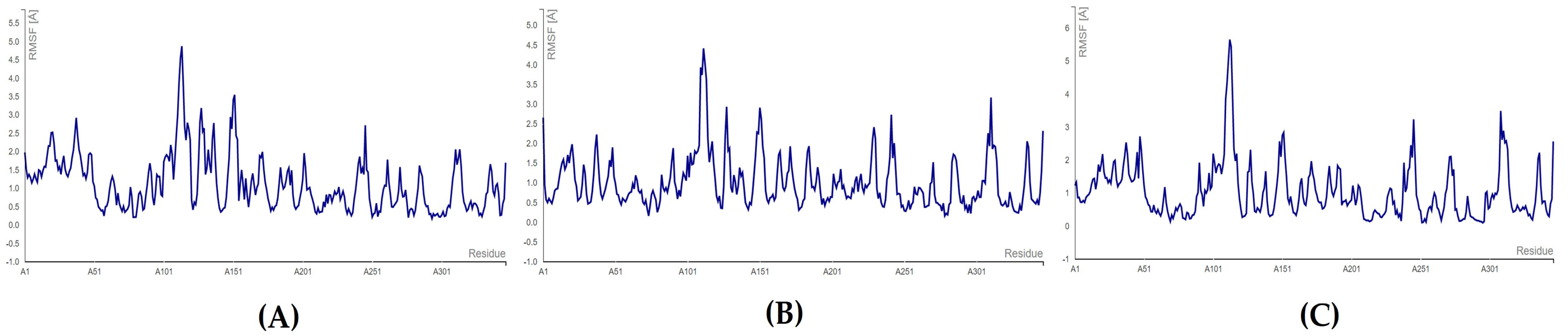
3.5. Toxicity Evaluation
3.5.1. ProTox-II Report
| ProTox-II Toxicity | ||||||
|---|---|---|---|---|---|---|
| Top Ligands | Toxicity Values | Probability | ||||
| LD50 (mg/kg) | Toxicity Class | Hepatotoxicity | Carcinogenicity | Immunotoxicity | Mutagenicity | |
| Bromophycoic acid B | 12 | 2 | Inactive | Inactive | Inactive | Inactive |
| Bromophycoic acid C | 12 | 2 | Inactive | Inactive | Inactive | Inactive |
| Bromophycolide E | 1000 | 4 | Inactive | Inactive | Active | Inactive |
| Bromophycolide H | 1000 | 4 | Inactive | Inactive | Active | Inactive |
| Bromophycolide P | 1000 | 4 | Inactive | Inactive | Active | Inactive |
| Bromophycolide R | 1000 | 4 | Inactive | Inactive | Active | Inactive |
| Bromophycolide S | 1000 | 4 | Inactive | Inactive | Active | Inactive |
| Floridoside | 23,000 | 6 | Inactive | Inactive | Inactive | Inactive |
| Thyrsenol A | 7 | 2 | Inactive | Inactive | Active | Inactive |
3.5.2. StopTox Report
| StopTox Acute Toxicity | |||||
|---|---|---|---|---|---|
| Top Ligands | Endpoints | ||||
| Inhalation | Oral | Dermal | Irritation and Corrosion | Skin Sensitization | |
| Bromophycoic acid B | Non-Toxic | Toxic | Non-Toxic | Eyes (-) Skin (-) | Non-sensitizer |
| Bromophycoic acid C | Non-Toxic | Toxic | Non-Toxic | Eyes (-) Skin (-) | Non-sensitizer |
| Bromophycolide E | Non-Toxic | Toxic | Non-Toxic | Eyes (-) Skin (-) | Sensitizer |
| Bromophycolide H | Non-Toxic | Non-Toxic | Non-Toxic | Eyes (-) Skin (-) | Sensitizer |
| Bromophycolide P | Non-Toxic | Non-Toxic | Non-Toxic | Eyes (-) Skin (-) | Sensitizer |
| Bromophycolide R | Toxic | Non-Toxic | Non-Toxic | Eyes (-) Skin (-) | Sensitizer |
| Bromophycolide S | Toxic | Non-Toxic | Non-Toxic | Eyes (-) Skin (-) | Sensitizer |
| Floridoside | Non-Toxic | Non-Toxic | Non-Toxic | Eyes (-) Skin (-) | Non-sensitizer |
| Thyrsenol A | Non-toxic | Toxic | Non-toxic | Eyes (-) Skin (-) | Non-sensitizer |
3.6. Pharmacokinetic Studies
3.6.1. Lipinski Framework (Ro5)
| Drug-Likeness Assessment | ||||||
|---|---|---|---|---|---|---|
| Top Ligands | Mol. Weight (g/mol) MW ≤ 500 | Rotatable Bonds RB ≤ 10 | H Bond Acceptors HBA ≤ 10 | H Bond Donors HBD ≤ 5 | C Log p Log p ≤ 5 | TPSA (Å2) ≤ 140 |
| Bromophycoic acid B | 521.48 | 5 | 5 | 3 | 4.78 | 86.99 |
| Bromophycoic acid C | 537.48 | 6 | 6 | 3 | 4.77 | 96.22 |
| Bromophycolide E | 584.38 | 1 | 4 | 2 | 5.79 | 66.76 |
| Bromophycolide H | 665.29 | 0 | 4 | 2 | 6.10 | 66.76 |
| Bromophycolide P | 584.38 | 0 | 4 | 1 | 5.83 | 55.76 |
| Bromophycolide R | 503.47 | 1 | 4 | 1 | 5.54 | 59.06 |
| Bromophycolide S | 584.38 | 1 | 4 | 1 | 5.87 | 59.06 |
| Floridoside | 254.23 | 5 | 8 | 6 | −2.53 | 139.84 |
| Thyrsenol A | 619.63 | 8 | 8 | 4 | 3.51 | 117.84 |
3.6.2. Swiss-ADME
| Swiss-ADME Output | |||||
|---|---|---|---|---|---|
| Top Ligands | Water Solubility | Bioavailability | GI Absorption | Absorption (%) | BBB Permeant |
| Bromophycoic acid B | Moderate | 0.56 | High | 78.98 | No |
| Bromophycoic acid C | Moderate | 0.56 | Low | 75.80 | No |
| Bromophycolide E | Poor | 0.17 | Low | 85.96 | No |
| Bromophycolide H | Poor | 0.17 | Low | 85.96 | No |
| Bromophycolide P | Poor | 0.17 | Low | 89.76 | No |
| Bromophycolide R | Poor | 0.17 | High | 88.62 | No |
| Bromophycolide S | Poor | 0.17 | Low | 88.62 | No |
| Floridoside | Soluble | 0.55 | Low | 60.75 | No |
| Thyrsenol A | Moderate | 0.55 | High | 68.34 | No |
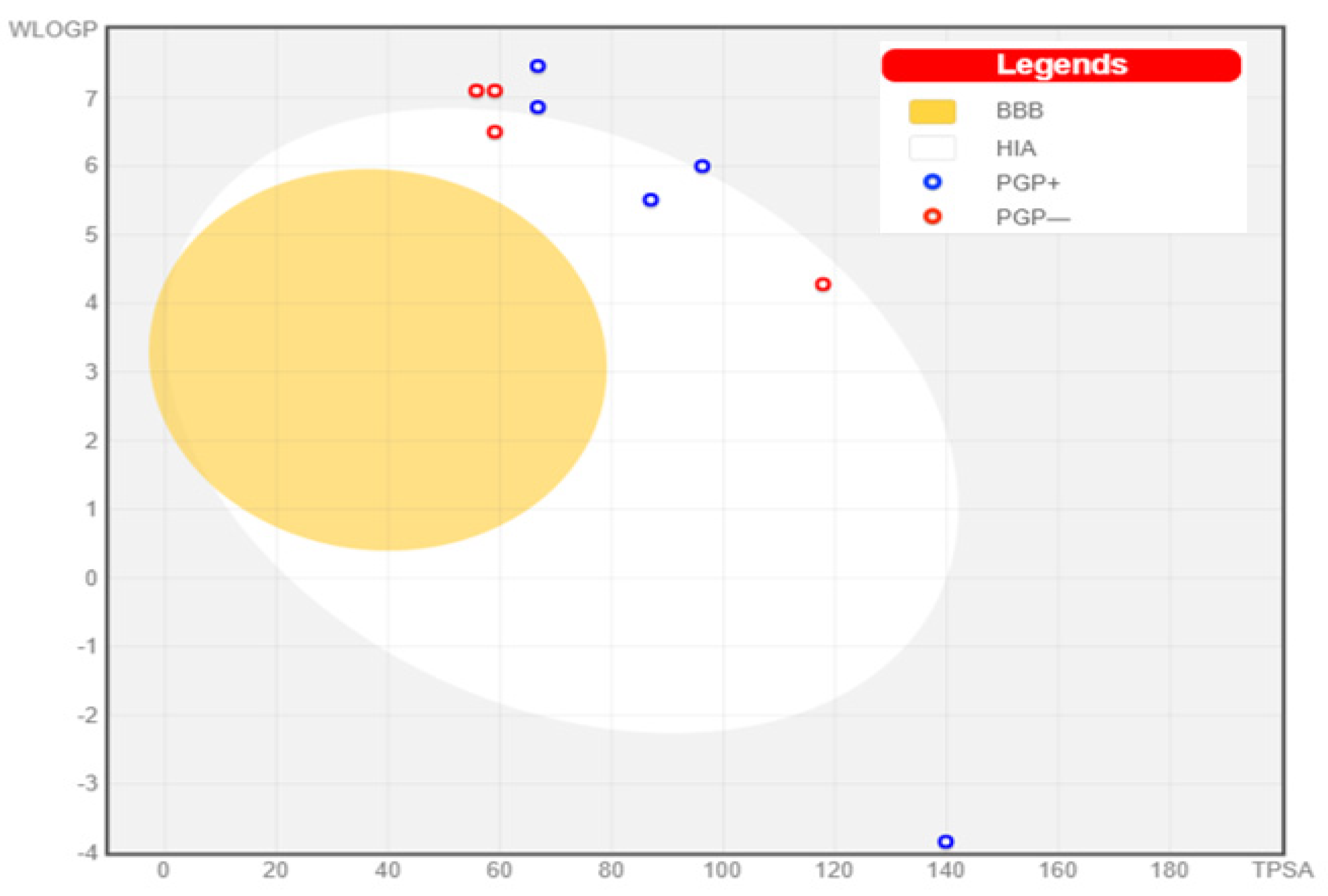
3.6.3. Bioactivity Evaluation
| PASS Online Program | |
|---|---|
| Top Ligands | Potential Bioactivity |
| Bromophycoic acid B | Antifungal, Histidine kinase inhibitor, Beta glucuronidase inhibitor, Antibacterial, Antioxidant, Antineoplastic, Interferon antagonist, Interferon gamma antagonist, Transcription factor NF kappa B stimulant, Immunostimulant |
| Bromophycoic acid C | Antifungal, Antiviral, Antioxidant, Anticancer, Antineoplastic, Beta glucuronidase inhibitor, Transcription factor NF kappa B stimulant |
| Bromophycolide E | Antineoplastic, Antibiotic, Antiviral, Antifungal, Beta glucuronidase inhibitor, Histidine kinase inhibitor, MMP9 expression inhibitor, Immunosuppressant, Respiratory analeptic, Antibacterial, Cytokine release inhibitor, Interleukin 10 antagonist, 1,3-Beta-glucan synthase inhibitor, Interferon gamma antagonist |
| Bromophycolide H | Antineoplastic, MMP9 expression inhibitor, Antifungal, Antibacterial, Antibiotic, Histidine kinase inhibitor, Cytokine release inhibitor, Interferon gamma antagonist, Interleukin 10 antagonist, 1,3-Beta-glucan synthase inhibitor |
| Bromophycolide P | Antineoplastic, Respiratory analeptic, Antibacterial, Antibiotic, Antifungal, MMP9 expression inhibitor, Beta glucuronidase inhibitor, Histidine kinase inhibitor, 1,3-Beta-glucan synthase inhibitor, Immunosuppressant, Interferon gamma antagonist, Cytokine release inhibitor, Expectorant, |
| Bromophycolide R | Antiviral, Antineoplastic alkaloid, Anticancer, Antifungal, Antibiotic, Antibacterial, Immunosuppressant, Transcription factor NF kappa B stimulant, Beta glucuronidase inhibitor, Histidine kinase inhibitor, 1,3-Beta-glucan synthase inhibitor, Respiratory analeptic, Anti-inflammatory, Cytokine release inhibitor, Interferon gamma antagonist, and an Interleukin 10 antagonist. |
| Bromophycolide S | Antiviral, Antineoplastic, Antibiotic, Anticancer, Antifungal, Immunosuppressant, Antibacterial, Transcription factor NF kappa B inhibitor, Histidine kinase inhibitor, Beta glucuronidase inhibitor, 1,3-Beta-glucan synthase inhibitor, Cytokine release inhibitor and an Interleukin 10 antagonist. |
| Floridoside | Antiviral, Anti-parasitic, Antifungal, Antifungal enhancer, Antibacterial, Anti-tuberculosic, Anticancer, Anti-infective, Antioxidant, Free radical scavenger, Anti-diabetic, Antineoplastic, Immunostimulant, Immunomodulator, Macrophage stimulant, Macrophage colony stimulating factor agonist, Respiratory analeptic, Transcription factor NF kappa B stimulant, Histamine release stimulant, Beta glucuronidase inhibitor, 1,3-Beta-glucan synthase inhibitor, Rhizopuspepsin inhibitor, GRP78 expression inhibitor, Mucolytic, Expectorant, Anti-inflammatory, Histamine release inhibitor, JAK2 expression inhibitor, Severe acute respiratory syndrome treatment, RdRp Inhibitor, Interferon gamma antagonist, Respiratory distress syndrome treatment, Cytokine release inhibitor, TNF expression inhibitor, Interleukin 2, 10, 12 agonist, Interleukin 1a, 4, 6 antagonist. |
| Thyrsenol A | Antineoplastic, Antifungal, Antiviral, Antiinflammatory, Antibacterial, Antibiotic |
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Puranik, A.; Lenehan, P.J.; Silvert, E.; Niesen, M.J.M.; Corchado-Garcia, J.; O’horo, J.C.; Virk, A.; Swift, M.D.; Halamka, J.; Badley, A.D.; et al. Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of Alpha and Delta variant prevalence. medRxiv, 2021; preprint. [Google Scholar] [CrossRef]
- Callaway, E. COVID drug drives viral mutations—And now some want to halt its use. Nature 2023, 614, 399. [Google Scholar] [CrossRef]
- Phillips, N. The coronavirus is here to stay—here’s what that means. Nature 2021, 590, 382–384. [Google Scholar] [CrossRef]
- Min, J.S.; Kwon, S.; Jin, Y.-H. SARS-CoV-2 RdRp Inhibitors Selected from a Cell-Based SARS-CoV-2 RdRp Activity Assay System. Biomedicines 2021, 9, 996. [Google Scholar] [CrossRef]
- Ahn, D.-G.; Choi, J.-K.; Taylor, D.R.; Oh, J.-W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012, 157, 2095–2104. [Google Scholar] [CrossRef]
- De Farias, S.T.; Dos Santos, A.P.D.S., Jr.; Rêgo, T.G.D.; José, M.V. Origin and Evolution of RNA-Dependent RNA Polymerase. Front. Genet. 2017, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Jockusch, S.; Tao, C.; Li, X.; Chien, M.; Kumar, S.; Morozova, I.; Kalachikov, S.; Russo, J.J.; Ju, J. Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci. Rep. 2020, 10, 16577. [Google Scholar] [CrossRef]
- Chien, M.; Anderson, T.K.; Jockusch, S.; Tao, C.; Li, X.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. [Google Scholar] [CrossRef]
- Ju, J.; Li, X.; Kumar, S.; Jockusch, S.; Chien, M.; Tao, C.; Morozova, I.; Kalachikov, S.; Kirchdoerfer, R.N.; Russo, J.J. Nucleotide analogues as inhibitors of SARS-CoV Polymerase. Pharmacol. Res. Perspect. 2020, 8, e00674. [Google Scholar] [CrossRef] [PubMed]
- Aftab, S.O.; Ghouri, M.Z.; Masood, M.U.; Haider, Z.; Khan, Z.; Ahmad, A.; Munawar, N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020, 18, 275. [Google Scholar] [CrossRef]
- Ahmad, J.; Ikram, S.; Ahmad, F.; Rehman, I.U.; Mushtaq, M. SARS-CoV-2 RNA Dependent RNA polymerase (RdRp)—A drug repurposing study. Heliyon 2020, 6, e04502. [Google Scholar] [CrossRef]
- Aktaş, A.; Tüzün, B.; Aslan, R.; Sayin, K.; Ataseven, H. New anti-viral drugs for the treatment of COVID-19 instead of favipiravir. J. Biomol. Struct. Dyn. 2020, 39, 7263–7273. [Google Scholar] [CrossRef]
- Alexpandi, R.; De Mesquita, J.F.; Pandian, S.K.; Ravi, A.V. Quinolines-Based SARS-CoV-2 3CLpro and RdRp Inhibitors and Spike-RBD-ACE2 Inhibitor for Drug-Repurposing Against COVID-19: An in silico Analysis. Front. Microbiol. 2020, 11, 1796. [Google Scholar] [CrossRef] [PubMed]
- Al-Masoudi, N.A.; Elias, R.S.; Saeed, B. Molecular Docking Studies of some Antiviral and Antimalarial Drugs Via Bindings to 3CL-Protease and Polymerase Enzymes of the Novel Coronavirus (SARS-CoV-2). Biointerface Res. Appl. Chem. 2020, 10, 6444–6459. [Google Scholar] [CrossRef]
- Ao, S.; Han, D.; Sun, L.; Wu, Y.; Liu, S.; Huang, Y. Identification of Potential Key Agents for Targeting RNA-Dependent RNA Polymerase of SARS-CoV-2 by Integrated Analysis and Virtual Drug Screening. Front. Genet. 2020, 11, 581668. [Google Scholar] [CrossRef] [PubMed]
- Calligari, P.; Bobone, S.; Ricci, G.; Bocedi, A. Molecular Investigation of SARS–CoV-2 Proteins and Their Interactions with Antiviral Drugs. Viruses 2020, 12, 445. [Google Scholar] [CrossRef]
- Da Silva, F.M.A.; Da Silva, K.P.A.; De Oliveira, L.P.M.; Costa, E.V.; Koolen, H.H.; Pinheiro, M.L.B.; De Souza, A.Q.L.; De Souza, A.D.L. Flavonoid glycosides and their putative human metabolites as potential inhibitors of the SARS-CoV-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp). Mem. Instit. Oswaldo Cruz. 2020, 115, e200207. [Google Scholar] [CrossRef]
- da Silva, J.K.R.; Figueiredo, P.L.B.; Byler, K.G.; Setzer, W.N. Essential Oils as Antiviral Agents, Potential of Essential Oils to Treat SARS-CoV-2 Infection: An In-Silico Investigation. Int. J. Mol. Sci. 2020, 21, 3426. [Google Scholar] [CrossRef]
- Elfiky, A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020, 248, 117477. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. J. Biomol. Struct. Dyn. 2021, 39, 3204–3212. [Google Scholar] [CrossRef] [PubMed]
- Indu, P.; Rameshkumar, M.R.; Arunagirinathan, N.; Al-Dhabi, N.A.; Arasu, M.V.; Ignacimuthu, S. Raltegravir, Indinavir, Tipranavir, Dolutegravir, and Etravirine against main protease and RNA-dependent RNA polymerase of SARS-CoV-2: A molecular docking and drug repurposing approach. J. Infect. Public. Health 2020, 13, 1856–1861. [Google Scholar] [CrossRef] [PubMed]
- Kandeel, M.; Kitade, Y.; Almubarak, A. Repurposing FDA-approved phytomedicines, natural products, antivirals and cell protectives against SARS-CoV-2 (COVID-19) RNA-dependent RNA polymerase. PeerJ 2020, 8, e10480. [Google Scholar] [CrossRef]
- Mohamed, T.A.; Elshamy, A.I.; Ibrahim, M.A.A.; Zellagui, A.; Moustafa, M.F.; Abdelrahman, A.H.M.; Ohta, S.; Pare, P.W.; Hegazy, M.-E.F. Carotane sesquiterpenes from Ferula vesceritensis: In silico analysis as SARS-CoV-2 binding inhibitors. RSC Adv. 2020, 10, 34541–34548. [Google Scholar] [CrossRef]
- Parvez, M.S.A.; Karim, M.A.; Hasan, M.; Jaman, J.; Karim, Z.; Tahsin, T.; Hasan, M.N.; Hosen, M.J. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int. J. Biol. Macromol. 2020, 163, 1787–1797. [Google Scholar] [CrossRef]
- Pokhrel, R.; Chapagain, P.; Siltberg-Liberles, J. Potential RNA-dependent RNA polymerase inhibitors as prospective therapeutics against SARS-CoV-2. J. Med. Microbiol. 2020, 69, 864–873. [Google Scholar] [CrossRef]
- Ruan, Z.; Liu, C.; Guo, Y.; He, Z.; Huang, X.; Jia, X.; Yang, T. SARS-CoV-2 and SARS-CoV: Virtual screening of potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12). J. Med. Virol. 2021, 93, 389–400. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Gordon, C.J.; Woolner, E.; Kocinkova, D.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020, 295, 16156. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Ahmed, S.; Mahtarin, R.; Ahmed, S.S.; Akter, S.; Islam, S.; Al Mamun, A.; Islam, R.; Hossain, N.; Ali, A.; Sultana, M.U.C.; et al. Investigating the binding affinity, interaction, and structure-activity-relationship of 76 prescription antiviral drugs targeting RdRp and Mpro of SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 39, 6290–6305. [Google Scholar] [CrossRef] [PubMed]
- Barage, S.; Karthic, A.; Bavi, R.; Desai, N.; Kumar, R.; Kumar, V.; Lee, K.W. Identification and characterization of novel RdRp and Nsp15 inhibitors for SARS-COV2 using computational approach. J. Biomol. Struct. Dyn. 2022, 40, 2557–2574. [Google Scholar] [CrossRef] [PubMed]
- Borquaye, L.S.; Gasu, E.N.; Ampomah, G.B.; Kyei, L.K.; Amarh, M.A.; Mensah, C.N.; Nartey, D.; Commodore, M.; Adomako, A.K.; Acheampong, P.; et al. Alkaloids from Cryptolepis sanguinolenta as Potential Inhibitors of SARS-CoV-2 Viral Proteins: An In Silico Study. BioMed Res. Int. 2020, 2020, 532–560. [Google Scholar] [CrossRef] [PubMed]
- Dwarka, D.; Agoni, C.; Mellem, J.J.; Soliman, M.E.; Baijnath, H. Identification of potential SARS-CoV-2 inhibitors from South African medicinal plant extracts using molecular modelling approaches. S. Afr. J. Bot. 2020, 133, 273–284. [Google Scholar] [CrossRef] [PubMed]
- El Hassab, M.A.; Shoun, A.A.; Al-Rashood, S.T.; Al-Warhi, T.; Eldehna, W.M. Identification of a New Potential SARS-COV-2 RNA-Dependent RNA Polymerase Inhibitor via Combining Fragment-Based Drug Design, Docking, Molecular Dynamics, and MM-PBSA Calculations. Front. Chem. 2020, 8, 584894. [Google Scholar] [CrossRef] [PubMed]
- Gul, S.; Ozcan, O.; Asar, S.; Okyar, A.; Barıs, I.; Kavakli, I.H. In silico identification of widely used and well-tolerated drugs as potential SARS-CoV-2 3C-like protease and viral RNA-dependent RNA polymerase inhibitors for direct use in clinical trials. J. Biomol. Struct. Dyn. 2021, 39, 6772–6791. [Google Scholar] [CrossRef]
- Gutierrez-Villagomez, J.M.; Campos-García, T.; Molina-Torres, J.; López, M.G.; Vázquez-Martínez, J. Alkamides and Piperamides as Potential Antivirals against the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Phys. Chem. Lett. 2020, 11, 8008–8016. [Google Scholar] [CrossRef]
- Kar, P.; Sharma, N.R.; Singh, B.; Sen, A.; Roy, A. Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: An in silico investigation. J. Biomol. Struct. Dyn. 2021, 39, 4774–4785. [Google Scholar] [CrossRef]
- Khan, A.; Khan, M.; Saleem, S.; Babar, Z.; Ali, A.; Khan, A.A.; Sardar, Z.; Hamayun, F.; Ali, S.S.; Wei, D.-Q. Phylogenetic Analysis and Structural Perspectives of RNA-Dependent RNA-Polymerase Inhibition from SARs-CoV-2 with Natural Products. Interdiscip. Sci. Comput. Life Sci. 2020, 12, 335–348. [Google Scholar] [CrossRef]
- Mutlu, O.; Ugurel, O.M.; Sariyer, E.; Ata, O.; Inci, T.G.; Ugurel, E.; Kocer, S.; Turgut-Balik, D. Targeting SARS-CoV-2 Nsp12/Nsp8 interaction interface with approved and investigational drugs: An in silico structure-based approach. J. Biomol. Struct. Dyn. 2022, 40, 918–930. [Google Scholar] [CrossRef]
- Narayanan, N.; Nair, D.T. Vitamin B12 may inhibit RNA-dependent-RNA polymerase activity of nsp12 from the SARS-CoV-2 virus. IUBMB Life 2020, 72, 2112–2120. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Oselladore, E.; Zagotto, G.; Memo, M.; Gianoncelli, A. A computational approach to drug repurposing against SARS-CoV-2 RNA dependent RNA polymerase (RdRp). J. Biomol. Struct. Dyn. 2022, 40, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Vora, J.; Patel, D.; Sinha, S.; Jha, P.C.; Shrivastava, N. Identification of natural inhibitors against prime targets of SARS-CoV-2 using molecular docking, molecular dynamics simulation and MM-PBSA approaches. J. Biomol. Struct. Dyn. 2022, 40, 3296–3311. [Google Scholar] [CrossRef]
- Singh, J.; Malik, D.; Raina, A. Computational investigation for identification of potential phytochemicals and antiviral drugs as potential inhibitors for RNA-dependent RNA polymerase of COVID-19. J. Biomol. Struct. Dyn. 2022, 40, 3492–3507. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Sk, M.F.; Sonawane, A.; Kar, P.; Sadhukhan, S. Plant-derived natural polyphenols as potential antiviral drugs against SARS-CoV-2 via RNA-dependent RNA polymerase (RdRp) inhibition: An in-silico analysis. J. Biomol. Struct. Dyn. 2021, 39, 6249–6264. [Google Scholar] [CrossRef] [PubMed]
- Elkarhat, Z.; Charoute, H.; Elkhattabi, L.; Barakat, A.; Rouba, H. Potential inhibitors of SARS-CoV-2 RNA dependent RNA polymerase protein: Molecular docking, molecular dynamics simulations and MM-PBSA analyses. J. Biomol. Struct. Dyn. 2022, 40, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Brunt, D.; Lakernick, P.M.; Wu, C. Discovering new potential inhibitors to SARS-CoV-2 RNA dependent RNA polymerase (RdRp) using high throughput virtual screening and molecular dynamics simulations. Sci. Rep. 2022, 12, 19986. [Google Scholar] [CrossRef]
- Olender, S.A.; Perez, K.K.; Go, A.S.; Balani, B.; Price-Haywood, E.G.; Shah, N.S.; Wang, S.; Walunas, T.L.; Swaminathan, S.; Slim, J.; et al. Remdesivir for Severe Coronavirus Disease 2019 (COVID-19) Versus a Cohort Receiving Standard of Care. Clin. Infect. Dis. 2021, 73, e4166–e4174. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdouh, A.; Bizanti, A.; Barbarawi, M.; Jabri, A.; Kumar, A.; Fashanu, O.E.; Khan, S.U.; Zhao, D.; Antar, A.A.; Michos, E.D. Remdesivir for the treatment of COVID-19: A systematic review and meta-analysis of randomized controlled trials. Contemp. Clin. Trials 2021, 101, 106272. [Google Scholar] [CrossRef]
- Dölken, L.; Stich, A.; Spinner, C.D. Remdesivir for Early COVID-19 Treatment of High-Risk Individuals Prior to or at Early Disease Onset—Lessons Learned. Viruses 2021, 13, 963. [Google Scholar] [CrossRef]
- Gottlieb, R.L.; Vaca, C.E.; Paredes, R.; Mera, J.; Webb, B.J.; Perez, G.; Oguchi, G.; Ryan, P.; Nielsen, B.U.; Brown, M.; et al. Early Remdesivir to Prevent Progression to Severe COVID-19 in Outpatients. N. Engl. J. Med. 2022, 386, 305–315. [Google Scholar] [CrossRef]
- Mahajan, L.; Singh, A. Gifty Clinical outcomes of using remdesivir in patients with moderate to severe COVID-19: A prospective randomised study. Indian J. Anaesth. 2021, 65 (Suppl. 1), S41–S46. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Binetti, E.; Borrazzo, C.; Cacciola, E.G.; Battistini, L.; Ceccarelli, G.; Mastroianni, C.M.; D’ettorre, G. Efficacy of Remdesivir-Containing Therapy in Hospitalized COVID-19 Patients: A Prospective Clinical Experience. J. Clin. Med. 2021, 10, 3784. [Google Scholar] [CrossRef] [PubMed]
- Yan, V.C.; Muller, F.L. Why Remdesivir Failed: Preclinical Assumptions Overestimate the Clinical Efficacy of Remdesivir for COVID-19 and Ebola. Antimicrob. Agents Chemother. 2021, 65, e0111721. [Google Scholar] [CrossRef] [PubMed]
- Ader, F.; Bouscambert-Duchamp, M.; Hites, M.; Peiffer-Smadja, N.; Poissy, J.; Belhadi, D.; Diallo, A.; Lê, M.-P.; Peytavin, G.; Staub, T.; et al. Remdesivir plus standard of care versus standard of care alone for the treatment of patients admitted to hospital with COVID-19 (DisCoVeRy): A phase 3, randomised, controlled, open-label trial. Lancet Infect. Dis. 2022, 22, 209–221. [Google Scholar] [CrossRef]
- Sun, D. Remdesivir for Treatment of COVID-19: Combination of Pulmonary and IV Administration May Offer Aditional Benefit. AAPS J. 2020, 22, 77, Erratum in AAPS J. 2020, 22, 102. [Google Scholar] [CrossRef]
- Martinez, D.R.; Schäfer, A.; Leist, S.R.; Li, D.; Gully, K.; Yount, B.; Feng, J.Y.; Bunyan, E.; Porter, D.P.; Cihlar, T.; et al. Prevention and therapy of SARS-CoV-2 and the B.1.351 variant in mice. Cell Rep. 2021, 36, 109450. [Google Scholar] [CrossRef]
- Taha, H.R.; Keewan, N.; Slati, F.; Al-Sawalha, N.A. Remdesivir: A Closer Look at Its Effect in COVID-19 Pandemic. Pharmacology 2021, 106, 462–468. [Google Scholar] [CrossRef]
- Matsuyama, S.; Kawase, M.; Nao, N.; Shirato, K.; Ujike, M.; Kamitani, W.; Shimojima, M.; Fukushi, S. The Inhaled Steroid Ciclesonide Blocks SARS-CoV-2 RNA Replication by Targeting the Viral Replication-Transcription Complex in Cultured Cells. J. Virol. 2020, 95, e01648-20. [Google Scholar] [CrossRef]
- Yang, J.-W.; Fan, L.-C.; Miao, X.-Y.; Mao, B.; Li, M.-H.; Lu, H.-W.; Liang, S.; Xu, J.-F. Corticosteroids for the treatment of human infection with influenza virus: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2015, 21, 956–963. [Google Scholar] [CrossRef]
- Kim, Y.; Jedrzejczak, R.; Maltseva, N.I.; Wilamowski, M.; Endres, M.; Godzik, A.; Michalska, K.; Joachimiak, A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020, 29, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, P.; Fang, L.; Hong, Y.; Zhu, X.; Wang, D.; Peng, G.; Xiao, S. Porcine deltacoronavirus nsp15 antagonizes interferon-β production independently of its endoribonuclease activity. Mol. Immunol. 2019, 114, 100–107. [Google Scholar] [CrossRef]
- El Sayed, K.A. Natural Products as Antiviral Agents. Stud. Nat. Prod. Chem. 2000, 24, 473–572. [Google Scholar] [CrossRef]
- Bzówka, M.; Mitusińska, K.; Raczyńska, A.; Samol, A.; Tuszyński, J.A.; Góra, A. Structural and Evolutionary Analysis Indicate That the SARS-CoV-2 Mpro Is a Challenging Target for Small-Molecule Inhibitor Design. Int. J. Mol. Sci. 2020, 21, 3099. [Google Scholar] [CrossRef]
- Kolarič, A.; Jukič, M.; Bren, U. Novel Small-Molecule Inhibitors of the SARS-CoV-2 Spike Protein Binding to Neuropilin 1. Pharmaceuticals 2022, 15, 165. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Z. Natural Products, Alone or in Combination with FDA-Approved Drugs, to Treat COVID-19 and Lung Cancer. Biomedicines 2021, 9, 689. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, N.; Yang, L.; Song, X.-Q. Bioactive natural products in COVID-19 therapy. Front. Pharmacol. 2022, 13, 926507. [Google Scholar] [CrossRef]
- Pokharkar, O.; Lakshmanan, H.; Zyryanov, G.V.; Tsurkan, M.V. Antiviral Potential of Antillogorgia americana and elisabethae Natural Products against nsp16–nsp10 Complex, nsp13, and nsp14 Proteins of SARS-CoV-2: An In Silico Investigation. Microbiol. Res. 2023, 14, 993–1019. [Google Scholar]
- Wang, D.; Huang, J.; Yeung, A.W.K.; Tzvetkov, N.T.; Horbańczuk, J.O.; Willschke, H.; Gai, Z.; Atanasov, A.G. The Significance of Natural Product Derivatives and Traditional Medicine for COVID-19. Processes 2020, 8, 937. [Google Scholar] [CrossRef]
- Ismail, M.M.; Alotaibi, B.S.; El-Sheekh, M.M. Therapeutic Uses of Red Macroalgae. Molecules 2020, 25, 4411. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.G.; Cadamuro, R.D.; Cabral, A.C.; Thaís da Silva, I.T.; Rodríguez-Lázaro, D.; Fongaro, G. Broad Spectrum Algae Compounds Against Viruses. Front. Microbiol. 2022, 12, 809296. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Riverol, A.; Piñón, R.A.; Morier, D.L.F.; Torres, L.Y.; Mendoza, L.D.; Barrio, A.G. Antiviral activity of an aqueous extract from the red alga Laurencia obtusa against influenza A and B viruses. Rev. Cub. Med. Trop. 2014, 66, 273–285. [Google Scholar]
- Gheda, S.F.; El-Adawi, H.I.; El-Deeb, N.M. Antiviral Profile of Brown and Red Seaweed Polysaccharides Against Hepatitis C Virus. Iran. J. Pharm. Res. IJPR 2016, 15, 483–491. [Google Scholar]
- Yu, X.-Q.; Jiang, C.-S.; Zhang, Y.; Sun, P.; Kurtán, T.; Mándi, A.; Li, X.-L.; Yao, L.-G.; Liu, A.-H.; Wang, B.; et al. Compositacins A–K: Bioactive chamigrane-type halosesquiterpenoids from the red alga Laurencia composita Yamada. Phytochemistry 2017, 136, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Hoshino, M.; Fujita, M.; Urban, S. Cycloelatanene A and B: Absolute configuration determination and structural revision by the crystalline sponge method. Chem. Sci. 2017, 8, 1547–1550. [Google Scholar] [CrossRef]
- Shaaban, M.; Abou-El-Wafa, G.S.E.; Golz, C.; Laatsch, H. New Haloterpenes from the Marine Red Alga Laurencia papillosa: Structure Elucidation and Biological Activity. Mar. Drugs 2021, 19, 35. [Google Scholar] [CrossRef]
- Masuda, M.; Kogame, K.; Arisawa, S.; Suzuki, M. Morphology and Halogenated Secondary Metabolites of Three Gran Canarian Species of Laurencia (Ceramiales, Rhodophyta). Bot. Mar. 1998, 41, 265–277. [Google Scholar] [CrossRef]
- Sentíes, A.; Díaz-Larrea, J.; Cassano, V.; Gil-Rodríguez, M.C.; Fujii, M.T. Laurencia marilzae (Ceramiales, Rhodophyta) from the Mexican Caribbean: A New Record for the Tropical Western Atlantic. Bull. Mar. Sci. 2011, 87, 681–686. [Google Scholar] [CrossRef]
- Suzuki, M.; Vairappan, C.S. Halogenated secondary metabolites from Japanese species of the red algal genus Laurencia (Rhodomelaceae, Ceramiales). Curr. Top. Phytochem. 2005, 7, 1–34. [Google Scholar]
- Wang, B.-G.; Gloer, J.B.; Ji, N.-Y.; Zhao, J.-C. Halogenated Organic Molecules of Rhodomelaceae Origin: Chemistry and Biology. Chem. Rev. 2013, 113, 3632–3685. [Google Scholar] [CrossRef]
- Suzuki, M.; Takahashi, Y.; Nakano, S.; Abe, T.; Masuda, M.; Ohnishi, T.; Noya, Y.; Seki, K.-I. An experimental approach to study the biosynthesis of brominated metabolites by the red algal genus Laurencia. Phytochemistry 2009, 70, 1410–1415. [Google Scholar] [CrossRef] [PubMed]
- Young, D.N.; Howard, B.M.; Fenical, W. Subcellular localization of brominated secondary metabolites in the red alga Laurencia snyderae. J. Phycol. 1980, 16, 182–185. [Google Scholar] [CrossRef]
- Vairappan, C.S.; Suzuki, M.; Abe, T.; Masuda, M. Halogenated metabolites with antibacterial activity from the Okinawan Laurencia species. Phytochemistry 2001, 58, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, M.T.; Vale, C.; Rauter, A.P. Halogenated Compounds from Marine Algae. Mar. Drugs 2010, 8, 2301–2317. [Google Scholar] [CrossRef] [PubMed]
- Juagdan, E.G.; Kalidindi, R.; Scheuer, P. Two new chamigranes from an hawaiian red alga Laurencia cartilaginea. Tetrahedron 1997, 53, 521–528. [Google Scholar] [CrossRef]
- König, G.M.; Wright, A.D. Laurencia rigida: Chemical Investigations of Its Antifouling Dichloromethane Extract. J. Nat. Prod. 1997, 60, 967–970. [Google Scholar] [CrossRef]
- Alarif, W.M.; Al-Lihaibi, S.S.; Abdel-Lateff, A.; Ayyad, S.-E.N. New Antifungal Cholestane and Aldehyde Derivatives from the Red Alga Laurencia papillosa. Nat. Prod. Commun. 2011, 6, 1821–1824. [Google Scholar] [CrossRef]
- Li, Y.-X.; Li, Y.; Qian, Z.J.; Kim, M.M.; Kim, S.K. In Vitro Antioxidant Activity of 5-HMF Isolated from Marine Red Alga Laurencia undulata in Free-Radical-Mediated Oxidative Systems. J. Microbiol. Biotechnol. 2009, 19, 1319–1327. [Google Scholar] [CrossRef]
- Topcu, G.; Aydogmus, Z.; Imre, S.; Gören, A.C.; Pezzuto, J.M.; Clement, J.A.; Kingston, D.G.I. Brominated Sesquiterpenes from the Red Alga Laurencia obtusa. J. Nat. Prod. 2003, 66, 1505–1508. [Google Scholar] [CrossRef]
- Davyt, D.; Fernandez, R.; Suescun, L.; Mombrú, A.W.; Saldaña, J.; Domínguez, L.; Coll, J.; Fujii, M.T.; Manta, E. New Sesquiterpene Derivatives from the Red Alga Laurencia scoparia. Isolation, Structure Determination, and Anthelmintic Activity. J. Nat. Prod. 2001, 64, 1552–1555. [Google Scholar] [CrossRef]
- Jung, W.-K.; Choi, I.; Oh, S.; Park, S.-G.; Seo, S.-K.; Lee, S.-W.; Lee, D.-S.; Heo, S.-J.; Jeon, Y.-J.; Je, J.-Y.; et al. Anti-asthmatic effect of marine red alga (Laurencia undulata) polyphenolic extracts in a murine model of asthma. Food Chem. Toxicol. 2009, 47, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Kurata, K.; Taniguchi, K.; Agatsuma, Y.; Suzuki, M. Diterpenoid feeding-deterrents from Laurencia saitoi. Phytochemistry 1998, 47, 363–369. [Google Scholar] [CrossRef]
- Dembitsky, V.M.; Tolstikov, G.A. Natural halogenated sesquiterpens from marine organisms, chemistry for sustainable development. Chem. Sustain. Dev. 2004, 12, 1–12. [Google Scholar]
- Suzuki, M.; Kurosawa, E.; Irie, T. Spirolaurenone, a new sesquiterpenoid containing bromine from Laurenciaglandulifera Kützing. Tetrahedron Lett. 1970, 11, 4995–4998. [Google Scholar] [CrossRef]
- Ji, N.-Y.; Li, X.-M.; Li, K.; Ding, L.-P.; Gloer, J.B.; Wang, B.-G. Diterpenes, Sesquiterpenes, and a C15-Acetogenin from the Marine Red Alga Laurencia mariannensis. J. Nat. Prod. 2007, 70, 1901–1905. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-D.; Miao, F.-P.; Li, K.; Ji, N.-Y. Sesquiterpenes and acetogenins from the marine red alga Laurencia okamurai. Fitoterapia 2012, 83, 518–522. [Google Scholar] [CrossRef]
- Liang, Y.; Li, X.-M.; Cui, C.-M.; Li, C.-S.; Sun, H.; Wang, B.-G. Sesquiterpene and Acetogenin Derivatives from the Marine Red Alga Laurencia okamurai. Mar. Drugs 2012, 10, 2817–2825. [Google Scholar] [CrossRef]
- Liang, Y.; Li, X.M.; Cui, C.M.; Li, C.S.; Wang, B.G. A new rearranged chamigrane sesquiterpene from Laurencia okamurai. Chin. Chem. Lett. 2009, 20, 190–192. [Google Scholar] [CrossRef]
- Li, X.-D.; Miao, F.-P.; Yin, X.-L.; Liu, J.-L.; Ji, N.-Y. Sesquiterpenes from the marine red alga Laurencia composita. Fitoterapia 2012, 83, 1191–1195. [Google Scholar] [CrossRef]
- Ji, N.-Y.; Li, X.-M.; Wang, B.-G. Sesquiterpenes and Other Metabolites from the Marine Red Alga Laurencia composita (Rhodomelaceae). Helv. Chim. Acta 2010, 93, 2281–2286. [Google Scholar] [CrossRef]
- Sims, J.J.; Lin, G.H.; Wing, R.M. Marine natural products X elatol, a halogenated sesquiterpene alcohol from the red alga Laurencia elata. Tetrahedron Lett. 1974, 15, 3487–3490. [Google Scholar] [CrossRef]
- Lhullier, C.; Donnangelo, A.; Caro, M.; Palermo, J.A.; Horta, P.A.; Falkenberg, M.; Schenkel, E.P. Isolation of elatol from Laurencia microcladia and its palatability to the sea urchin Echinometra lucunter. Biochem. Syst. Ecol. 2009, 37, 254–259. [Google Scholar] [CrossRef]
- González, A.; Darias, J.; Díaz, A.; Fourneron, J.; Martín, J.; Pérez, C. Evidence for the biogenesis of halogenated chamigrenes from the red alga Laurencia obtusa. Tetrahedron Lett. 1976, 17, 3051–3054. [Google Scholar] [CrossRef]
- Vairappan, C.S. Potent antibacterial activity of halogenated metabolites from Malaysian red algae, Laurencia majuscula (Rhodomelaceae, Ceramiales). Biomol. Eng. 2003, 20, 255–259. [Google Scholar] [CrossRef]
- Dos Santos, A.O.; Veiga-Santos, P.; Ueda-Nakamura, T.; Filho, B.P.D.; Sudatti, D.B.; Bianco, .M.; Pereira, R.C.; Nakamura, C.V. Effect of Elatol, Isolated from Red Seaweed Laurencia dendroidea, on Leishmania amazonensis. Mar. Drugs 2010, 8, 2733–2743. [Google Scholar] [CrossRef] [PubMed]
- Desoti, V.C.; Lazarin-Bidóia, D.; Sudatti, D.B.; Pereira, R.C.; Alonso, A.; Ueda-Nakamura, T.; Dias Filho, B.P.; Nakamura, C.V.; De Oliveira Silva, S. Trypanocidal Action of (−)-Elatol Involves an Oxidative Stress Triggered by Mitochondria Dysfunction. Mar. Drugs 2012, 10, 1631–1646. [Google Scholar] [CrossRef]
- Campos, A.; Souza, C.B.; Lhullier, C.; Falkenberg, M.; Schenkel, E.P.; Ribeiro-Do-Valle, R.M.; Siqueira, J.M. Anti-tumour effects of elatol, a marine derivative compound obtained from red algae Laurencia microcladia. J. Pharm. Pharmacol. 2012, 64, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.A.; Urban, S. Phytochemical studies of the southern Australian marine alga, Laurencia elata. Phytochemistry 2011, 72, 2081–2089. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2003, 20, 1–48. [Google Scholar] [CrossRef]
- Dorta, E.; Díaz-Marrero, A.R.; Cueto, M.; D’croz, L.; Maté, J.L.; Darias, J. Chamigrenelactone, a polyoxygenated sesquiterpene with a novel structural type and devoid of halogen from Laurencia obtusa. Tetrahedron Lett. 2004, 45, 7065–7068. [Google Scholar] [CrossRef]
- Brito, I.; Cueto, M.; Díaz-Marrero, A.R.; Darias, J.; San Martín, A. Oxachamigrenes, New Halogenated Sesquiterpenes from Laurencia obtusa. J. Nat. Prod. 2002, 65, 946–948. [Google Scholar] [CrossRef] [PubMed]
- Gonzaález, A.; Martín, J.; Martín, V.; Norte, M.; Pérez, R. Biomimetic approach to the synthesis of rhodolaureol and rhodolauradiol. Tetrahedron Lett. 1982, 23, 2395–2398. [Google Scholar] [CrossRef]
- Suzuki, M.; Kurosawa, E.; Furusaki, A. The Structure and Absolute Stereochemistry of a Halogenated Chamigrene Derivative from the Red Alga Laurencia Species. Bull. Chem. Soc. Jpn. 1988, 61, 3371–3373. [Google Scholar] [CrossRef]
- Ji, N.-Y.; Li, X.-M.; Li, K.; Wang, B.-G. Halogenated Sesquiterpenes from the Marine Red Alga Laurencia saitoi (Rhodomelaceae). Helv. Chim. Acta 2009, 92, 1873–1879. [Google Scholar] [CrossRef]
- Howard, B.M.; Fenical, W. Structures and chemistry of two new halogen-containing chamigrene derivatives from laurencia. Tetrahedron Lett. 1971, 16, 1687–1690. [Google Scholar] [CrossRef]
- Jongaramruong, J.; Blackman, A.J.; Skelton, B.W.; White, A.H. Chemical Relationships between the Sea Hare Aplysia parvula and the Red Seaweed Laurencia filiformis from Tasmania. Aust. J. Chem. 2002, 55, 275–280. [Google Scholar] [CrossRef]
- Suzuki, M.; Daitoh, M.; Vairappan, C.S.; Abe, T.; Masuda, M. Novel Halogenated Metabolites from the Malaysian Laurencia pannosa. J. Nat. Prod. 2001, 64, 597–602. [Google Scholar] [CrossRef]
- Suescun, L.; Mombrú, A.W.; Mariezcurrena, R.A.; Davyt, D.; Fernández, R.; Manta, E. Two natural products from the algae Laurencia scoparia. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 2001, 57, 286–288. [Google Scholar] [CrossRef]
- Francisco, M.E.Y.; Erickson, K.L. Ma’iliohydrin, a Cytotoxic Chamigrene Dibromohydrin from a Philippine Laurencia Species. J. Nat. Prod. 2001, 64, 790–791. [Google Scholar] [CrossRef]
- Da Silva Machado, F.L.D.S.; Ventura, T.L.B.; Gestinari, L.M.d.S.; Cassano, V.; Resende, J.A.L.C.; Kaiser, C.R.; Lasunskaia, E.B.; Muzitano, M.F.; Soares, A.R. Sesquiterpenes from the Brazilian Red Alga Laurencia dendroidea. J. Agardh. Molecules 2014, 19, 3181–3192. [Google Scholar] [CrossRef]
- Rucker, R.; Kretzuschmar, U. 9-Aristolen-1a-ol and 1,2,9,10-tetradehydroaristolane, new aristolane type sesquiterpenes. Liebigs Ann. Chem. 1971, 748, 214–217. [Google Scholar] [CrossRef]
- Ji, N.-Y.; Li, X.-M.; Ding, L.-P.; Wang, B.-G. Aristolane Sesquiterpenes and Highly Brominated Indoles from the Marine Red Alga Laurencia similis (Rhodomelaceae). Helv. Chim. Acta 2007, 90, 385–391. [Google Scholar] [CrossRef]
- Li, C.-S.; Li, X.-M.; Cui, C.-M.; Wang, B.-G. Brominated Metabolites from the Marine Red Alga Laurencia similis. Z. Naturforsch. 2010, 65, 87–89. [Google Scholar] [CrossRef]
- Su, S.; Sun, W.-S.; Wang, B.; Cheng, W.; Liang, H.; Zhao, Y.-Y.; Zhang, Q.-Y.; Wu, J. A novel brominated cuparene-derived sesquiterpene ether from the red alga Laurencia sp. J. Asian Nat. Prod. Res. 2010, 12, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Kamada, T.; Vairappan, C.S. New Bioactive Secondary Metabolites from Bornean Red Alga, Laurencia similis (Ceramiales). Nat. Prod. Commun. 2013, 8, 287–288. [Google Scholar] [CrossRef]
- Fraga, B.M. Natural sesquiterpenoids. Nat. Prod. Rep. 1997, 14, 145–162. [Google Scholar] [CrossRef]
- Kladi, M.; Vagias, C.; Papazafiri, P.; Furnari, G.; Serio, D.; Roussis, V. New sesquiterpenes from the red alga Laurencia microcladia. Tetrahedron 2007, 63, 7606–7611. [Google Scholar] [CrossRef]
- Yu, X.-Q.; He, W.-F.; Liu, D.-Q.; Feng, M.-T.; Fang, Y.; Wang, B.; Feng, L.-H.; Guo, Y.-W.; Mao, S.-C. A seco-laurane sesquiterpene and related laurane derivatives from the red alga Laurencia okamurai Yamada. Phytochemistry 2014, 103, 162–170. [Google Scholar] [CrossRef]
- Davyt, D.; Fernandez, R.; Suescun, L.; Mombrú, A.W.; Saldaña, J.; Dominguez, L.; Fujii, M.T.; Manta, E. Bisabolanes from the red alga Laurencia scoparia. J. Nat. Prod. 2006, 69, 1113–1116. [Google Scholar] [CrossRef]
- De Carvalho, L.T.; Fujii, M.T.; Roque, N.F.; Kato, M.J.; Lago, J.H.G. Aldingenin A, new brominated sesquiterpene from red algae Laurencia aldingensis. Tetrahedron Lett. 2003, 44, 2637–2640. [Google Scholar] [CrossRef]
- de Carvalho, L.R.; Fujii, M.T.; Roque, N.F.; Lago, J.H.G. Aldingenin derivatives from the red alga Laurencia aldingensis. Phytochemistry 2006, 67, 1331–1335. [Google Scholar] [CrossRef]
- Kladi, M.; Vagias, C.; Furnari, G.; Moreau, D.; Roussakis, C.; Roussis, V. Cytotoxic cuparene sesquiterpenes from Laurencia microcladia. Tetrahedron Lett. 2005, 46, 5723–5726. [Google Scholar] [CrossRef]
- Mao, S.-C.; Guo, Y.-W. A Laurane Sesquiterpene and Rearranged Derivatives from the Chinese Red Alga Laurencia okamurai Yamada. J. Nat. Prod. 2006, 69, 1209–1211. [Google Scholar] [CrossRef]
- Srikrishna, A.; Khan, I.; Babu, R.R.; Sajjanshetty, A. The first total synthesis of (±)-laurokamurene B. Tetrahedron 2007, 63, 12616–12620. [Google Scholar] [CrossRef]
- Irie, T.; Suzuki, T.; Yasunari, Y.; Kurosawa, E.; Masamune, T. Laurene, a sesquiterpene hydrocarbon from Laurencia species. Tetrahedron 1969, 25, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Wratten, S.J.; Faulkner, D.J. Metabolites of the red alga Laurencia subopposita. J. Org. Chem. 1977, 42, 3343–3349. [Google Scholar] [CrossRef]
- Alarif, W.M.; Al-Lihaibi, S.S.; Ayyad, S.-E.N.; Abdel-Rhman, M.H.; Badria, F.A. Laurene-type sesquiterpenes from the Red Sea red alga Laurencia obtusa as potential antitumor–antimicrobial agents. Eur. J. Med. Chem. 2012, 55, 462–466. [Google Scholar] [CrossRef]
- Yamamura, S.; Hirata, Y. Structures of aplysin and aplysinol, naturally occurring bromo-compounds. Tetrahedron 1963, 19, 1485–1496. [Google Scholar] [CrossRef]
- Elzen, G.W.; Williams, H.J.; Vinson, S.B. Isolation, identification and bioassay of cotton synomones mediating searching behavior by parasitoidCampoletis sonorensis. J. Chem. Ecol. 1984, 10, 1251–1264. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.-C.; Guo, Y.-W. Cuparene-Derived Sesquiterpenes from the Chinese Red Alga Laurencia okamurai Yamada. Helv. Chim. Acta 2005, 88, 1034–1039. [Google Scholar] [CrossRef]
- Goldsmith, D.J.; John, T.K.; Kwong, C.D.; Painter, G.R. Preparation and rearrangement of trichothecane-like compounds. Synthesis of aplysin and filiformin. J. Org. Chem. 1980, 45, 3989–3993. [Google Scholar] [CrossRef]
- Shizuri, Y.; Yamada, K. Laurebiphenyl, a dimeric sesquiterpene of the cyclolaurane-type from the red alga Laurencia nidifica. Phytochemistry 1985, 24, 1385–1386. [Google Scholar] [CrossRef]
- Liang, Y.; Li, X.-M.; Li, C.-S.; Sun, H.; Wang, B.-G. Laurane-, Cyclolaurane-, and Cuparane-type Sesquiterpenes from the Marine Red Alga Laurencia okamurai. Nat. Prod. Commun. 2014, 9, 323–324. [Google Scholar] [CrossRef]
- Sun, J.; Han, L.; Shi, D.; Ma, M.; Li, S.; Wang, S.; Han, L.; Yang, Y.; Fan, X.; Shi, J.; et al. Sesquiterpenes from Red Alga Laurencia tristicha. J. Nat. Prod. 2008, 68, 915–919, Erratum in J. Nat. Prod. 2008, 71, 296. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, H.; Nagamochi, M.; Ishibashi, H.; Fukumoto, K. A remarkable substituent effect on the enantioselectivity of tandem asymmetric epoxidation and enantiospecific ring expansion of cyclopropylidene alcohols: A new enantiocontrolled synthesis of (-)-debromoaplysin and (-)-aplysin. J. Org. Chem. 1994, 59, 74–79. [Google Scholar] [CrossRef]
- Ji, N.-Y.; Li, X.-M.; Li, K.; Ding, L.-P.; Wang, B.-G. Laurane-derived sesquiterpenes from the marine red alga Laurencia tristicha (Rhodomelaceae). Nat. Prod. Res. 2008, 22, 715–718. [Google Scholar] [CrossRef]
- Kazlauskas, R.; Murphy, P.; Quinn, R.; Wells, R. New laurene derivatives from Laurencia filiformis. Aust. J. Chem. 1976, 29, 2533–2539. [Google Scholar] [CrossRef]
- Dias, D.A.; White, J.M.; Urban, S. Laurencia Filiformis: Phytochemical Profiling by Conventional and HPLC-NMR Approaches. Nat. Prod. Commun. 2009, 4, 157–172. [Google Scholar] [CrossRef]
- Appleton, D.R.; Babcock, R.C.; Copp, B.R. Novel tryptophan-derived dipeptides and bioactive metabolites from the sea hare Aplysia dactylomela. Tetrahedron 2001, 57, 10181–10189. [Google Scholar] [CrossRef]
- König, G.M.; Wright, A.D. New C15 Acetogenins and Sesquiterpenes from the Red Alga Laurencia sp. cf. L. gracilis. J. Nat. Prod. 1994, 57, 477–485. [Google Scholar] [CrossRef]
- Kladi, M.; Xenaki, H.; Vagias, C.; Papazafiri, P.; Roussis, V. New cytotoxic sesquiterpenes from the red algae Laurencia obtusa and Laurencia microcladia. Tetrahedron 2006, 62, 182–189. [Google Scholar] [CrossRef]
- Iliopoulou, D.; Roussis, V.; Pannecouque, C.; De Clercq, E.; Vagias, C. Halogenated sesquiterpenes from the red alga Laurencia obtusa. Tetrahedron 2002, 58, 6749–6755. [Google Scholar] [CrossRef]
- Aydoğmuş, Z.; Imre, S.; Ersoy, L.; Wray, V. Halogenated secondary metabolites from Laurencia Obtusa. Nat. Prod. Res. 2004, 18, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.R.; Erickson, K.L. Austradiol acetate and austradiol diacetate, 4,6-dihydroxy-(+)-selinane derivatives from an Australian Laurencia sp. J. Org. Chem. 1982, 47, 3917–3921. [Google Scholar] [CrossRef]
- Kazlauskas, R.; Murphy, P.; Wells, R.; Daly, J.; Oberhansli, W. Heterocladol, a halogenated selinane sesquiterpene of biosynthetic significance from the red alga Laurencia filiformis: Its isolation, crystal structure andabsolute configuration. Aust. J. Chem. 1977, 30, 2679–2687. [Google Scholar] [CrossRef]
- Guella, G.; Skropeta, D.; Mancini, I.; Pietra, F. The First 6,8-Cycloeudesmane Sesquiterpene from a Marine Organism: The Red Seaweed Laurencia microcladia from the Baia di Calenzana, Elba Island. Z. Für Naturforschung B 2002, 57, 1147–1151. [Google Scholar] [CrossRef]
- Suzuki, M.; Takahashi, Y.; Mitome, Y.; Itoh, T.; Abe, T.; Masuda, M. Brominated metabolites from an Okinawan Laurencia intricata. Phytochemistry 2002, 60, 861–867. [Google Scholar] [CrossRef]
- Reward, B.M.; Fenical, W. α- and β-snyderol; new bromo-monocyclic sesquiterpenes from the seaweed Laurencia. Tetrahedron Lett. 1976, 17, 41–44. [Google Scholar] [CrossRef]
- Su, H.; Shi, D.-Y.; Li, J.; Guo, S.-J.; Li, L.-L.; Yuan, Z.-H.; Zhu, X.-B. Sesquiterpenes from Laurencia similis. Molecules 2009, 14, 1889–1897. [Google Scholar] [CrossRef]
- Kuniyoshi, M.; Marma, M.S.; Higa, T.; Bernardinelli, G.; Jefford, C.W. New Bromoterpenes from the Red Alga Laurencia luzonensis. J. Nat. Prod. 2001, 64, 696–700. [Google Scholar] [CrossRef]
- Kuniyoshi, M.; Wahome, P.G.; Miono, T.; Hashimoto, T.; Yokoyama, M.; Shrestha, K.L.; Higa, T. Terpenoids from Laurencia luzonensis. J. Nat. Prod. 2005, 68, 1314–1317. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yuan, Z.-H.; Li, J.; Guo, S.-J.; Deng, L.-P.; Han, L.-J.; Zhu, X.-B.; Shi, D.-Y. Sesquiterpenes from the Marine Red Alga Laurencia saitoi. Helv. Chim. Acta 2009, 92, 1291–1297. [Google Scholar] [CrossRef]
- Makhanu, D.S.; Yokoyama, M.; Miono, T.; Maesato, T.; Maedomari, M.; Wisespongpand, P.; Kuniyoshi, M. New sesquiterpenes from the Okinawan red alga Laurencia luzonensis. Bull. Fac. Sci. Univ. Ryukyus. 2006, 81, 115–120. [Google Scholar]
- Vairappan, C.S.; Kamada, T.; Lee, W.-W.; Jeon, Y.-J. Anti-inflammatory activity of halogenated secondary metabolites of Laurencia snackeyi (Weber-van Bosse) Masuda in LPS-stimulated RAW 264.7 macrophages. J. Appl. Phycol. 2013, 25, 1805–1813. [Google Scholar] [CrossRef]
- Wijesinghe, W.; Kim, E.-A.; Kang, M.-C.; Lee, W.-W.; Lee, H.-S.; Vairappan, C.S.; Jeon, Y.-J. Assessment of anti-inflammatory effect of 5β-hydroxypalisadin B isolated from red seaweed Laurencia snackeyi in zebrafish embryo in vivo model. Environ. Toxicol. Pharmacol. 2014, 37, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yuan, Z.; Li, J.; Guo, S.; Han, L.; Zhu, X.; Shi, D. [Studies on chemical constituents of Laurencia saitoi]. Zhongguo Zhong Yao Za Zhi 2009, 34, 871–874. [Google Scholar]
- Wright, A.D.; Goclik, E.; König, G.M. Three New Sesquiterpenes from the Red Alga Laurencia perforata. J. Nat. Prod. 2003, 66, 435–437. [Google Scholar] [CrossRef]
- Vairappan, C.S.; Suzuki, M.; Ishii, T.; Okino, T.; Abe, T.; Masuda, M. Antibacterial activity of halogenated sesquiterpenes from Malaysian Laurencia spp. Phytochemistry 2008, 69, 2490–2494. [Google Scholar] [CrossRef]
- Erickson, K.L.; Beutler, J.A.; Gray, G.N.; Cardellina, J.H.; Boyd, M.R. Majapolene A, a Cytotoxic Peroxide, and Related Sesquiterpenes from the Red Alga Laurencia majuscula. J. Nat. Prod. 1995, 58, 1848–1860. [Google Scholar] [CrossRef]
- Monde, K.; Taniguchi, T.; Miura, N.; Vairappan, C.S.; Suzuki, M. Absolute configurations of brominated sesquiterpenes determined by vibrational circular dichroism. Chirality 2006, 18, 335–339. [Google Scholar] [CrossRef]
- Da Silva, F.L.M.; Pacienza-Lima, W.; Rossi-Bergmann, B.; de Souza, L.M.G.; Fujii, M.T.; Campos de Paula, J.; Costa, S.S.; Lopes, N.P.; Kaiser, C.R.; Soares, A.R. Antileishmanial Sesquiterpenes from the Brazilian Red Alga Laurencia dendroidea. Planta Med. 2010, 77, 733–735. [Google Scholar] [CrossRef]
- Al-Massarani, S. Phytochemical and Biological Properties of Sesquiterpene Constituents From the Marine Red Seaweed Laurencia: A Review. Nat. Prod. Chem. Res. 2014, 2, 147. [Google Scholar] [CrossRef]
- Wang, B.; Svetlov, V.; Wolf, Y.I.; Koonin, E.V.; Nudler, E.; Artsimovitch, I. Allosteric Activation of SARS-CoV-2 RNA-Dependent RNA Polymerase by Remdesivir Triphosphate and Other Phosphorylated Nucleotides. mBio 2021, 12, e0142321. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information Protein Database. Available online: http://www.ncbi.nlm.nih.gov/protein/ (accessed on 10 March 2023).
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Pence, H.E.; Williams, A. ChemSpider: An Online Chemical Information Resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Lyu, C.; Chen, T.; Qiang, B.; Liu, N.; Wang, H.; Zhang, L.; Liu, Z. CMNPD: A comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 2020, 49, D509–D515. [Google Scholar] [CrossRef] [PubMed]
- Biovia, D.S. Discovery Studio. Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 15 December 2022).
- Avogadro: An Open-Source Molecular Builder and Visualization Tool. Version 1.95. Available online: http://avogadro.cc/ (accessed on 23 November 2022).
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Kuriata, A.; Gierut, A.M.; Oleniecki, T.; Ciemny, M.P.; Kolinski, A.; Kurcinski, M.; Kmiecik, S. CABS-flex 2.0: A web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 2018, 46, W338–W343. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Borba, J.V.; Alves, V.M.; Braga, R.C.; Korn, D.R.; Overdahl, K.; Silva, A.C.; Hall, S.U.; Overdahl, E.; Braga, R.; Kleinstreuer, N.; et al. STopTox: An in Silico Alternative to Animal Testing for Acute Systemic and Topical Toxicity. Environ. Health Perspect. 2022, 130, 27012. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Abraham, M.H.; Le, J.; Hersey, A.; Luscombe, C.N.; Beck, G.; Sherborne, B.; Cooper, I. Rate-Limited Steps of Human Oral Absorption and QSAR Studies. Pharm. Res. 2002, 19, 1446–1457. [Google Scholar] [CrossRef]
- Husain, A.; Ahmad, A.; Alam Khan, S.; Asif, M.; Bhutani, R.; Al-Abbasi, F.A. Synthesis, molecular properties, toxicity and biological evaluation of some new substituted imidazolidine derivatives in search of potent anti-inflammatory agents. Saudi Pharm. J. 2016, 24, 104–114. [Google Scholar] [CrossRef]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the Biological Activity Spectra of Organic Compounds Using the Pass Online Web Resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Begum, F.; Srivastava, A.K.; Ray, U. Repurposing nonnucleoside antivirals against SARS-CoV2 NSP12 (RNA dependent RNA polymerase): In silico-molecular insight. Biochem. Biophys. Res. Commun. 2021, 571, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Mosayebnia, M.; Bozorgi, A.H.; Rezaeianpour, M.; Kobarfard, F. In silico prediction of SARS-CoV-2 main protease and polymerase inhibitors: 3D-Pharmacophore modelling. J. Biomol. Struct. Dyn. 2022, 40, 6569–6586. [Google Scholar] [CrossRef] [PubMed]
- Tungary, E.; Wahjudi, M.; Kok, T. Secondary Metabolites of Various Indonesian Medicinal Plants as SARS-CoV-2 Inhibitors: In Silico Study. MPI Media Pharm. Indones. 2022, 4, 136–146. [Google Scholar] [CrossRef]
- Ikeguchi, M.; Ueno, J.; Sato, M.; Kidera, A. Protein Structural Change Upon Ligand Binding: Linear Response Theory. Phys. Rev. Lett. 2005, 94, 078102. [Google Scholar] [CrossRef]
- Agoni, C. The Binding of Remdesivir to SARS-CoV-2 RNA-Dependent RNA Polymerase May Pave The Way Towards the Design of Potential Drugs for COVID-19 Treatment. Curr. Pharm. Biotechnol. 2021, 22, 1520–1537. [Google Scholar] [CrossRef]
- Mishra, A.; Rathore, A.S. RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2. J. Biomol. Struct. Dyn. 2022, 40, 6039–6051. [Google Scholar] [CrossRef]
- Arora, S.; Lohiya, G.; Moharir, K.; Shah, S.; Yende, S. Identification of Potential Flavonoid Inhibitors of the SARS-CoV-2 Main Protease 6YNQ: A Molecular Docking Study. Digit. Chin. Med. 2020, 3, 239–248. [Google Scholar] [CrossRef]
- Umar, A.K. Flavonoid compounds of buah merah (Pandanus conoideus Lamk) as a potent SARS-CoV-2 main protease inhibitor: In silico approach. Futur. J. Pharm. Sci. 2021, 7, 158–159. [Google Scholar] [CrossRef]
- Ortega, J.T.; Jastrzebska, B.; Rangel, H.R. Omicron SARS-CoV-2 Variant Spike Protein Shows an Increased Affinity to the Human ACE2 Receptor: An In Silico Analysis. Pathogens 2021, 11, 45. [Google Scholar] [CrossRef]
- Katuwal, S.; Upadhyaya, S.R.; Marahatha, R.; Shrestha, A.; Regmi, B.P.; Khadayat, K.; Basnet, S.; Basnyat, R.C.; Parajuli, N. In Silico Study of Coumarins: Wedelolactone as a Potential Inhibitor of the Spike Protein of the SARS-CoV-2 Variants. J. Trop. Med. 2023, 2023, 4771745. [Google Scholar] [CrossRef]
- Pokharkar, O.; Lakshmanan, H.; Zyryanov, G.; Tsurkan, M. In Silico Evaluation of Antifungal Compounds from Marine Sponges against COVID-19-Associated Mucormycosis. Mar. Drugs 2022, 20, 215. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef] [PubMed]
- Cron, R.Q.; Caricchio, R.; Chatham, W.W. Calming the cytokine storm in COVID-19. Nat. Med. 2021, 27, 1674–1675. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pokharkar, O.; Anumolu, H.; Zyryanov, G.V.; Tsurkan, M.V. Natural Products from Red Algal Genus Laurencia as Potential Inhibitors of RdRp and nsp15 Enzymes of SARS-CoV-2: An In Silico Perspective. Microbiol. Res. 2023, 14, 1020-1048. https://doi.org/10.3390/microbiolres14030069
Pokharkar O, Anumolu H, Zyryanov GV, Tsurkan MV. Natural Products from Red Algal Genus Laurencia as Potential Inhibitors of RdRp and nsp15 Enzymes of SARS-CoV-2: An In Silico Perspective. Microbiology Research. 2023; 14(3):1020-1048. https://doi.org/10.3390/microbiolres14030069
Chicago/Turabian StylePokharkar, Omkar, Harshavardhan Anumolu, Grigory V. Zyryanov, and Mikhail V. Tsurkan. 2023. "Natural Products from Red Algal Genus Laurencia as Potential Inhibitors of RdRp and nsp15 Enzymes of SARS-CoV-2: An In Silico Perspective" Microbiology Research 14, no. 3: 1020-1048. https://doi.org/10.3390/microbiolres14030069
APA StylePokharkar, O., Anumolu, H., Zyryanov, G. V., & Tsurkan, M. V. (2023). Natural Products from Red Algal Genus Laurencia as Potential Inhibitors of RdRp and nsp15 Enzymes of SARS-CoV-2: An In Silico Perspective. Microbiology Research, 14(3), 1020-1048. https://doi.org/10.3390/microbiolres14030069








