Abstract
Carbapenem-resistant Klebsiella pneumoniae (CRKP)-associated infections have become a major concern and life-threatening worldwide. Understanding the epidemiology of CRKP using a reliable molecular technology can help to develop an effective infection control policies. In the western region of Saudi Arabia, there are no sufficient data on the prevalence of CRKP and its carbapenem-resistant determinants. Therefore, this study aimed to determine the molecular epidemiology of CRKP and identify the most common carbapenemase genes. In the current study, a total of 191 CRKP isolates were collected and obtained from clinical specimens of patients at King Fahad Armed Forces Hospital (KFAFH), Jeddah, Saudi Arabia. All isolates that were resistant or intermediately susceptible to either of the carbapenem antimicrobials (imipenem, meropenem, or ertapenem) were included. All CRKP showed resistance to ceftazidime, cefepime, and piperacillin/tazobactam, whereas low (14%) and moderate (37.7%) levels of resistance were reported against tigecycline and colistin, respectively. The most common carbapenemase genes identified were blaOXA-48 (n = 157 [82.2%]), followed by blaNDM in 27 (14%) isolates. The blaVIM and blaKPC were reported in only one isolate each and no blaIMP producers were detected among all tested isolates. The high prevalence of OXA-48 among K. pneumoniae isolates reported in the current study may reflect that OXA-48 has become an endemic in Saudi Arabian hospitals. The second major finding was that the identification of CRKP co-harbors both blaNDM and blaOXA-48, and such isolates can be threating for healthcare societies (patients and healthcare workers) due to their high level of resistance to carbapenems. These results suggest that the use of molecular diagnostic methods and proper surveillance programs are required to monitor and control the spread of all multidrug-resistant (MDR) bacteria, including CRKP. Therefore, further research is recommended to expand the study and further analyze the genotyping of the most common clones of CRKP in other hospitals in the western regions of Saudi Arabia.
1. Introduction
Over the last decade, the nosocomial outbreaks of multi-drug resistant (MDR) bacterial isolates in Gram-negative bacilli (GNB), particularly carbapenem-resistant Klebsiella pneumoniae (CRKP), have been considered a potential threat around the world. The healthcare-associated infections of these antimicrobial-resistant pathogens have become significantly life-threatening and difficult to treat. This can also result in a higher cost for the health care system to control these infections.
The CRKP isolates belong to the most significant extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae that are capable of breaking down the carbapenems, described as the last treatment option for most infections of MDR Gram-negative bacilli. The mechanism of carbapenem resistance is mainly due to the production of carbapenemase enzymes which are classified into class A (KPC), class B (VIM, IMP, and NDM), and class D (OXA-48) [1]. These emerged CRKP classes have been extensively reported in K. pneumoniae associated with serious infections [1,2].
The ability of transmissible plasmid-encoded carbapenemase enzymes (named as K. pneumoniae carbapenemase) among Gram-negative bacilli is now a global health issue that keeps rising [3,4]. Since the carbapenems are very stable and can resist hydroxylation caused by beta-lactamase hydrolysis, these antimicrobials are currently used as the first line of treating severe infections caused by ESBL-producing K. pneumoniae [5,6,7]. However, the overuse of carbapenems has led to the emergence of CRKP and MDR organisms. These pathogens have the ability to produce carbapenemase enzymes that resist multiple antimicrobials, resulting in a major health issue as the treatment plan becomes more challenging [2,8,9].
Some pathogenic bacterial strains may exhibit mild carbapenem resistance, particularly those that harbour KPC-1 (K. pneumoniae carbapenemase-1), while high-level resistance to carbapenems has been found to be only in strains that experience the loss of outer membrane porins due to the presence of KPC-2 as well as KPC-3 [8,9,10,11]. One particular KPC-2-producing or KPC-3-producing K. pneumoniae clone (known as clonal complex (CC) 258 is now widely distributed globally. This clone usually bears the KPC gene together with several other attained antimicrobial resistance-conferring genes [10,11]. Other carbapenemases were also identified within K. pneumoniae strains including metallo-beta-lactamases (MBLs), such as IMP, VIM, and NDM, as well as expanded spectrum oxacillinase OXA-48 [10,11,12,13,14,15]. The New Delhi metallo-beta-lactamase-1 (NDM-1) within K. Pneumoniae strains was firstly identified in India. NDM-1-producing strains of K. pneumoniae are now spread across the European region, most areas of Africa, Asia, and North America [12]. The OXA-48-producing K. pneumoniae was initially reported in Turkey and has now spread to other Mediterranean, Middle Eastern, and European nations [15,16]. Among the three major carbapenemases, which have been observed in K. pneumoniae, KPC-producing strains have been implicated in the spread of nosocomial infections [17]. In contrast, the NDM and OXA-48 strains have been widely linked to hospital- and community-acquired infections [17,18]. Previous studies suggested that the gene accountable for the production of the OXA-48 enzyme is known to have the ability to disseminate between different enterobacterial species far more frequently than KPC and NDM genes [15,16,17,18,19,20].
There are several methods that are employed for the detection of carbapenemases in bacteria. These methods include either phenotypic-based or molecular-based detection methods which vary in their sensitivity, specificity, advantages, and disadvantages [20,21,22,23,24]. One of the recent molecular methods used to detect carbapenemases genes is multiplex real-time PCR, which is known to be highly specific, effective, and prominent for the identification of carbapenemase-producing bacteria [24,25]. Such a method has enabled the detection of the spread of carbapenemase-producing K. pneumoniae that has been globally recognized in several countries around the world. Numerous recent reports have addressed the risk of emerging MDR Gram-negative bacteria, including CRE, CRP, and CRKP [25,26,27,28].
In Saudi Arabia, various studies reported the prevalence of CRKP using molecular advanced methods [29,30,31]. However, profound investigations are still required to obtain a sufficient understanding of the surveillance and infection control of the CRKP in Saudi Arabia. Jeddah city in the western region is considered to be the main gateway of the holy places in Makkah and Madinah which are visited yearly by millions of Muslims from different countries. These huge number of visitors can significantly contribute to the spreading of MDR bacteria and their clones, not only in the holy cities but also in the whole western region. The prevalence and molecular epidemiology of CRKP, particularly in the western region of Saudi Arabia, has not been thoroughly reviewed. Therefore, the main aim of this study is to determine the common nosocomial isolates of carbapenemase-producing K. pneumoniae in Jeddah, Saudi Arabia, using an advanced molecular method. This can eventually encourage health care facilities to improve their policies of the infection control via rapid detection of MDR bacterial pathogens.
2. Materials and Methods
2.1. Study Design
The cross-sectional study was conducted at King Fahad Armed Forces Hospital (KFAFH) in Jeddah, Saudi Arabia. In total, 191 K. pneumoniae isolates were collected between April and July 2018. Ethical approval was obtained from the KFAFH committee (Number: REC 242). The inclusion criteria of the study include all K. pneumoniae isolates that were resistant or intermediately susceptible to either of the carbapenem antimicrobials (imipenem, meropenem, or ertapenem) and sent to a microbiology laboratory. However, all Carbapenem-susceptible K. pneumoniae and duplicate samples were excluded from the study.
2.2. Sample Collection
Different clinical specimens (blood, sputum, urine, wound swabs, intravascular catheter tips, tissue, and body fluids) were collected from KFAFH. All specimens were cultured on sheep blood agar and MacConkey agar plates. The plates were incubated overnight in a 37 °C incubator. Mixed bacterial growth from the original cultured agar was inoculated into MacConkey agar to select pure isolated colonies. Pure colonies of all bacterial isolates were inoculated in thioglycollate broth medium (OXOID, code: CM0173) with 20% (v/v) glycerol in 1.5 mL Eppendorf tubes, followed by storage samples at −80 °C for further analysis.
2.3. Microbiological Identification and Antimicrobial Susceptibility Testing of CRKP Isolates
The identification and antimicrobial susceptibility testing (AST) procedures in the microbiology laboratory were carried out using the VITEK® 2 automated identification system (bioMérieux). Briefly, 3 mL of 0.45% NaCl sterile saline was transferred into a sterile clear plastic test tube. A sterile disposable plastic loop was used to transfer an adequate number of pure isolated colonies into the saline tube. The tube was vortexed to obtain a homogenized suspension with a density between 0.50 and 0.63 McFarland, checked by VITEK® 2 DensiCHEK™. The VITEK® 2 Gram-negative identification card (GN) from (bioMérieux) for Gram-negative bacteria was used to identify the isolates. Then, AST-N291 and AST-N292 cards (bioMérieux) were used for the AST, according to the manufacturer’s instructions of the VITEK® 2 system.
The AST cards were used to establish the antimicrobial’s minimum inhibitory concentration (MIC) and the interpretation (sensitive, intermediate, or resistant) of each bacterial isolate. This can establish the antibiogram of all tested CRKP isolates. The susceptibility tests for several antimicrobial agents were performed, including imipenem, meropenem, ertapenem, ampicillin, amoxicillin/clavulanic acid, ceftazidime, cefepime, amikacin, gentamicin, ciprofloxacin, levofloxacin, piperacillin/tazobactam, trimethoprim/sulfamethoxazole, colistin, and tigecycline.
2.4. Control Strains
In our study, we used K. pneumoniae NCTC 13440, 13438, 13442, and 13443 for VIM-1, KPC, OXA-48, and NDM-1. E. coli NCTC 13476 as a positive control for IMP. Additionally, E. coli NCTC 10418 was used as a negative control.
2.5. Molecular Detection of Carbapenemase Genes of CRKP Using Multiplex-PCR
The multiplex-PCR was used for the identification of common carbapenemase genes in K. pneumoniae. Five pairs of primers of the most common carbapenemase genes (blaNDM, blaOXA-48, blaKPC, blaIMP, and blaVIM) were obtained from Macrogen (Seoul, South Korea) (Table S1). The primers were optimized at the same annealing temperature, as Zarakolu et al. recommended (Zarakolu, Eser et al., 2016). First, the DNA template of all samples was prepared and extracted from the overnight cultures that were cultivated from the isolates stored in −80 °C for further multiplex-PCR analysis. Two colonies from the cultures were placed into 1.5 mL sterile Eppendorf tubes with 200 μL of sterile distilled water containing 10% Chelex 100 (HiMedia). The cell suspension was vortexed thoroughly and then dry-incubated at 96 °C for 15 min in Eppendorf ThermoMixer® C. The mixed suspension was cooled on an ice tray for 2 min and the centrifugation was then carried out for 5 min at 13,000× g to separate bacterial DNA from the other cell components. The resulting supernatant (2 μL) containing the bacterial DNA was used in the PCR reaction.
The multiplex-PCR was performed to amplify multiple gene targets in a single PCR reaction, as described by Zarakolu et al. (2016). The PCR reaction comprised 25 μL of the final volume of the PCR reaction, which was made up of 0.5 μL of sterile RNase-free water, 2 μL of the DNA template, 1 μL (2 μM) of each primer (blaNDM, blaOXA-48, blaKPC, blaIMP, and blaVIM genes, which have forward and reverse primers each, in a total of 10 primers), along with 12.5 μL of the Taq PCR master mix (Qiagen, Germany).
An Eppendorf Mastercycler® pro S thermal cycler was used for the multiplex PCR reaction. The PCR reaction was started as an initial denaturation at 95 °C for 5 min followed by 36 cycles. Each cycle involved three steps of 94 °C (45 s) for denaturation, 53 °C (45 s) for annealing, and 72 °C (1 min) for extension. These cycles were ended with a final extension step at 72 °C for 6 min. The electrophoresis system (BIO-RAD) with 1X TAE buffer was used to immerse the agarose gel. Then, 7 μL of the 6X DNA loading dye (NORGEN BIOTEK CORP, Thorold, ON, Canada) was added to the resulting PCR amplicon tube and mixed gently. Then, 8 μL of the PCR tubs was loaded to the immersed agarose gel, and 8 μL of the positive and negative controls was loaded. The gel was ran for 55 min at 100 Volts. A Syngene G:BOX system was used for gel electrophoresis to visualize the resulting PCR products for each testing bacteria.
2.6. Statistical Analysis
All resulting data were stored and analyzed using an appropriate statistical program (IBM SPSS® software, Version 24). The descriptive statistics of the data and variables are represented as percentages and frequencies.
3. Results
3.1. The Demographic Characteristics of Patients with CRKP Infections
A total of 191 K. pneumoniae isolates that showed low sensitivity to carbapenems were obtained from patients’ specimens that were sent to the microbiology laboratory at the KFAFH hospital in Jeddah, Saudi Arabia. A 54.5% (n = 104) of the specimens were collected from males and 45.5% (n = 87) were females. The ages of the patients ranged from 10 months to 97 years old, and the mean age was 68.14 ± 16.785 years. In addition, these results were divided into age groups: <1 year = 1% (n = 2), 1 to 19 years = 0% (n = 0), 20 to 39 years = 6.8% (n = 13), 40 to 59 years = 11.6% (n = 22), 60 to 79 years = 59.7% (n = 114), and 80 to 99 years = 20.9% (n = 40) (Table 1).

Table 1.
The demographic characteristics of patients (gender and age groups).
The majority (33%) of CRKP isolates were collected from sputum samples (n = 63), followed by urine with 25% (n = 48), blood with 17.3% (n = 33), wound swabs with 17.3% (n = 33), intravascular catheter tips with 4.2% (n = 8), body fluids with 1.6% (n = 3), and tissues with 1.6% (n = 3) (Figure S1). The samples were collected from several departments in KFAFH with 75.4% (n = 144) of the samples from inpatients (hospital-admitted patients) and 24.6% (n = 47) from outpatient departments (OPDs) and emergency room (ER) departments. The majority of isolates were recovered from medical wards (n = 78, 40.8%), about 23.6% of isolated were obtained from the intensive care unit (ICU) (n = 45), followed by 13.6% (n = 26) and 11% (n = 21) were isolated from the ER and the OPD, respectively. The minority of isolates were recovered from both the surgical ward and VIP suites (n = 7, 3.7%), followed by 2.6% of isolated were from the cardiac care unit (CCU) (n = 5), and only 1% of isolates (n = 2) were from the pediatric ward (Figure S2).
3.2. Antimicrobial Susceptibility Testing (AST)
All the K. pneumoniae isolates (n = 191) showed a full resistance to ceftazidime, cefepime, and piperacillin–tazobactam. Moreover, ciprofloxacin, levofloxacin, tobramycin, amikacin, and gentamicin showed high levels of resistance, with approximately 98.4%, 97.9%, 94.8%, 82.7%, and 77%, respectively. Furthermore, there was a low level of resistance represented by tigecycline and colistin, showing 14.1% and 37.7%, respectively.
Regarding the carbapenems, our results showed a high level of resistance for meropenem and ertapenem, showing 94.8% (n = 181) and 81.7% (n = 156), respectively. Additionally, the remaining isolates for the meropenem test account for 5.2% (n = 10), showing intermediate results. Moreover, 2.6% (n = 5) of ertapenem was intermediate and 15.7% (n = 30) was not tested. In contrast, imipenem showed 49.7% (n = 95) resistance results and 50.3% (n = 96) intermediate results for all the isolates with no susceptible result (Figure 1).
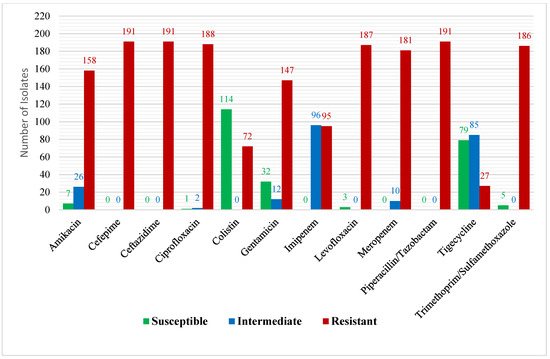
Figure 1.
Antimicrobial profiles of carbapenem-resistant K. pneumoniae isolates.
3.3. The Molecular Detection of Carbapenemase Genes of CRKP Isolates
Table 2 shows the descriptive frequency of genes, revealed by gel electrophoresis (Figure S3). The blaOXA-48 gene was reported in 157 (82.2%), which represents the highest percentage compared to other carbapenemases. It is also seen that the blaNDM gene was identified in 27 (14.1%) isolates. The blaVIM and blaKPC genes were reported in one isolate (0.5%) each, whereas all tested isolates were negative for the blaIMP gene. Additionally, fifteen K. pneumoniae isolates showed to have both blaOXA-48 and blaNDM genes (7.9%) (Table 3), and only one isolate was positive for the blaNDM and blaKPC genes. There were five K. pneumoniae isolates showed negative results to all tested carbapenemases genes.

Table 2.
The descriptive analysis of the carbapenemase genes from CRKP isolates.

Table 3.
The MIC of K. pneumoniae isolates that co-harbored blaOXA-48 and blaNDM carbapenemase genes.
3.4. The Correlation between Carbapenemase Genes and Antimicrobial Profiles
Table 4 shows the Spearman’s correlation coefficient which shows the degree of relationship which exists between genes and antimicrobials. This equally provides the direction of relationship between the variables in the study. As evidenced, it is clear that the blaOXA-48 gene and antimicrobials revealed a weak positive correlation coefficient (0.143) that is statistically significant at the 0.05 alpha level (p-value 0.049 < 0.05). Secondly, the blaNDM gene and antimicrobials revealed a weak negative correlation coefficient (−0.072) that is not statistically significant at the 0.05 alpha level (p-value 0.325 > 0.05). Thirdly, the blaVIM gene and antimicrobials revealed a weak negative correlation coefficient (−0.032) that is not statistically significant at the 0.05 alpha level (p-value 0.662 > 0.05). Lastly, the blaKPC gene and antimicrobials revealed a weak negative correlation coefficient (−0.107) that is not statistically significant at the 0.05 alpha level (p-value 0.141 > 0.010).

Table 4.
The correlation coefficient between carbapenemase genes and antimicrobials.
3.5. The Correlation between Carbapenemase Genes and Age Groups, Gender, Specimen
Types and Hospital Departments
Table 5 shows the Spearman’s correlation coefficient which shows the degree of relationship existing between genes and patients age groups, gender, specimen types, and hospital departments. As evidenced, the blaOXA-48 gene revealed weak positive correlation coefficients with age and gender (0.095 and 0.096, respectively) which are not statistically significant at the 0.05 alpha level (p-value 0.192 and 0.187 > 0.05), while the blaOXA-48 showed negative correlation coefficients with the sample type and ward (−0.148 and −0.212, respectively) which are statistically significant (p-value 0.041 and 0.003 < 0.05). Secondly, the blaNDM gene revealed weak negative correlation coefficients with age and gender (−0.086 and −0.009, respectively) which are not statistically significant at the 0.05 alpha level (p-value 0.237 and 0.90 > 0.05), while the blaNDM gene showed positive correlation coefficients with the sample type and ward (0.113 and 0.148, respectively), showing that the relationship between the sample type and blaNDM is not significant, while ward and blaNDM showed a significant relationship to be significant. Thirdly, the blaVIM gene revealed weak negative correlation coefficients with age, gender, and ward (−0.104, −0.066, and −0.078, respectively) that are not statistically significant at the 0.05 alpha level (p-value 0.153, 0.362, and 0.285 > 0.05), while the blaVIM gene showed a weak positive correlation with the sample type (0.092) that is also not statistically significance. Lastly, the blaKPC gene revealed weak negative correlation coefficients with age and gender (−0.002 and −0.066, respectively) that is not statistically significant at the 0.05 alpha level (p-value 0.975 and 0.362 > 0.05), and the blaKPC gene showed positive correlation coefficients with the sample type and ward (0.037 and 0.052, respectively) that are not statistically significant at the 0.05 alpha level (p-value 0.608 and 0.473 > 0.05).

Table 5.
The correlation coefficient between carbapenemase genes and patients’ age groups, gender, specimen types, and hospital departments.
4. Discussion
The emergence of nosocomial carbapenem-resistant K. pneumoniae (CRKP)-associated infections have become major threat to health care system worldwide. This has led to a significant increase in morbidity and mortality rates in health care settings globally. The CRKP can hydrolyze carbapenems by producing many carbapenemase enzymes such as KPC, NDM, IMP, VIM, and OXA-48 [5,9,29]. These enzymes have been identified among K. pneumoniae isolates and have been reported in different countries [10,29]. At the national level, there are some studies that have identified the dissemination of CRKP, particularly OXA-48- and NDM-producing isolates in Saudi Arabia [30,31,32]. There is no doubt that understanding the epidemiology behavior of CRKP using a reliable molecular technology can enhance the development of effective control measures to control the spread of such isolates.
The western region of Saudi Arabia is a global destination for millions of Muslim pilgrims visiting holy places from all around the world every year. To our knowledge, in the western region of Saudi Arabia, there are no sufficient data on the prevalence of CRKP and its carbapenem-resistant determinants. Therefore, this study was intended to determine the prevalence of CRKP and identify the most common carbapenemase genes in K. pneumoniae isolated from KFAFH, which is one of the largest hospitals in Jeddah, the western region of Saudi Arabia. In addition, this study may support the detection of demographic changes between CRKP isolates recovered from Jeddah and other cities of Saudi Arabia that were conducted in comparable studies.
In this study, one of the most significant risk factors associated with CRKP was patient old age considering that more than 80% of the CRKP isolates were obtained from elderly patients over 60 years old. Conversely, CRKP isolates in patients less than 20 years old were only 1% of the total CRKP isolates. This finding is consistent with (Al-Zahrani and Alasiri 2018) findings [30] which significantly showed that old age is a risk factor associated with CRKP isolation in their study area. This finding was also previously reported by Jiao et al. (2015) who found that older age was a risk factor for CRKP infection and colonization [28].
Furthermore, the majority of CRKP samples in this study were isolated from hospital-admitted patients. Medical ward patients were the most susceptible patients to CRKP infections (40.8% of all isolates and 54.2% of the inpatients), followed by intensive care unit (ICU) patients (23.6% of all isolates and 31.25% of the inpatients) and ER patients (13.6% of all isolates). Similarly, other studies have reported that CRKP isolates were highly associated with ICU-admitted patients [30,31]. However, admitted patients can be transferred to other wards based on their health status. Although the observation of the current study does not clearly distinguish whether CRKP isolates have hospital or community origin, these results may support the hypothesis that MDR bacteria, including CRKP, have become common residents in various hospital environments, particularly ICUs. The automated method (Vitek 2 system) was used for antimicrobial susceptibility testing (AST) as a routine test in KFAFH to determine the antimicrobial susceptibility pattern and MICs of K. pneumoniae isolates. All isolates were subjected to the antimicrobial susceptibility test against carbapenems, mainly imipenem and meropenem. Notably, meropenem showed a higher resistance level (94.8%) than imipenem (49.7%). However, no susceptible isolate was observed for both antimicrobials (meropenem and imipenem). In contrast, a previous study conducted in Riyadh hospitals revealed that around 21 of K. pneumoniae isolates displayed high level of resistance rates (42.9%) for both imipenem and meropenem [33]. Imipenem susceptibility results in their study showed slight similarities with our findings. However, our isolates showed higher meropenem resistance rates (94.8%), compared to the study of Al-Agamy et al. (2018) [33], which reported 42.9%. Similarly, Al-Zahrani and Alasiri showed that 52% of 54 CRKP isolates (recovered from the Southern region of Saudi Arabia-Asir province) were imipenem-resistant and 59% were meropenem-resistant. They also reported that five isolates of CRKP were meropenem-susceptible [30]. In the current study, all 191 isolates were highly resistant to both third- and fourth-generation cephalosporins (ceftazidime and cefepime), and piperacillin-tazobactam. These results are in agreement with those obtained by Zarakolu et al. in Turkey and Al-Zahrani and Alasiri. In Saudi Arabia, carbapenemase enzymes, such as KPC, NDM, IMP, VIM, and OXA-48, have been described as the most important types among Enterobacteriaceae, including K. pneumoniae [30,31]. These enzymes have recently been identified and reported in many countries around the world. For example, NDM- and OXA-48-producing K. pneumoniae were firstly identified in India in 2009 and Turkey in 2008, respectively, and then disseminated globally [33,34]. Thereafter, OXA-48- and NDM-positive isolates of K. pneumoniae have been isolated and described as the major carbapenemase determinants in Enterobacteriaceae from many Gulf countries, including Saudi Arabia [29,30,31,32,33,34,35,36]. In our study, we also found that most CRKP isolates 82.2% (n = 157) were positive for OXA-48, followed by 14.1% (n = 27) of isolates harboring the blaNDM. These findings could support previous research which links the wide spread of OXA-48- and NDM-producing K. pneumoniae in our countries, and the receiving huge number of visitors, pilgrims, and migrant workers from OXA-48- and NDM-endemic countries, such as Pakistan, Turkey, and India (Al-Zahrani & Alasiri, 2018). In the current study, only one VIM-positive and one KPC-positive K. pneumoniae were isolated, and no IMP-positive K. pneumoniae isolate was detected. Althoug, VIM-positive K. pneumoniae has been reported in some Mediterranean and Middle East countries, including Saudi Arabia (Tzouvelekis et al., 2012), the isolation of VIM-producing K. pneumoniae in Saudi Arabia remains low. Moreover, KPC-producing K. pneumoniae is predominant in North and South America, and only a few isolates have been reported in Saudi Arabia [37].
The most important clinically relevant finding was that fifteen (7.9%) CRKP isolates co-harbored blaOXA-48 and blaNDM carbapenemase genes. This can be explained as plasmids that harbor blaNDM have been linked to carry other carbapenemase genes (e.g., blaVIM, blaOXA-48) in previous studies [30,35]. Such isolates are more likely to exhibit high level of resistance to carbapenems and their infections are usually associated with high mortality rate [31,38,39].
In conclusion, CRKP isolates are among the most important nosocomial pathogens, leading to outbreaks in healthcare settings around the world. There is no doubt that the accurate identification of CRKP is very necessary to control the spread of such infections. Therefore, the molecular detection of CRKP and other carbapenemase producers are considered the fastest and most accurate method for the rapid characterization of these MDR pathogens. The findings of this study suggest that it is important to screen any isolate that showed reduced susceptibility to carbapenems due to carbapenemase production using phenotypic and molecular detection methods.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres13040054/s1, Table S1: Oligonucleotide sequences of primers used in the study.; Figure S1: Distribution of various specimen types collected.; Figure S2: Distribution of hospital departments from which samples were collected.; Figure S3: Representation of the multiplex-PCR patterns of various carbapenemase genes of K. pneumoniae isolates shown on gel electrophoresis.
Author Contributions
W.A. is the principle investigator (PI) and supervisor for the MSc student A.A.-J., who wrote and edited the draft of the manuscript. A.A.-J. is a MSc research student who performed all the experiments under the supervision of W.A., and I.A.-Z. wrote the thesis under supervision. I.A.-Z. is the Co-PI and supervisor for the MSc student A.A.-J. All authors have read and agreed to the published version of the manuscript.
Funding
The project was funded by Deanship of scientific research, King Abdulaziz University, Jeddah, Saudi Arabia. The grant number: G-615-290-38.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of KFAFH.
Conflicts of Interest
The authors declare that they have no conflicts of interest related to this study.
References
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and mechanisms of resistance of extensively drug resistant Gram-negative bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef]
- Chung, P.Y. The emerging problems of Klebsiella pneumoniae infections: Carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 2016, 363, fnw219. [Google Scholar] [CrossRef]
- Vardakas, K.Z.; Matthaiou, D.K.; Falagas, M.E.; Antypa, E.; Koteli, A.; Antoniadou, E. Characteristics, risk factors and outcomes of carbapenem-resistant Klebsiella pneumoniae infections in the intensive care unit. J. Infect. 2015, 70, 592–599. [Google Scholar] [CrossRef]
- Viau, R.; Frank, K.M.; Jacobs, M.R.; Wilson, B.; Kaye, K.; Donskey, C.J.; Bonomo, R.A. Intestinal carriage of carbapenemase-producing organisms: Current status of surveillance methods. Clin. Microbiol. Rev. 2016, 29, 1–27. [Google Scholar] [CrossRef]
- Pitout, J.D.; Nordmann, P.; Poirel, L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef]
- Lee, C.-R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef]
- Lee, C.-R.; Park, K.S.; Lee, J.H.; Jeon, J.H.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. The threat of carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-HvKP). Biomed. Res. 2018, 29, 2438–2441. [Google Scholar]
- De Rosa, F.G.; Corcione, S.; Cavallo, R.; Di Perri, G.; Bassetti, M. Critical issues for Klebsiella pneumoniae KPC-carbapenemase producing K. pneumoniae infections: A critical agenda. Future Microbiol. 2015, 10, 283–294. [Google Scholar] [CrossRef]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef]
- Alhazmi, W.A. Review on the Carbapenem Resistance Mechanisms of Klebsiella pneumoniae. J. Pharm. Res. Int. 2021, 33, 352–357. [Google Scholar] [CrossRef]
- Chen, L.; Mathema, B.; Chavda, K.D.; DeLeo, F.R.; Bonomo, R.A.; Kreiswirth, B.N. Carbapenemase-producing Klebsiella pneumoniae: Molecular and genetic decoding. Trends Microbiol. 2014, 22, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L.; Walsh, T.R.; Livermore, D.M. The emerging NDM carbapenemases. Trends Microbiol. 2011, 19, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Makena, A.; Düzgün, A.Ö.; Brem, J.; McDonough, M.A.; Rydzik, A.M.; Abboud, M.I.; Schofield, C.J. Comparison of Verona integron-borne metallo-β-lactamase (VIM) variants reveals differences in stability and inhibition profiles. Antimicrob. Agents Chemother. 2016, 60, 1377–1384. [Google Scholar] [CrossRef]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Severin, J. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.; Peirano, G.; Kock, M.M.; Strydom, K.-A.; Matsumura, Y. The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 2019, 33, e00102-19. [Google Scholar] [CrossRef]
- Zarakolu, P.; Eser, O.K.; Aladag, E.; Al-Zahrani, I.A.; Day, K.M.; Atmaca, O.; Akova, M. Epidemiology of carbapenem-resistant Klebsiella pneumoniae colonization: A surveillance study at a Turkish university hospital from 2009 to 2013. Diagn. Microbiol. Infect. Dis. 2016, 85, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Seibert, G.; Hörner, R.; Meneghetti, B.H.; Righi, R.A.; Forno, N.L.F.D.; Salla, A. Nosocomial infections by Klebsiella pneumoniae carbapenemase producing enterobacteria in a teaching hospital. Einstein 2014, 12, 282–286. [Google Scholar] [CrossRef][Green Version]
- Dortet, L.; Poirel, L.; Nordmann, P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. BioMed Res. Int. 2014, 2014, 249856. [Google Scholar] [CrossRef]
- Erdem, F.; Oncul, O.; Aktas, Z. Characterization of resistance genes and polymerase chain reaction-based replicon typing in carbapenem-resistant Klebsiella pneumoniae. Microb. Drug Resist. 2019, 25, 551–557. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 2014, 20, 821–830. [Google Scholar] [CrossRef]
- Nodari, C.S.; Gales, A.C.; Barth, A.L.; Magagnin, C.M.; Zavascki, A.P.; Carvalhaes, C.G. Detection of OXA-370 directly from rectal swabs and blood culture vials using an immunochromatographic assay. J. Microbiol. Methods 2017, 139, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.; Gupta, R.; Raut, S.S.; Nataraj, G.; Mehta, P.R. Carba NP as a simpler, rapid, cost-effective, and a more sensitive alternative to other phenotypic tests for detection of carbapenem resistance in routine diagnostic laboratories. J. Lab. Physicians 2017, 9, 100. [Google Scholar] [CrossRef]
- Wareham, D.W.; Phee, L.M.; Abdul Momin, M.H.F. Direct detection of carbapenem resistance determinants in clinical specimens using immunochromatographic lateral flow devices. J. Antimicrob. Chemother. 2018, 73, 1997–1998. [Google Scholar] [CrossRef] [PubMed]
- Pavelkovich, A.; Balode, A.; Edquist, P.; Egorova, S.; Ivanova, M.; Kaftyreva, L.; Naaber, P. Detection of carbapenemase-producing enterobacteriaceae in the baltic countries and st. Petersburg area. BioMed Res. Int. 2014, 2014, 548960. [Google Scholar] [CrossRef] [PubMed]
- Al-Zahrani, I.A. Routine detection of carbapenem-resistant Gram-negative bacilli in clinical laboratories: A review of current challenges. Saudi Med. J. 2018, 39, 861. [Google Scholar] [CrossRef]
- Hoxha, A.; Kärki, T.; Giambi, C.; Montano, C.; Sisto, A.; Bella, A.; Pedna, M. Attributable mortality of carbapenem-resistant Klebsiella pneumoniae infections in a prospective matched cohort study in Italy, 2012–2013. J. Hosp. Infect. 2016, 92, 61–66. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017, 215 (Suppl. 1), S28–S36. [Google Scholar] [CrossRef]
- Jiao, Y.; Qin, Y.; Liu, J.; Li, Q.; Dong, Y.; Shang, Y.; Huang, Y.; Liu, R. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection/colonization and predictors of mortality: A retrospective study. Pathog. Glob. Health 2015, 109, 68–74. [Google Scholar] [CrossRef]
- Girmenia, C.; Serrao, A.; Canichella, M. Epidemiology of carbapenem resistant Klebsiella pneumoniae infections in mediterranean countries. Mediterr. J. Hematol. Infect. Dis. 2016, 8, e2016032. [Google Scholar] [CrossRef]
- Shibl, A.; Al-Agamy, M.; Memish, Z.; Senok, A.; Khader, S.A.; Assiri, A. The emergence of OXA-48-and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 2013, 17, e1130–e1133. [Google Scholar] [CrossRef]
- Al-Zahrani, I.A.; Alasiri, B.A. The emergence of carbapenem-resistant Klebsiella pneumoniae isolates producing OXA-48 and NDM in the Southern (Asir) province, Saudi Arabia. Saudi Med. J. 2018, 39, 23. [Google Scholar] [CrossRef] [PubMed]
- Uz Zaman, T.; Alrodayyan, M.; Albladi, M.; Aldrees, M.; Siddique, M.I.; Aljohani, S.; Balkhy, H.H. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect. Dis. 2018, 18, 205. [Google Scholar] [CrossRef] [PubMed]
- Quale, J.; Spelman, D.; Hooper, D.; Bloom, A. Overview of Carbapenemase Producing Gram-Negative Bacilli; UpToDate: Waltham, MA, USA, 2016; Volume 21. [Google Scholar]
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Jamal, W.Y.; Albert, M.J.; Rotimi, V.O. High prevalence of New Delhi metallo-β-lactamase-1 (NDM-1) producers among carbapenem-resistant Enterobacteriaceae in Kuwait. PLoS ONE 2016, 11, e0152638. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; Aljallal, A.; Radwan, H.H.; Shibl, A.M. Characterization of carbapenemases, ESBLs, and plasmid-mediated quinolone determinants in carbapenem-insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J. Infect. Public Health 2018, 11, 64–68. [Google Scholar] [CrossRef]
- Al-Qadheeb, N.S.; Althawadi, S.; Alkhalaf, A.; Hosaini, S.; Alrajhi, A.A. Evolution of tigecycline resistance in Klebsiella pneumoniae in a single patient. Ann. Saudi Med. 2010, 30, 404–407. [Google Scholar] [CrossRef][Green Version]
- Lau, M.Y.; Teng, F.E.; Chua, K.H.; Ponnampalavanar, S.; Chong, C.W.; Abdul Jabar, K.; The, C.S.J. Molecular Characterization of Carbapenem Resistant Klebsiella pneumoniae in Malaysia Hospital. Pathogens 2021, 10, 279. [Google Scholar] [CrossRef]
- Lumbreras-Iglesias, P.; Rodicio, M.R.; Valledor, P.; Suárez-Zarracina, T.; Fernández, J. High-Level Carbapenem Resistance among OXA-48-Producing Klebsiella pneumoniae with Functional OmpK36 Alterations: Maintenance of Ceftazidime/Avibactam Susceptibility. Antibiotics 2021, 10, 1174. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).