Association Between Angiotensinogen Gene M235T and Renin–Angiotensin System Insertion/Deletion Variants and Risk of Cardiovascular Disease in North African and Middle Eastern Populations: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Outcome Measurement
2.3. Data Sources and Search Strategy
2.4. Data Extraction
2.5. Quality Assessment
2.6. Statistical Analysis
3. Results
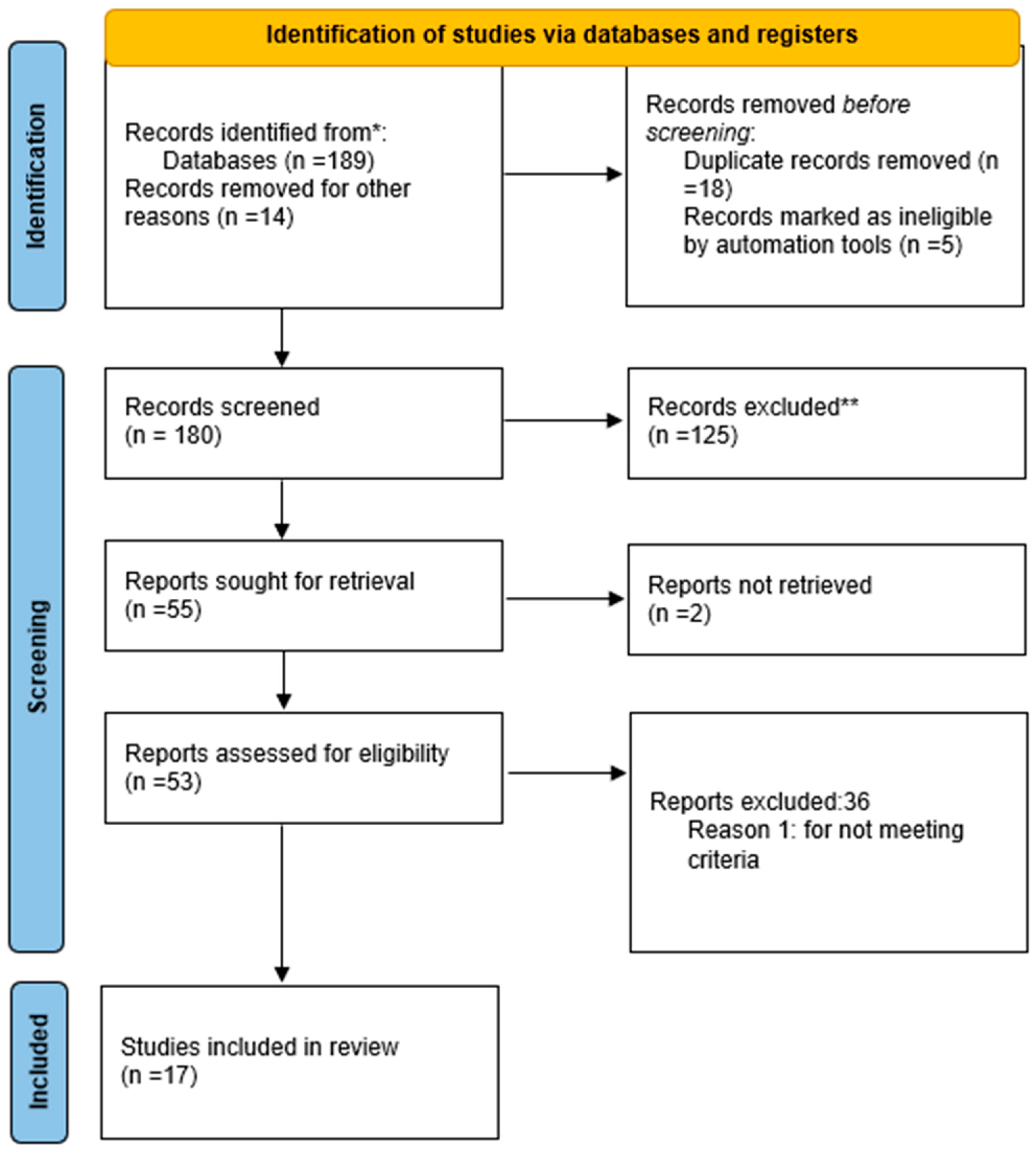
3.1. Study Selection
3.2. Study Characteristics
3.3. Risk of Bias
3.4. Heterogeneity Test
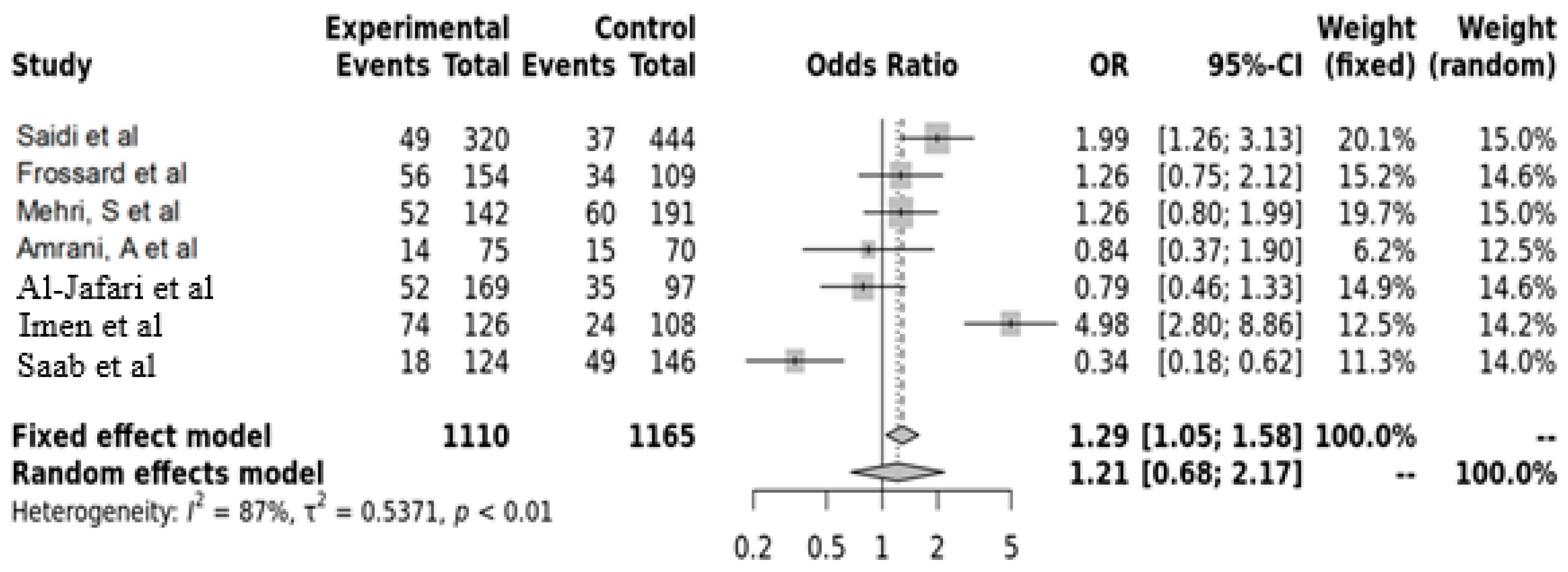
3.5. AGT Gene MT237 Polymorphism with Hypertension
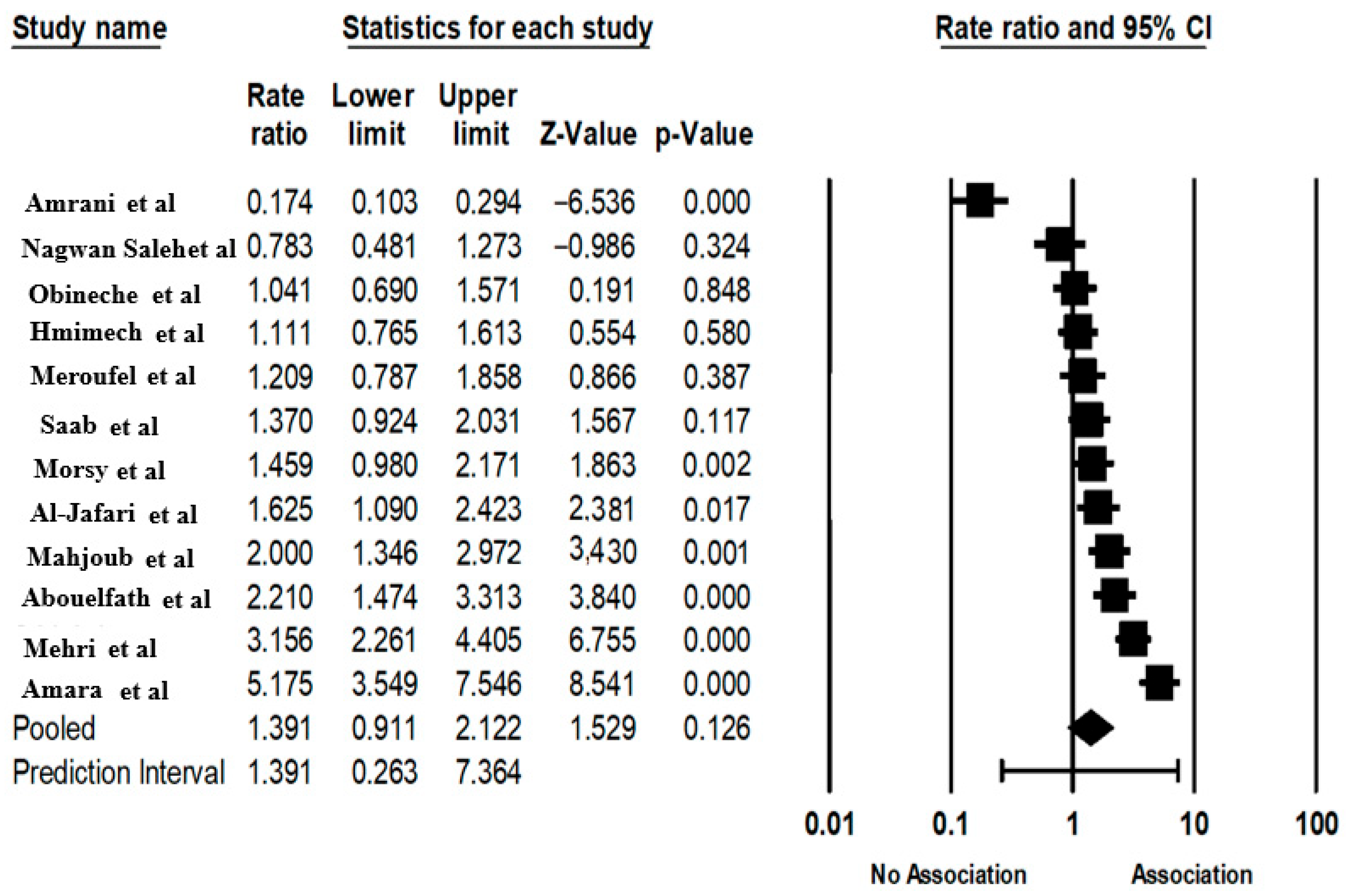
3.6. The ACE Gene INSERTION/DELETION Polymorphism and CVD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CVD: | cardiovascular disease |
| AGT: | angiotensinogen gene |
| ACE: | angiotensin converter enzyme |
| CAD: | coronary artery disease |
References
- Malekpour, M.-R.; Abbasi-Kangevari, M.; Ghamari, S.-H.; Khanali, J.; Heidari-Foroozan, M.; Moghaddam, S.S.; Azangou-Khyavy, M.; Rezazadeh-Khadem, S.; Rezaei, N.; Shobeiri, P.; et al. The burden of metabolic risk factors in North Africa and the Middle East, 1990–2019. eClinicalMedicine 2023, 60, 102022. [Google Scholar] [CrossRef] [PubMed]
- Ben Abdelaziz, A.; Melki, S.; Ben Abdelaziz, A.; Ben Salem, K.; Soulimane, A.; Serhier, Z.; Dahdi, S.A. Profile and evolution of the Global Burden of Morbidity in the Maghreb (Tunisia, Morocco, Algeria). The Triple burden of morbidity. Tunis Med. 2018, 96, 760–773. [Google Scholar] [PubMed]
- Fgaier, M. Health economic studies using transferred costs from the Middle East and North Africa region: A protocol for a systematic review of the methodology and reporting quality. In Proceedings of the 2022 IEEE 20th Jubilee International Symposium on Intelligent Systems and Informatics (SISY), Subotica, Serbia, 15–17 September 2022. [Google Scholar]
- Tadmouri, G.O.; Ali, M.T.A.; Ali, S.A.H.; Khaja, N.A. CTGA: The database for genetic disorders in Arab populations. Nucleic Acids Res. 2006, 34, D602. [Google Scholar] [CrossRef]
- Tadmouri, G.O.; Sastry, K.S.; Chouchane, L. Arab gene geography: From population diversities to personalized medical genomics. Glob. Cardiol. Sci. Pract. 2015, 2014, 54. [Google Scholar] [CrossRef]
- Al-Gazali, L.; Hamamy, H.; Al-Arrayad, S. Genetic disorders in the Arab world. BMJ 2006, 333, 831–834. [Google Scholar] [CrossRef]
- Scott, E.M.; Halees, A.; Itan, Y.; Spencer, E.G.; He, Y.; Azab, M.A.; Gabriel, S.B.; Belkadi, A.; Boisson, B.; Abel, L.; et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 2016, 48, 1071–1076. [Google Scholar] [CrossRef]
- Stogiannis, D.; Siannis, F.; Androulakis, E. Heterogeneity in meta-analysis: A comprehensive overview. Int. J. Biostat. 2024, 20, 169–199. [Google Scholar] [CrossRef]
- Borenstein, M. How to understand and report heterogeneity in a meta-analysis: The difference between I-squared and prediction intervals. Integr. Med. Res. 2023, 12, 101014. [Google Scholar] [CrossRef] [PubMed]
- Martorell-Marugan, J.; Toro-Dominguez, D.; Alarcon-Riquelme, M.E.; Carmona-Saez, P. MetaGenyo: A web tool for meta-analysis of genetic association studies. BMC Bioinform. 2017, 18, 563. [Google Scholar] [CrossRef]
- Abouelfath, R.; Habbal, R.; Laaraj, A.; Khay, K.; Harraka, M.; Nadifi, S. ACE insertion/deletion polymorphism is positively associated with resistant hypertension in Morocco. Gene 2018, 658, 178–183. [Google Scholar] [CrossRef]
- Saleh, N.Y.; Salem, S.S.; Fotoh, W.M.A.; Soliman, S.E.; Abo-Haded, H.M. Angiotensin-converting enzyme insertion/deletion (ACE I/D) gene polymorphism in Egyptian children with congenital heart disease. Birth Defects Res. 2020, 112, 963–969. [Google Scholar] [CrossRef]
- Meroufel, D.N.; Médiène-Benchekor, S.; Dumont, J.; Benhamamouch, S.; Amouyel, P.; Brousseau, T. A study on the polymorphisms of the renin–angiotensin system pathway genes for their effect on blood pressure levels in males from Algeria. J. Renin-Angiotensin-Aldosterone Syst. 2014, 15, 1–6. [Google Scholar] [CrossRef]
- Morsy, M.-M.F.; Abdelaziz, N.A.M.; Boghdady, A.M.; Ahmed, H.; Abu Elfadl, E.M.; Ismail, M.A. Angiotensin converting enzyme DD genotype is associated with development of rheumatic heart disease in Egyptian children. Rheumatol. Int. 2009, 31, 17–21. [Google Scholar] [CrossRef]
- Mahjoub, S.; Mehri, S.; Bousaada, R.; Ouarda, F.; Zaroui, A.; Zouari, B.; Mechmeche, R.; Hammami, M.; Ben Arab, S. Association of ACE I/D polymorphism in Tunisian patients with dilated cardiomyopathy. J. Renin-Angiotensin-Aldosterone Syst. 2010, 11, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Mehri, S.; Mahjoub, S.; Hammami, S.; Zaroui, A.; Frih, A.; Betbout, F.; Mechmeche, R.; Hammami, M. Renin-angiotensin system polymorphisms in relation to hypertension status and obesity in a Tunisian population. Mol. Biol. Rep. 2012, 39, 4059–4065. [Google Scholar] [CrossRef] [PubMed]
- Amara, A.; Mrad, M.; Sayeh, A.; Lahideb, D.; Layouni, S.; Haggui, A.; Fekih-Mrissa, N.; Haouala, H.; Nsiri, B. The Effect of ACE I/D Polymorphisms Alone and With Concomitant Risk Factors on Coronary Artery Disease. Clin. Appl. Thromb. Hemost. 2018, 24, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Hmimech, W.; Diakite, B.; Korchi, F.; Baghdadi, D.; Idrissi, H.T.J.H.; Haboub, M.; Habbal, R.; Nadifi, S. Impact of I/D polymorphism of angiotensin-converting enzyme (ACE) gene on myocardial infarction susceptibility among young Moroccan patients. BMC Res. Notes 2017, 10, 763. [Google Scholar] [CrossRef]
- Amrani, A.; Hamed, M.B.B.; Talebbendiab, F.M. Association study between some renin-angiotensin system gene variants and essential hypertension in a sample of Algerian population: Case control study. Ann. Biol. Clin. 2015, 73, 557–563. [Google Scholar] [CrossRef]
- Al-Jafari, A.A.; Daoud, M.S.; Ataya, F.S. Renin-angiotensin system gene polymorphisms and coronary artery disease in Saudi patients with diabetes mellitus. Int. J. Clin. Exp. Pathol. 2017, 10, 10505–10514. [Google Scholar]
- Saab, Y.; Gard, P.; Overall, A. The association of hypertension with renin-angiotensin system gene polymorphisms in the Lebanese population. J. Renin-Angiotensin-Aldosterone Syst. 2011, 12, 588–594. [Google Scholar] [CrossRef]
- Obineche, E.N.; Frossard, P.M.; Bokhari, A.M. An association study of five genetic loci and left ventricular hypertrophy amongst Gulf Arabs. Hypertension Research. 2001, 24, 635–639. [Google Scholar] [CrossRef]
- Saidi, S.; Mallat, S.G.; Almawi, W.Y.; Mahjoub, T. Association between renin–angiotensin–aldosterone system genotypes and haplotypes and risk of ischemic stroke of atherosclerotic etiology. Acta Neurol. Scand. 2009, 119, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Imen, T.; Grissa, M.H.; Boubaker, H.; Beltaief, K.; Messous, S.; Tounsi, N.; Slimani, A.; Khouloud, C.; Bouida, W.; Boukef, R.; et al. AGT M235t polymorphism and heart failure in a cohort of Tunisian population: Diagnostic and prognostic value. Int. J. Clin. Exp. Med. 2015, 8, 16346–16351. [Google Scholar] [PubMed]
- Frossard, P.M.; Hill, S.H.; Elshahat, Y.I.; Obineche, E.N.; Bokhari, A.M.; Lestringant, G.G.; John, A.; Abdulle, A.M. Associations of angiotensinogen gene mutations with hypertension and myocardial infarction in a gulf population. Clin Genet. 1998, 54, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Koch, W.; Kastrati, A.; Mehilli, J.; Böttiger, C.; von Beckerath, N.; Schömig, A. Insertion/deletion polymorphism of the angiotensin I-converting enzyme gene is not associated with restenosis after coronary stent placement. Circulation 2000, 102, 197–202. [Google Scholar] [CrossRef]
- Rigat, B.; Hubert, C.; Alhenc-Gelas, F.; Cambien, F.; Corvol, P.; Soubrier, F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Investig. 1990, 86, 1343–1346. [Google Scholar] [CrossRef]
- Reiter, L.M.; Christensen, D.L.; Gjesing, A.P. Renin angiotensinogen system gene polymorphisms and essential hypertension among people of West African descent: A systematic review. J. Hum. Hypertens. 2016, 30, 467–478. [Google Scholar] [CrossRef]
- Al-Bayati, M.M.; Al-Nahi, A.S. Effect of ACE I/D polymorphism in cardiovascular disease patients. Int. J. Health Sci. 2022, 6, 9671–9676. [Google Scholar] [CrossRef]
- Borai, I.H.; Hassan, N.S.; Shaker, O.G.; Ashour, E.; Badrawy, M.E.; Fawzi, O.M.; Mageed, L. Synergistic effect of ACE and AGT genes in coronary artery disease. Beni-Suef Univ. J. Basic Appl. Sci. 2018, 7, 111–117. [Google Scholar] [CrossRef]
- Younes, S.; Shi, Z.; Zayed, H. Genetic Variations Associated with Coronary Artery Disease and Myocardial Infarction in the Arab World: A Systematic Review and Meta-Analysis. Highlights in BioScience [Internet]. 22 July 2020. Available online: http://highlightsin.org/index.php/bioscience/article/view/30 (accessed on 12 December 2023).
- Akopyan, A.A.; Kirillova, K.I.; Strazhesko, I.D.; Samokhodskaya, L.M.; Orlova, Y.A. Association of AGT, ACE, NOS3, TNF, MMP9, CYBA polymorphism with subclinical arterial wall changes. Kardiologiia 2021, 61, 57–65. [Google Scholar] [CrossRef]
- Shahid, M.; Rehman, K.; Akash, M.S.H.; Suhail, S.; Kamal, S.; Imran, M.; Assiri, M.A. Genetic Polymorphism in Angiotensinogen and Its Association with Cardiometabolic Diseases. Metabolites 2022, 12, 1291. [Google Scholar] [CrossRef]
- Gao, T.; Huang, L.; Fu, Q.; Bai, Y. Association of polymorphisms in the AGT gene (M235T, T174M) with ischemic stroke in the Chinese population. J. Renin-Angiotensin-Aldosterone Syst. 2015, 16, 681–686. [Google Scholar] [CrossRef]
- Wang, S.; Dai, Y.; Chen, L.; Dong, Z.; Chen, Y.; Li, C.; Zhong, X.; Lin, W.; Zhang, J.; Aalto-Setala, K. Genetic Polymorphism of Angiotensin Converting Enzyme and Risk of Coronary Restenosis after Percutaneous Transluminal Coronary Angioplasties: Evidence from 33 Cohort Studies. PLoS ONE 2013, 8, e75285. [Google Scholar] [CrossRef][Green Version]
- Li, X.; Li, Q.; Wang, Y.; Li, Y.; Ye, M.; Ren, J.; Wang, Z. AGT gene polymorphisms (M235T, T174M) are associated with coronary heart disease in a Chinese population. J. Renin Angiotensin Aldosterone Syst. 2013, 14, 354–359. [Google Scholar] [CrossRef]
- Fajar, J.K.; Pikir, B.S.; Sidarta, E.P.; Saka, P.N.B.; Akbar, R.R.; Tamara, F.; Mayasari, E.D.; Gunawan, A.; Heriansyah, T. The genes polymorphism of angiotensinogen (AGT) M235T and AGT T174M in patients with essential hypertension: A meta-analysis. Gene Rep. 2019, 16, 100421. [Google Scholar] [CrossRef]
- Staessen, J.A.; Ginocchio, G.; Wang, J.G.; Saavedra, A.P.; Soubrier, F.; Vlietinck, R.; Fagard, R. Genetic variability in the renin-angiotensin system: Prevalence of alleles and genotypes. J. Cardiovasc. Risk 1997, 4, 401–422. [Google Scholar] [CrossRef]

| Authors | Country | Year | DISEASE | Case | Control | Case | Control | Begg Test | Egger Test | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | Aa | aa | AA | Aa | aa | |||||||||
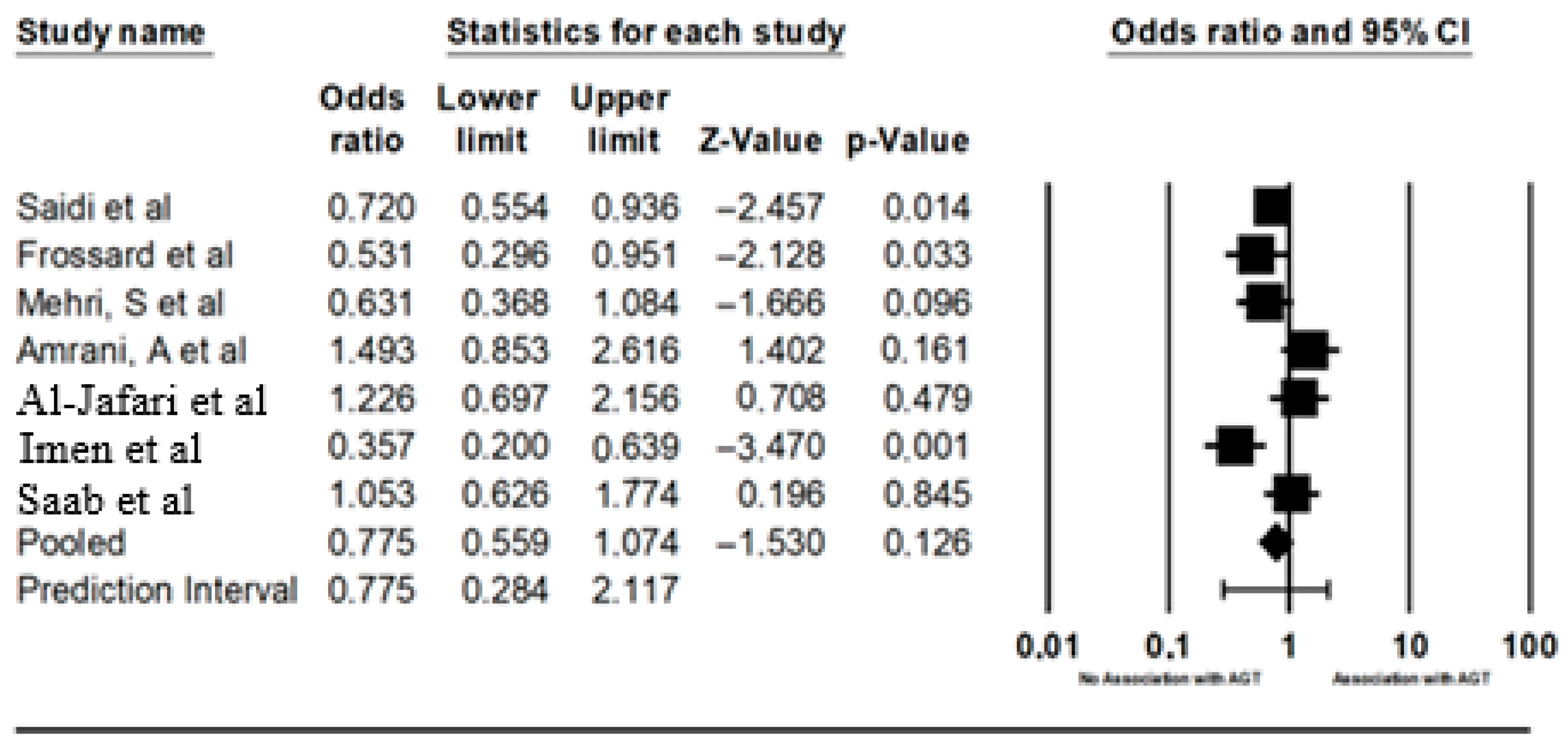
| AGT M273M | Saidi et al. [23] | TUNISIA | 2008 | Ischemia | 320 | 444 | 49 | 149 | 122 | 37 | 172 | 235 | 0.4882 | 0.4882 |
| Frossard, et al. [25] | ABUDHABI | 1998 | Hypertension | 154 | 109 | 56 | 74 | 24 | 34 | 43 | 32 | 0.0278 | 0.0621 | |
| Mehri, S et al. [16] | TUNISIA | 2011 | Hypertension | 142 | 191 | 52 | 67 | 23 | 60 | 82 | 49 | 0.0556 | 0.0778 | |
| Amrani, A et al. [19] | ALGERIA | 2015 | Hypertension | 75 | 70 | 14 | 13 | 48 | 15 | 25 | 30 | 0.0355 | 0.0621 | |
| Al-Jafari, et al. [20] | SAUDI ARABIA | 2017 | Hypertension | 169 | 97 | 52 | 70 | 47 | 35 | 40 | 22 | 0.1147 | 0.1338 | |
| Imen et al. [24] | TUNSIAN | 2015 | Heart failure | 126 | 108 | 74 | 32 | 20 | 24 | 36 | 48 | 0.0019 | 0.0133 | |
| Saab, Y. et al. [21] | LIBANON | 2012 | Hypertension | 124 | 146 | 18 | 72 | 34 | 49 | 59 | 38 | 0.0237 | 0.0621 | |
| ACE | Abouelfath, R et al. [11] | Morocco | 2018 | Resistant hypertension | 88 | 110 | 41 | 26 | 21 | 26 | 40 | 44 | 0.008 | 0.024 |
| Saleh, N.Y. et al. [12] | Egyptian | 2020 | Congenital heart disease | 70 | 70 | 25 | 34 | 11 | 26 | 40 | 4 | 0.0249 | 0.0598 | |
| Meroufel, D.N. et al. [13] | Algeria | 2014 | Hypertension | 91 | 113 | 48 | 36 | 7 | 54 | 47 | 12 | 0.7114 | 0.8555 | |
| Morsy, M.-M.F. et al. [14] | Egyptian | 2011 | CVD | 139 | 79 | 37 | 59 | 43 | 11 | 39 | 29 | 0.7129 | 0.8555 | |
| Mahjoub, S. et al. [15] | Tunisian | 2010 | Dilated cardiomyopathy | 76 | 151 | 26 | 38 | 12 | 22 | 83 | 46 | 0.1162 | 0.2324 | |
| Mehri, S. et al. [16] | Tunisian | 2010 | CVD | 119 | 238 | 64 | 41 | 14 | 51 | 106 | 81 | 0.1433 | 0.2457 | |
| Amara, A. et al. [17] | Tunisian | 2016 | Coronary artery disease | 145 | 300 | 110 | 32 | 3 | 95 | 147 | 58 | 0.9329 | 0.9329 | |
| Hmimech, W. et al. [18] | Morocco | 2017 | Myocardial infraction | 140 | 182 | 87 | 45 | 8 | 113 | 52 | 17 | 0.005 | 0.02 | |
| Enyioma N. OBINECHE et al. [22] | Abbu Dhabi emirate | 2001 | CVD | 92 | 100 | 28 | 58 | 6 | 30 | 62 | 8 | 0.0024 | 0.0144 | |
| Amrani, A et al. [19] | Algeria | 2015 | Resistant hypertension | 75 | 70 | 10 | 40 | 25 | 43 | 25 | 2 | 0.4652 | 0.6978 | |
| Al-Jafari, A.A. et al. [20] | SAUDI ARABIA | 2017 | Coronary artery disease | 169 | 97 | 111 | 43 | 15 | 54 | 26 | 17 | 0.0002 | 0.0024 | |
| Saab, Y. et al. [21] | LIBANEESE | 2012 | Hypertension | 124 | 146 | 78 | 37 | 9 | 76 | 58 | 12 | 0.842 | 0.9185 | |
| Model | Test of Association | Test of Heterogeneity | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p-Value | Model | p-Value | I2 | p-Value (Egger’s Test) | ||
| ACE ID | Allele contrast (A vs. a) | 1.3904 | [0.9110; 2.1221] | 0.1264 | Random | 0 | 0.9238 | 0.0018 |
| Recessive model (AA vs. Aa + aa) | 1.5587 | [0.9202; 2.6401] | 0.098 | Random | 0 | 0.9044 | 0.086 | |
| Dominant model (AA + Aa vs. aa) | 1.5116 | [0.9123; 2.5047] | 0.108 | Random | 0 | 0.7678 | 0.1272 | |
| Over-dominant (Aa vs. AA + aa) | 0.799 | [0.6114; 1.0441] | 0.1002 | Random | 0.0009 | 0.6525 | 0.0545 | |
| (AA vs. aa) | 1.8062 | [0.8963; 3.6401] | 0.098 | Random | 0 | 0.8538 | 0.0293 | |
| (AA vs. Aa) | 1.4802 | [0.9328; 2.3490] | 0.095 | Random | 0 | 0.8574 | 0.1293 | |
| (Aa vs. aa) | 1.2645 | [0.8631; 1.8524] | 0.228 | Random | 0.0158 | 0.5288 | 0.1701 | |
| AGT M273T | Allele contrast (A vs. a) | 1.3171 | [1.1658; 1.4881] | 9.73 × 10−6 | Fixed | 0 | 0.9122 | 0.5076 |
| Recessive model (AA vs. Aa + aa) | 1.2852 | [1.0485; 1.5754] | 0.015681 | Fixed | 0 | 0.8749 | 0.5584 | |
| Dominant model (AA + Aa vs. aa) | 1.5160 | [1.2593; 1.8252] | 1.11 × 10−5 | Fixed | 0 | 0.838 | 0.523 | |
| Over-dominant (Aa vs. AA + aa) | 1.2042 | [1.0144; 1.4295] | 0.033678 | Fixed | 0.0045 | 0.6811 | 0.1335 | |
| (AA vs. aa) | 1.6291 | [1.2763; 2.0793] | 8.88 × 10−5 | Fixed | 0 | 0.8767 | 0.3505 | |
| (AA vs. Aa) | 1.1017 | [0.8824; 1.3756] | 0.392214 | Fixed | 0 | 0.8067 | 0.8789 | |
| (Aa vs. aa) | 1.4493 | [1.1849; 1.7727] | 0.000305 | Fixed | 0.0023 | 0.7061 | 0.3579 | |
| Effect Size and 95% Interval | Test of Null (2-Tail) | Prediction Interval | Between Studies | Other Heterogeneity Statistics | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Number of Studies | Point Estimate | Lower Limit | Upper Limit | Z-Value | p-Value | Lower Limit | Upper Limit | Tau | Tau Sq | Q-Value | df (Q) | p-Value | I-Squared (%) |
| Fixed | 7 | 0.763 | 0.642 | 0.906 | −3.08231 | 2.05 × 10−3 | _ | _ | _ | _ | 1,838,854 | 6 | 5.33 × 10−3 | 67.370 |
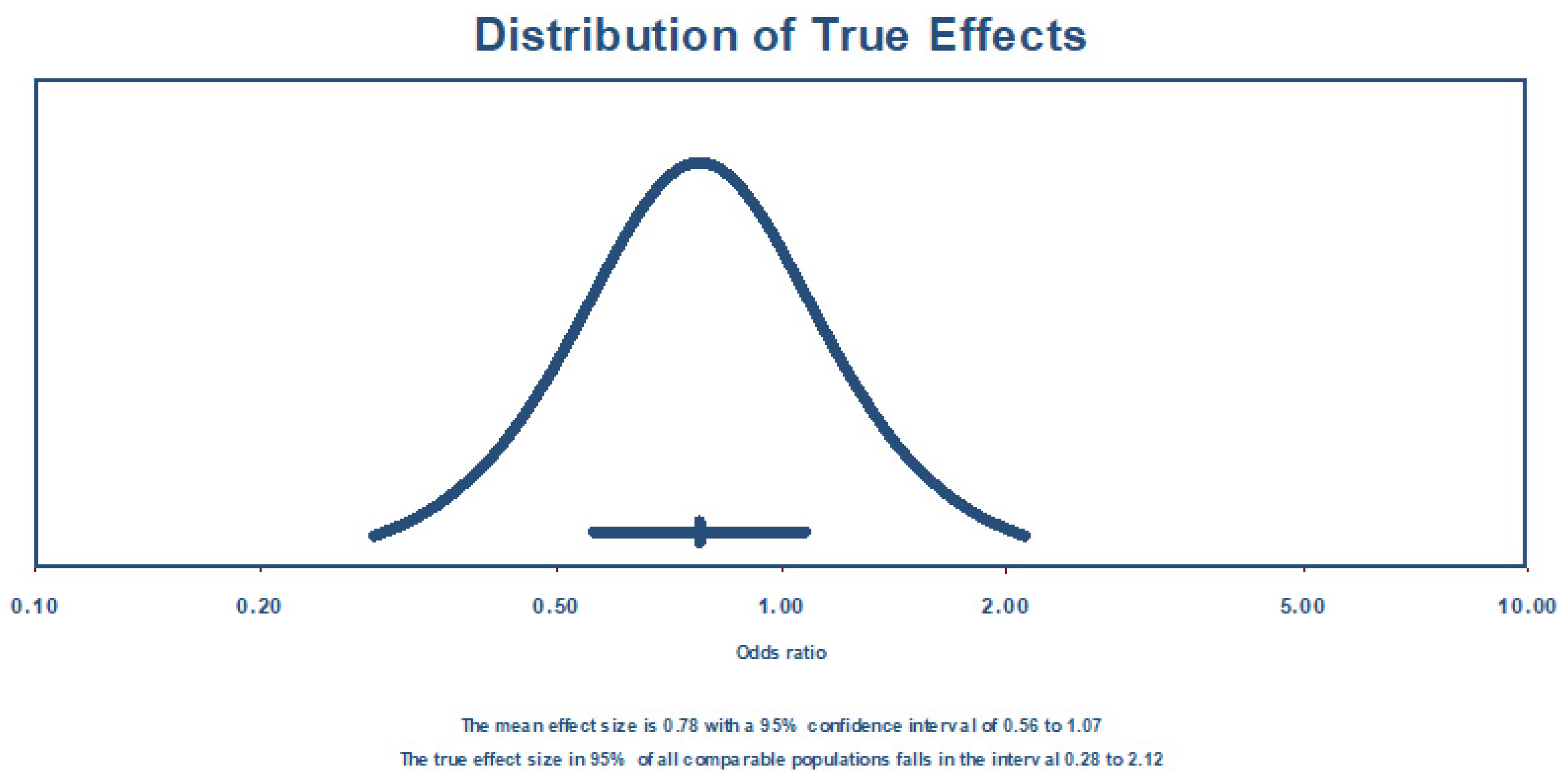
| Random effects | 7 | 0.775 | 0.559 | 1.074 | −1.5301 | 0.125993 | 0.28383 | 2.117001 | 0.35362 | 0.12505 | _ | _ | _ | _ |
| Fixed | 12 | 1.567 | 1.395 | 1.761 | 7.576372 | 3.55 × 10−14 | _ | _ | _ | _ | 1,440,533 | 11 | 0 | 92.363 |
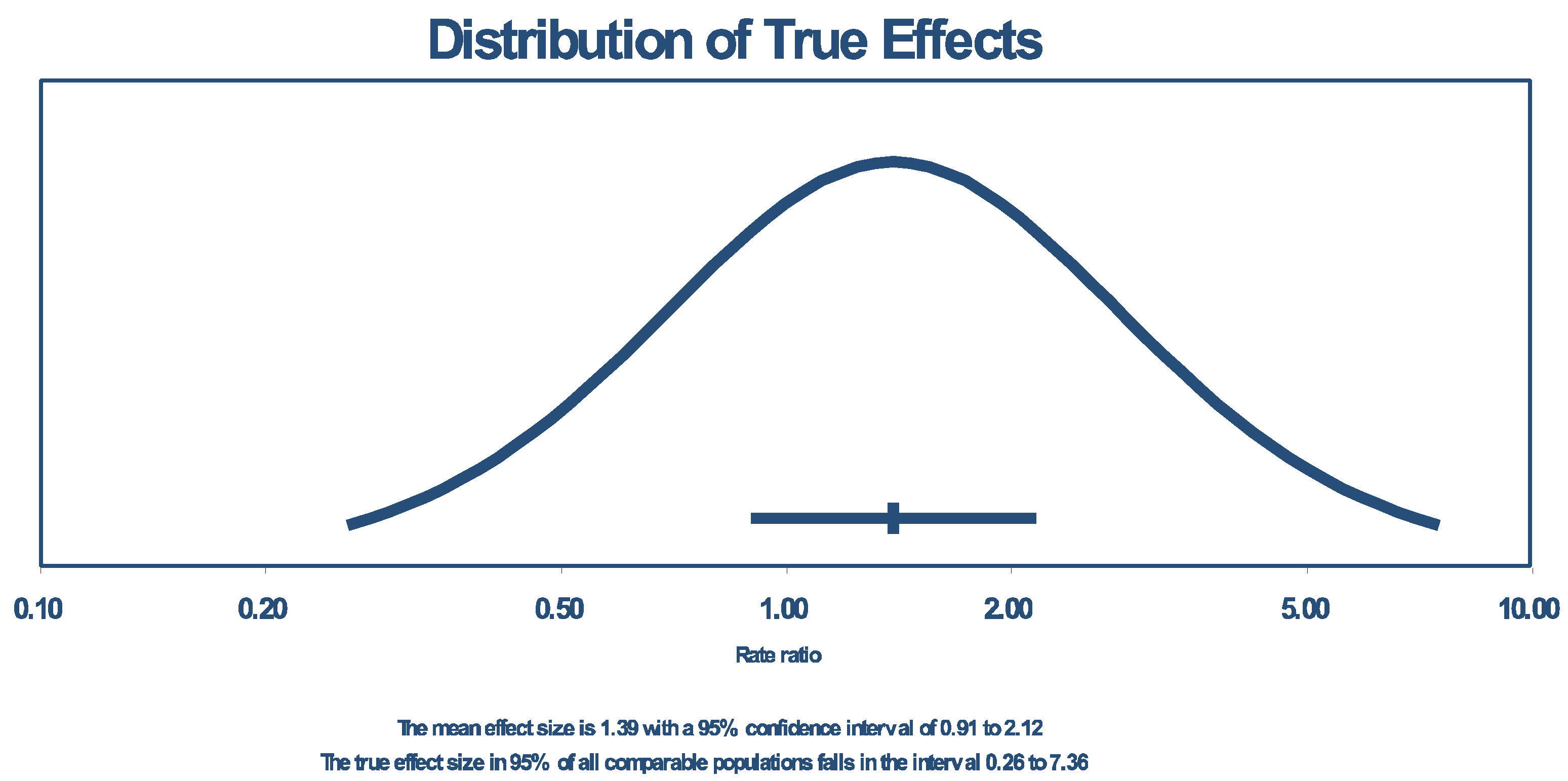
| Random effects | 12 | 1.390 | 0.911 | 2.121 | 1.529486 | 0.126144 | 0.26256 | 7.364047 | 0.71640 | 0.51323 | _ | _ | _ | _ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Mansouri, R.; Dehbi, H.; Habbal, R. Association Between Angiotensinogen Gene M235T and Renin–Angiotensin System Insertion/Deletion Variants and Risk of Cardiovascular Disease in North African and Middle Eastern Populations: A Systematic Review and Meta-Analysis. Cardiogenetics 2025, 15, 23. https://doi.org/10.3390/cardiogenetics15030023
El Mansouri R, Dehbi H, Habbal R. Association Between Angiotensinogen Gene M235T and Renin–Angiotensin System Insertion/Deletion Variants and Risk of Cardiovascular Disease in North African and Middle Eastern Populations: A Systematic Review and Meta-Analysis. Cardiogenetics. 2025; 15(3):23. https://doi.org/10.3390/cardiogenetics15030023
Chicago/Turabian StyleEl Mansouri, Rajaa, Hind Dehbi, and Rachida Habbal. 2025. "Association Between Angiotensinogen Gene M235T and Renin–Angiotensin System Insertion/Deletion Variants and Risk of Cardiovascular Disease in North African and Middle Eastern Populations: A Systematic Review and Meta-Analysis" Cardiogenetics 15, no. 3: 23. https://doi.org/10.3390/cardiogenetics15030023
APA StyleEl Mansouri, R., Dehbi, H., & Habbal, R. (2025). Association Between Angiotensinogen Gene M235T and Renin–Angiotensin System Insertion/Deletion Variants and Risk of Cardiovascular Disease in North African and Middle Eastern Populations: A Systematic Review and Meta-Analysis. Cardiogenetics, 15(3), 23. https://doi.org/10.3390/cardiogenetics15030023