TNNC1 Gene Mutation in Ebstein’s Anomaly and Left Ventricular Hypertrabeculation: A Case Report of a New Causative Mutation?
Abstract
1. Introduction
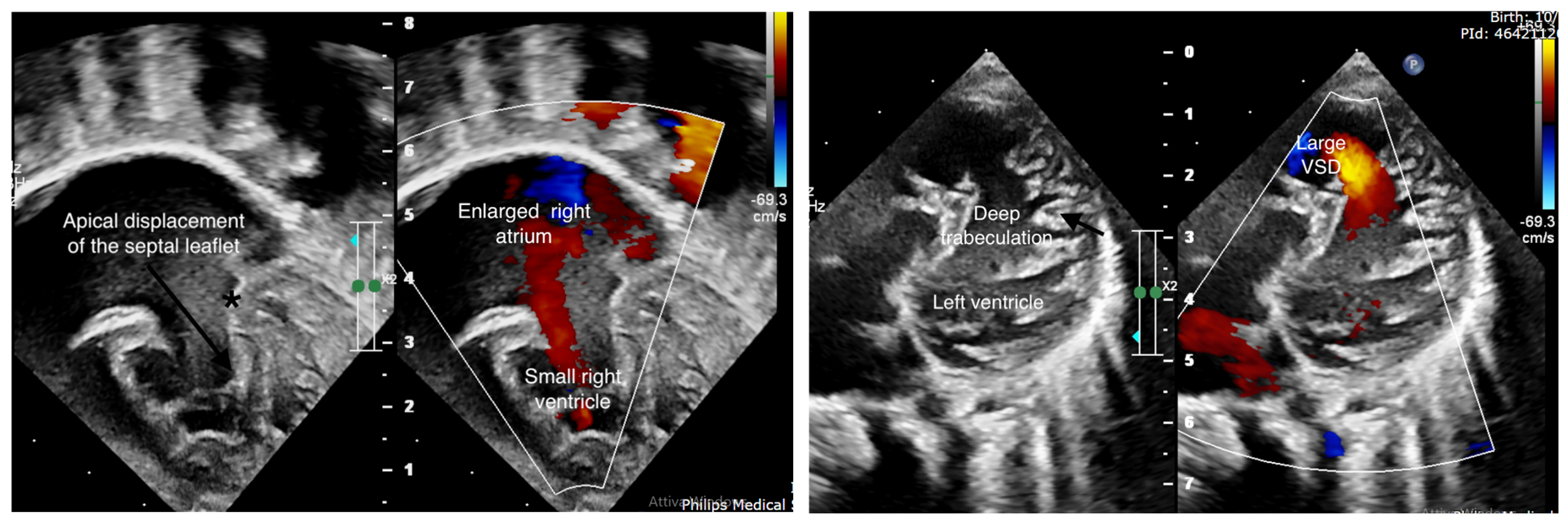
2. Case Report
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| C | Epicardial compacted layer |
| Ca2+ | Calcium |
| cTnC | Troponin C1 |
| EA | Ebstein’s anomaly |
| LV | Left ventricle |
| LVNC | Left ventricular noncompaction |
| NC | Endocardial noncompacted layer |
References
- Lupo, P.J.; Langlois, P.H.; Mitchell, L.E. Epidemiology of Ebstein anomaly: Prevalence and patterns in Texas, 1999-2005. Am. J. Med. Genet. A 2011, 155A, 1007–1014. [Google Scholar] [CrossRef]
- Attenhofer Jost, C.H.; Connolly, H.M.; Dearani, J.A.; Edwards, W.D.; Danielson, G.K. Ebstein’s anomaly. Circulation 2007, 115, 277–285. [Google Scholar] [CrossRef]
- Thareja, S.K.; Frommelt, M.A.; Lincoln, J.; Lough, J.W.; Mitchell, M.E.; Tomita-Mitchell, A. A Systematic Review of Ebstein’s Anomaly with Left Ventricular Noncompaction. J. Cardiovasc. Dev. Dis. 2022, 9, 115. [Google Scholar] [CrossRef]
- Krieger, E.V.; Valente, A.M. Diagnosis and management of ebstein anomaly of the tricuspid valve. Curr. Treat. Options Cardiovasc. Med. 2012, 14, 594–607. [Google Scholar] [CrossRef]
- Vermeer, A.M.; van Engelen, K.; Postma, A.V.; Baars, M.J.; Christiaans, I.; De Haij, S.; Klaassen, S.; Mulder, B.J.; Keavney, B. Ebstein anomaly associated with left ventricular noncompaction: An autosomal dominant condition that can be caused by mutations in MYH7. Am. J. Med. Genet. C Semin. Med. Genet. 2013, 163C, 178–184. [Google Scholar] [CrossRef]
- Aras, D.; Tufekcioglu, O.; Ergun, K.; Ozeke, O.; Yildiz, A.; Topaloglu, S.; Deveci, B.; Sahin, O.; Kisacik, H.L.; Korkmaz, S. Clinical features of isolated ventricular noncompaction in adults long-term clinical course, echocardiographic properties, and predictors of left ventricular failure. J. Card. Fail. 2006, 12, 726–733. [Google Scholar] [CrossRef]
- Stähli, B.E.; Gebhard, C.; Biaggi, P.; Klaassen, S.; Valsangiacomo Buechel, E.; Attenhofer Jost, C.H.; Jenni, R.; Tanner, F.C.; Greutmann, M. Left ventricular noncompaction: Prevalence in congenital heart disease. Int. J. Cardiol. 2013, 167, 2477–2481. [Google Scholar] [CrossRef]
- Budde, B.S.; Binner, P.; Waldmüller, S.; Höhne, W.; Blankenfeldt, W.; Hassfeld, S.; Brömsen, J.; Dermintzoglou, A.; Wieczorek, M.; May, E.; et al. Noncompaction of the ventricular myocardium is associated with a de novo mutation in the beta-myosin heavy chain gene. PLoS ONE 2007, 2, e1362. [Google Scholar] [CrossRef]
- Nijak, A.; Alaerts, M.; Kuiperi, C.; Corveleyn, A.; Suys, B.; Paelinck, B.; Saenen, J.; Van Craenenbroeck, E.; Van Laer, L.; Loeys, B.; et al. Left ventricular noncompaction with Ebstein anomaly attributed to a TPM1 mutation. Eur. J. Med. Genet. 2018, 61, 8–10. [Google Scholar] [CrossRef]
- Sicko, R.J.; Browne, M.L.; Rigler, S.L.; Druschel, C.M.; Liu, G.; Fan, R.; Romitti, P.A.; Caggana, M.; Kay, D.M.; Brody, L.C.; et al. Genetic Variants in Isolated Ebstein Anomaly Implicated in Myocardial Development Pathways. PLoS ONE 2016, 11, e0165174. [Google Scholar] [CrossRef]
- Neu, A.; Eiselt, M.; Paul, M.; Sauter, K.; Stallmeyer, B.; Isbrandt, D.; Schulze-Bahr, E. A homozygous SCN5A mutation in a severe, recessive type of cardiac conduction disease. Hum. Mutat. 2010, 31, E1609–E1621. [Google Scholar] [CrossRef]
- Carlston, C.M.; Bleyl, S.B.; Andrews, A.; Meyers, L.; Brown, S.; Bayrak-Toydemir, P.; Bale, J.F.; Botto, L.D. Expanding the genetic and clinical spectrum of the NONO-associated X-linked intellectual disability syndrome. Am. J. Med. Genet. 2019, 179, 792–796. [Google Scholar] [CrossRef]
- Samudrala, S.S.K.; North, L.M.; Stamm, K.D.; Earing, M.G.; Frommelt, M.A.; Willes, R.; Tripathi, S.; Dsouza, N.R.; Zimmermann, M.T.; Mahnke, D.K.; et al. Novel KLHL26 variant associated with a familial case of Ebstein’s anomaly and left ventricular noncompaction. Mol. Genet. Genom. Med. 2020, 8, e1152. [Google Scholar] [CrossRef]
- McGee, M.; Warner, L.; Collins, N. Ebstein’s Anomaly, Left Ventricular Noncompaction, and Sudden Cardiac Death. Case Rep. Cardiol. 2015, 2015, 854236. [Google Scholar] [CrossRef]
- Pignatelli, R.H.; Texter, K.M.; Denfield, S.W.; Grenier, M.A.; Altman, C.A.; Ayres, N.A.; Chandra-Bose Reddy, S. LV Noncompaction in Ebstein’s anomaly in infants and outcomes. JACC Cardiovasc. Imaging 2014, 7, 207. [Google Scholar] [CrossRef]
- Hirono, K.; Hata, Y.; Ibuki, K.; Yoshimura, N. Familial Ebstein’s anomaly, left ventricular noncompaction, and ventricular septal defect associated with an MYH7 mutation. J. Thorac. Cardiovasc. Surg. 2014, 148, e223–e226. [Google Scholar] [CrossRef]
- Pavlova, M.; Fouron, J.C.; Drblik, S.P.; van Doesburg, N.H.; Bigras, J.L.; Smallhorn, J.; Harder, J.; Robertson, M. Factors affecting the prognosis of Ebstein’s anomaly during fetal life. Am. Heart J. 1998, 135, 1081–1085. [Google Scholar] [CrossRef]
- Torigoe, F.; Ishida, H.; Ishii, Y.; Ishii, R.; Narita, J.; Kawazu, Y.; Kayatani, F.; Inamura, N. Fetal echocardiographic prediction score for perinatal mortality in tricuspid valve dysplasia and Ebstein’s anomaly. Ultrasound Obstet. Gynecol. 2020, 55, 226–232. [Google Scholar] [CrossRef]
- Stöllberger, C.; Wegner, C.; Finsterer, J. Fetal Ventricular Hypertrabeculation/Noncompaction: Clinical Presentation, Genetics, Associated Cardiac and Extracardiac Abnormalities and Outcome. Pediatr. Cardiol. 2015, 36, 1319–1326. [Google Scholar] [CrossRef]
- Zhang, W.; Dai, X.; Liu, H.; Li, L.; Zhou, S.; Zhu, Q.; Chen, J. Case report: Prenatal diagnosis of fetal noncompaction cardiomyopathy with bradycardia accompanied by de novo CALM2 mutation. Front. Pediatr. 2022, 10, 1012600. [Google Scholar] [CrossRef]
- Gong, H.; Lyu, X.; Wang, Q.; Hu, M.; Zhang, X. Endothelial to mesenchymal transition in the cardiovascular system. Life Sci. 2017, 184, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, J.; Yutzey, K.E. Molecular and developmental mechanisms of congenital heart valve disease. Birth Defects Res. A Clin. Mol. Teratol. 2011, 91, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Postma, A.V.; van Engelen, K.; van de Meerakker, J.; Rahman, T.; Probst, S.; Baars, M.J.; Bauer, U.; Pickardt, T.; Sperling, S.R.; Berger, F.; et al. Mutations in the sarcomere gene MYH7 in Ebstein anomaly. Circ. Cardiovasc. Genet. 2011, 4, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Li, M.X.; Hwang, P.M. Structure and function of cardiac troponin C (TNNC1): Implications for heart failure, cardiomyopathies, and troponin modulating drugs. Gene 2015, 571, 153–166. [Google Scholar] [CrossRef]
- Reinoso, T.R.; Landim-Vieira, M.; Shi, Y.; Johnston, J.R.; Chase, P.B.; Parvatiyar, M.S.; Landstrom, A.P.; Pinto, J.R.; Tadros, H.J. A comprehensive guide to genetic variants and post-translational modifications of cardiac troponin C. J. Muscle Res. Cell Motil. 2021, 42, 323–342. [Google Scholar] [CrossRef]
- Landim-Vieira, M.; Johnston, J.R.; Ji, W.; Mis, E.K.; Tijerino, J.; Spencer-Manzon, M.; Jeffries, L.; Hall, E.K.; Panisello-Manterola, D.; Khokha, M.K.; et al. Familial dilated cardiomyopathy associated with a novel combination of compound heterozygous TNNC1 variants. Front. Physiol. 2020, 10, 1612. [Google Scholar] [CrossRef]
- Ploski, R.; Rydzanicz, M.; Ksiazczyk, T.M.; Franaszczyk, M.; Pollak, A.; Kosinska, J.; Michalak, E.; Stawinski, P.; Ziolkowska, L.; Bilinska, Z.T.; et al. Evidence for troponin C (TNNC1) as a gene for autosomal recessive restrictive cardiomyopathy with fatal outcome in infancy. Am. J. Med. Genet. A 2016, 170, 3241–3248. [Google Scholar] [CrossRef]
- Tadros, H.J.; Life, C.S.; Garcia, G.; Pirozzi, E.; Jones, E.G.; Datta, S.; Parvatiyar, M.S.; Chase, P.B.; Allen, H.D.; Kim, J.J.; et al. Meta-analysis of cardiomyopathy-associated variants in troponin genes identifies loci and intragenic hot spots that are associated with worse clinical outcomes. J. Mol. Cell Cardiol. 2020, 142, 118–125. [Google Scholar] [CrossRef]
- Wu, H.; Lee, J.; Vincent, L.G.; Wang, Q.; Gu, M.; Lan, F.; Churko, J.M.; Sallam, K.I.; Matsa, E.; Sharma, A.; et al. Epigenetic Regulation of Phosphodiesterases 2A and 3A Underlies Compromised β-Adrenergic Signaling in an iPSC Model of Dilated Cardiomyopathy. Cell Stem cell. 2015, 17, 89–100. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raso, I.; Chillemi, C.; Prontera, G.; Laoreti, A.; Cattaneo, E.; Calcaterra, V.; Zuccotti, G.V.; Mannarino, S. TNNC1 Gene Mutation in Ebstein’s Anomaly and Left Ventricular Hypertrabeculation: A Case Report of a New Causative Mutation? Cardiogenetics 2025, 15, 24. https://doi.org/10.3390/cardiogenetics15030024
Raso I, Chillemi C, Prontera G, Laoreti A, Cattaneo E, Calcaterra V, Zuccotti GV, Mannarino S. TNNC1 Gene Mutation in Ebstein’s Anomaly and Left Ventricular Hypertrabeculation: A Case Report of a New Causative Mutation? Cardiogenetics. 2025; 15(3):24. https://doi.org/10.3390/cardiogenetics15030024
Chicago/Turabian StyleRaso, Irene, Claudia Chillemi, Giorgia Prontera, Arianna Laoreti, Elisa Cattaneo, Valeria Calcaterra, Gian Vincenzo Zuccotti, and Savina Mannarino. 2025. "TNNC1 Gene Mutation in Ebstein’s Anomaly and Left Ventricular Hypertrabeculation: A Case Report of a New Causative Mutation?" Cardiogenetics 15, no. 3: 24. https://doi.org/10.3390/cardiogenetics15030024
APA StyleRaso, I., Chillemi, C., Prontera, G., Laoreti, A., Cattaneo, E., Calcaterra, V., Zuccotti, G. V., & Mannarino, S. (2025). TNNC1 Gene Mutation in Ebstein’s Anomaly and Left Ventricular Hypertrabeculation: A Case Report of a New Causative Mutation? Cardiogenetics, 15(3), 24. https://doi.org/10.3390/cardiogenetics15030024







