Genetic Testing for Patients with Cardiomyopathies: The INDACO Study—Towards a Cardiogenetic Clinic
Abstract
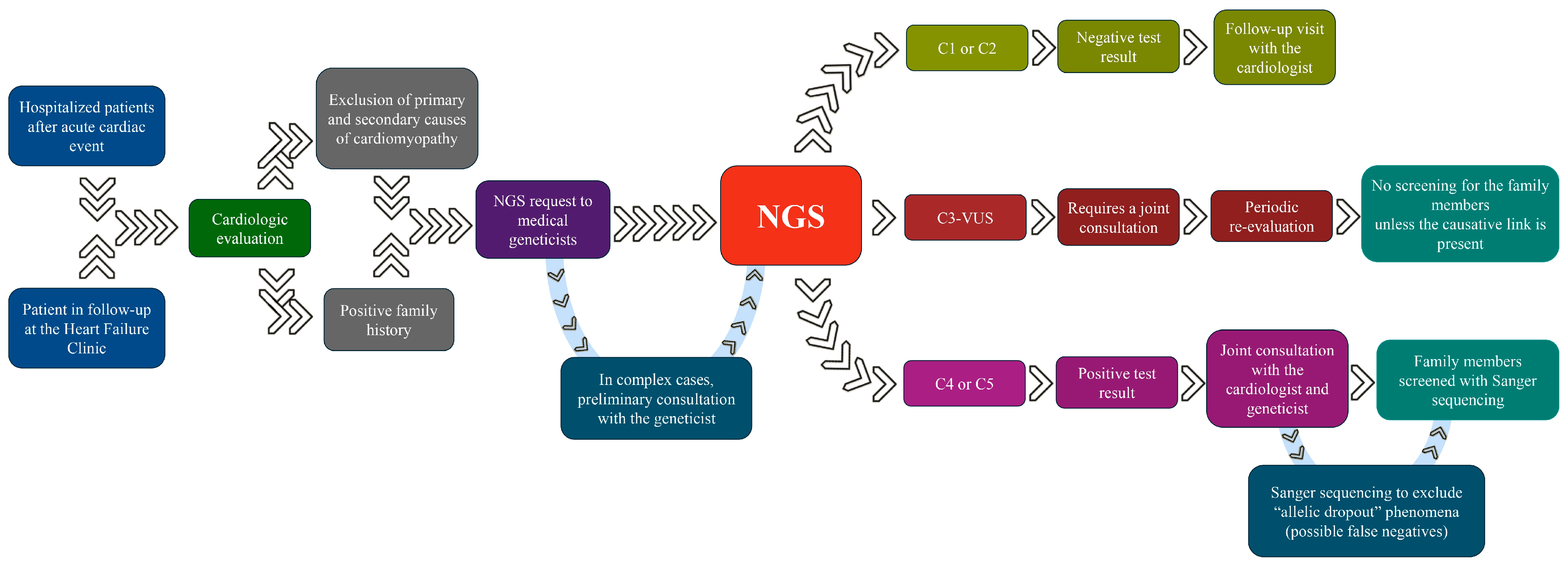
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McKenna, W.J.; Judge, D.P. Epidemiology of the inherited cardiomyopathies. Nat. Rev. Cardiol. 2021, 18, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Girolami, F.; Vergaro, G.; Pieroni, M.; Passantino, S.; Giannotti, G.; Grippo, G.; Canale, M.L.; Favilli, S.; Cappelli, F.; Olivotto, I.; et al. Percorso clinico proposto dall’ANMCO Toscana per la diagnosi genetica delle cardiomiopatie in un sistema assistenziale in rete. G. Ital. Cardiol. 2020, 21, 926–934. [Google Scholar] [CrossRef]
- Wilde, A.A.M.; Semsarian, C.; Márquez, M.F.; Sepehri Shamloo, A.; Ackerman, M.J.; Ashley, E.A.; Sternick, E.B.; Barajas-Martinez, H.; Behr, E.R.; Bezzina, C.R.; et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the state of genetic testing for cardiac diseases. EP Eur. 2022, 24, 1307–1367. [Google Scholar] [CrossRef]
- Quarta, G.; Muir, A.; Pantazis, A.; Syrris, P.; Gehmlich, K.; Garcia-Pavia, P.; Ward, D.; Sen-Chowdhry, S.; Elliott, P.M.; McKenna, W.J. Familial Evaluation in Arrhythmogenic Right Ventricular Cardiomyopathy. Circulation 2011, 123, 2701–2709. [Google Scholar] [CrossRef] [PubMed]
- Girolami, F.; Gozzini, A.; Pálinkás, E.D.; Ballerini, A.; Tomberli, A.; Baldini, K.; Marchi, A.; Zampieri, M.; Passantino, S.; Porcedda, G.; et al. Genetic Testing and Counselling in Hypertrophic Cardiomyopathy: Frequently Asked Questions. J. Clin. Med. 2023, 12, 2489. [Google Scholar] [CrossRef] [PubMed]
- Ommen, S.R.; Mital, S.; Burke, M.A.; Day, S.M.; Deswal, A.; Elliott, P.; Evanovich, L.L.; Hung, J.; Joglar, J.A.; Kantor, P.; et al. 2020 AHA/ACC Guideline for the Diagnosis and Treatment of Patients With Hypertrophic Cardiomyopathy: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2020, 76, 3022–3055. [Google Scholar] [CrossRef]
- Girolami, F.; Frisso, G.; Benelli, M.; Crotti, L.; Iascone, M.; Mango, R.; Mazzaccara, C.; Pilichou, K.; Arbustini, E.; Tomberli, B.; et al. Contemporary genetic testing in inherited cardiac disease: Tools, ethical issues, and clinical applications. J. Cardiovasc. Med. 2018, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Vaisitti, T.; Sorbini, M.; Callegari, M.; Kalantari, S.; Bracciamà, V.; Arruga, F.; Vanzino, S.B.; Rendine, S.; Togliatto, G.; Giachino, D.; et al. Clinical exome sequencing is a powerful tool in the diagnostic flow of monogenic kidney diseases: An Italian experience. J. Nephrol. 2021, 34, 1767–1781. [Google Scholar] [CrossRef]
- Shestak, A.G.; Bukaeva, A.A.; Saber, S.; Zaklyazminskaya, E.V. Allelic Dropout Is a Common Phenomenon That Reduces the Diagnostic Yield of PCR-Based Sequencing of Targeted Gene Panels. Front. Genet. 2021, 12, 620337. [Google Scholar] [CrossRef]
- Musunuru, K.; Hershberger, R.E.; Day, S.M.; Klinedinst, N.J.; Landstrom, A.P.; Parikh, V.N.; Prakash, S.; Semsarian, C.; Sturm, A.C.; American Heart Association Council on Genomic and Precision Medicine; et al. Genetic Testing for Inherited Cardiovascular Diseases: A Scientific Statement From the American Heart Association. Circ. Genom. Precis. Med. 2020, 13, E000067. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Kumar, S.; Elliott, P.; Kalman, J.M.; Fatkin, D. Arrhythmic Genotypes in Familial Dilated Cardiomyopathy: Implications for Genetic Testing and Clinical Management. Heart Lung Circ. 2019, 28, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Mazzaccara, C.; Lombardi, R.; Mirra, B.; Barretta, F.; Esposito, M.V.; Uomo, F.; Caiazza, M.; Monda, E.; Losi, M.A.; Limongelli, G.; et al. Next-Generation Sequencing Gene Panels in Inheritable Cardiomyopathies and Channelopathies: Prevalence of Pathogenic Variants and Variants of Unknown Significance in Uncommon Genes. Biomolecules 2022, 12, 1417. [Google Scholar] [CrossRef] [PubMed]
- Cirino, A.L.; Lakdawala, N.K.; McDonough, B.; Conner, L.; Adler, D.; Weinfeld, M.; O’Gara, P.; Rehm, H.L.; Machini, K.; Lebo, M.; et al. A Comparison of Whole Genome Sequencing to Multigene Panel Testing in Hypertrophic Cardiomyopathy Patients. Circ. Cardiovasc. Genet. 2017, 10, e001768. [Google Scholar] [CrossRef] [PubMed]
- Bagnall, R.D.; Ingles, J.; Dinger, M.E.; Cowley, M.J.; Ross, S.B.; Minoche, A.E.; Lal, S.; Turner, C.; Colley, A.; Rajagopalan, S.; et al. Whole Genome Sequencing Improves Outcomes of Genetic Testing in Patients With Hypertrophic Cardiomyopathy. J. Am. Coll. Cardiol. 2018, 72, 419–429. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESCGuidelines for the diagnosis treatment of acute chronic heart failure: Developed by the Task Force for the diagnosis treatment of acute chronic heart failure of the European Society of Cardiology (ESC) With the special contribution of the Heart Failure Association (HFA) of the E.S.C. Eur. Heart J. 2021, 42, 3599–3726. [Google Scholar] [CrossRef] [PubMed]
- Arbelo, E.; Protonotarios, A.; Gimeno, J.R.; Arbustini, E.; Barriales-Villa, R.; Basso, C.; Bezzina, C.R.; Biagini, E.; Blom, N.A.; de Boer, R.A.; et al. 2023 ESC Guidelines for the management of cardiomyopathies: Developed by the task force on the management of cardiomyopathies of the European Society of Cardiology (ESC). Eur. Heart J. 2023, 44, 3503–3626. [Google Scholar] [CrossRef] [PubMed]
- Lilley, M.; Christian, S.; Blumenschein, P.; Chan, S.; Somerville, M. A centralized approach to out-of-province genetic testing leads to cost savings: The Alberta experience. Clin. Genet. 2013, 84, 373–377. [Google Scholar] [CrossRef] [PubMed]
| Panel of 64 Genes DCM and ARVD | Panel of 50 Genes for HCM | Panel of 17 Genes for LVNC |
|---|---|---|
| ABCC9; ACTA1; ACTC1; ACTN2; ALMS1; ANK2; ANKRD1; BAG3; CRYAB; CSRP3; DES; DMD; DNAJC19; DOLK; DSC2; DSG2; DSP; EMD; EYA4; FHL1; FHL2; FKRP; FKTN; GATAD1; GLA; HFE; ILK; JUP; LAMA4; LAMP2; LDB3; LMNA; MYBPC3; MYH6; MYH7; MYL2; MYL3; MYPN; NEXN; NKX2-5; PDLIM3; PKP2; PLN; PRDM16; RAF1; RBM20; RYR2; SCN1B; SCN5A; SDHA; SGCD; TAZ; TBX5; TCAP; TGFB3; TMEM43; TNNC1; TNNI3; TNNT2; TPM1; TTN; TTR; TXNRD2; VCL. | ABCC9; ACTC1; ACTN2; BAG3; BRAF; CACNA1C; CALR3; CAV3; COX15; CRYAB; CSRP3; DES; FHL1; FXN; GAA; GLA; HRAS; JPH2; KLF10; LAMP2; LDB3; MAP2K1; MAP2K2; MYBPC3; MYH6; MYH7; MYL2; MYL3; MYLK2; MYO6; MYOZ2; MYPN; NEXN; NRAS; PLN; PRKAG2; PTPN11; RAF1; SHOC2; SLC25A4; SOS1; TCAP; TNNC1; TNNI3; TNNT2; TPM1; TRIM63; TTN; TTR; VCL | ACTC1; CASQ2; DNAJC19; DTNA; HCN4; LDB3; MIB1; MYBPC3; MYH7; NKX2-5; PRDM16; RYR2; TAZ; TBX5; TNNT2; TPM1; TTN. |
| Panel of 12 Genes for Amyloidosis | Panel of 58 Genes for HCM | Panel of 79 Genes for DCM and ARVD |
|---|---|---|
| APOA1, APOA2, APOC2, APOC3, B2M, FGA, GLA, GSN, LYZ, MEFV, NLRP3, TTR | ABCC9, ACADVL, ACTC1, ACTN2, AGL, ATAD3A, BAG3, BRAF, CACNA1C, CALR3, CAV3, COX15, CRYAB, CSRP3, DES, FHL1, FHOD3, FLNC, FXN, GAA, GLA, GYG1, HRAS, JPH2, KLF10, LAMP2, LDB3, LZTR1, MAP2K1, MAP2K2, MYBPC3, MYH6, MYH7, MYL2, MYL3, MYLK2, MYO6, MYOZ2, MYPN, NEXN, NRAS, PLN, PRKAG2, PTPN11, RAF1, RIT1, SHOC2, SLC25A4, SOS1, TCAP, TNNC1, TNNI3, TNNT2, TPM1, TRIM63, TTN, TTR, VCL | ABCC9, ACTA1, ACTC1, ACTN2, ALMS1, ANK2, ANKRD1, BAG3, CDH2, CRYAB, CSRP3, CTNNA3, DES, DMD, DNAJC19, DOLK, DSC2, DSG2, DSP, EMD, EPG5, EYA4, FHL1, FHL2, FKRP, FKTN, FLNC, GATAD1, GLA, HAMP, HFE, HFE2, IDH2, ILK, JUP, LAMA4, LAMP2, LDB3, LMNA, MYBPC3, MYH6, MYH7, MYL2, MYL3, MYPN, NEXN, NKX2-5, PDLIM3, PKP2, PLN, PRDM16, PSEN1, PSEN2, RAF1, RBM20, RYR2, SCN1B, SCN5A, SDHA, SGCD, SLC40A1, SLC6A6, SPEG, TAZ, TBX5, TCAP, TFR2, TGFB3, TMEM43, TNNC1, TNNI3, TNNI3K, TNNT2, TPM1, TTN, TTR, TXNRD2, VCL, XK |
| Patients (n = 170) | |
|---|---|
| Age, mean (SD), y | 64 (15) |
| Sex Males Females | 111 (60.3%) 59 (39.7%) |
| Positive family history | 81 (44%) |
| Relative with a known variant | 27 (15.9%) |
| Clinical suspicion DCM HCM ARVD LVNC Amyloidosis | 81 (33.8%) 66 (38.8%) 34 (20%) 3 (1.8%) 26 (15.1%) |
| Clinical Suspicion | Reference Sequence | Mutated Gene | Variant | Interpretation |
|---|---|---|---|---|
| DCM | NM_004281.4 | BAG3 | c.1534delC p.(Ser513fs*53) | C4 |
| NM_005572.3 | LMNA | c.569G>A (p.Arg190Gln) | C4 | |
| NM_000256.3 | MYBPC3 | c.1224-80G>A | C4 | |
| NM_000257.4 | MYH7 | c.2134C>T p.(Arg712Cys) | C4 | |
| NM_005633.4 | SOS1 | c.755T>C; p. (lle252Thr) | C4 | |
| NM_001267550.2 | TTN | c.49870C>T; p.(Arg16624*) | C4 | |
| c.74724_74730dupTCCTGGT; p.Pro24911fs*23 | C4 | |||
| c.93166C>T; p.Arg31056* | C4 | |||
| c.93202G>T; p.(Glu31086*) | C4 | |||
| HCM | NM_000256.3 | MYBPC3 | c.1828G>C p.Asp610His | C4 |
| c.3617delG p.Gly1206fs*31 | C4 | |||
| NM_000257.4 | MYH7 | c.1484T>C; p.(Val495Ala) | C4 | |
| c.2123G>C p.(Gly708Ala) | C5 | |||
| c.2631G>T; p.(Met877Ile) | C5 | |||
| NM_000363.5 | TNNI3 | c.557G>A (p.Arg186Gln) | C4 | |
| ARVD | NM_001458.5 | FLNC | c.3937C>T p.(Arg1313*) | C5 |
| NM_004415.4 | DSP | c.3337C>T [p.(Arg1113*)] | C5 | |
| c.3889C>T p.(Gln1297*) | C4 | |||
| NM_ 004949.5 | DSC2 | c.268G>T (p.Glu90*) | C5 | |
| NM_004281.4 | BAG3 | c.1534delC p.(Ser513fs*53) | C4 | |
| Amyloidosis | NM_000371.4 | TTR | c.250T>C p.(Phe84Leu) (alias p.Phe64Leu) | C5 |
| Clinical Suspect | Positivity Rate for C4 and C5 Variants | Positivity Rate for C3-VUS, C4 and C5 Variants |
|---|---|---|
| DCM | 26.3% | 63.1% |
| HCM | 26% | 52.1% |
| ARVD | 28.6% | 92.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianco, M.; Giordano, N.; Gazzola, V.; Biolè, C.; Nangeroni, G.; Lazzero, M.; Brach del Prever, G.M.; Mioli, F.; Gobello, G.; Mousavi, A.H.; et al. Genetic Testing for Patients with Cardiomyopathies: The INDACO Study—Towards a Cardiogenetic Clinic. Cardiogenetics 2024, 14, 122-131. https://doi.org/10.3390/cardiogenetics14030010
Bianco M, Giordano N, Gazzola V, Biolè C, Nangeroni G, Lazzero M, Brach del Prever GM, Mioli F, Gobello G, Mousavi AH, et al. Genetic Testing for Patients with Cardiomyopathies: The INDACO Study—Towards a Cardiogenetic Clinic. Cardiogenetics. 2024; 14(3):122-131. https://doi.org/10.3390/cardiogenetics14030010
Chicago/Turabian StyleBianco, Matteo, Noemi Giordano, Valentina Gazzola, Carloalberto Biolè, Giulia Nangeroni, Maurizio Lazzero, Giulia Margherita Brach del Prever, Fiorenza Mioli, Giulia Gobello, Amir Hassan Mousavi, and et al. 2024. "Genetic Testing for Patients with Cardiomyopathies: The INDACO Study—Towards a Cardiogenetic Clinic" Cardiogenetics 14, no. 3: 122-131. https://doi.org/10.3390/cardiogenetics14030010
APA StyleBianco, M., Giordano, N., Gazzola, V., Biolè, C., Nangeroni, G., Lazzero, M., Brach del Prever, G. M., Mioli, F., Gobello, G., Mousavi, A. H., Guidante, M., Deaglio, S., Giachino, D. F., & Chinaglia, A. (2024). Genetic Testing for Patients with Cardiomyopathies: The INDACO Study—Towards a Cardiogenetic Clinic. Cardiogenetics, 14(3), 122-131. https://doi.org/10.3390/cardiogenetics14030010






